Abstract
Bisphenol A (BPA), a ubiquitous environmental endocrine disruptor, is considered an obesogen. However, its role in the hypothalamic control of energy balance remains largely unexplored. Because disruption of the circadian clock is tightly associated with metabolic consequences, we explored how BPA affects the components of the molecular circadian clock in the feeding-related neurons of the hypothalamus. In immortalized POMC and NPY/AgRP-expressing hypothalamic cell lines and primary culture, we describe how BPA significantly alters mRNA expression of circadian clock genes Bmal1,Per2, and Rev-Erbα. Furthermore, we use newly generated Bmal1-knockout (KO) hypothalamic cell lines to link the BPA-induced neuropeptide dysregulation to the molecular clock. Specifically, BPA increased Npy, Agrp, and Pomc mRNA expression in wild type hypothalamic cells, whereas the increase in Npy, but not Agrp or Pomc, was abolished in cell lines lacking BMAL1. In line with this increase, BPA led to increased BMAL1 binding to the Npy promotor, potentially increasing Npy transcription. In conclusion, we show that BPA-mediated dysregulation of the circadian molecular clock is linked to the deleterious effects of BPA on neuropeptide expression. Furthermore, we describe hypothalamic Bmal1-KO cell lines to study the role of BMAL1 in hypothalamic responses to metabolic, hormonal, and environmental factors.
Appropriate control of energy homeostasis depends on a properly regulated circadian system (1). The suprachiasmatic nucleus (SCN) is the body’s central clock system and lesions in this region lead to disruptions in the rhythm of food intake (2). Changes in the timing of feeding have been linked to weight gain in both animal models (3) and epidemiological studies (4, 5). The SCN, entrained by light, is responsible for maintaining the rhythm of clocks in all other regions, including the arcuate nucleus (ARC) of the hypothalamus, an area critical for the regulation of energy balance (6). Factors including a high-fat diet can disrupt the circadian clock in this region, uncoupling it from the SCN (1, 7). Other obesity-promoting factors, such as environmental chemicals (8), acting at the level of the hypothalamus, may also disrupt this synchrony.
Bisphenol A (BPA) is an environmental endocrine disrupting chemical detected in 90% to 95% of urine samples (9, 10). BPA is considered an obesogen, a class of exogenous factors that dysregulate lipid homeostasis and energy balance, predisposing people to weight gain (8). In humans, urinary BPA concentrations are positively correlated with increased body mass index scores and comorbidities, such as insulin resistance and type 2 diabetes (9, 11–13). Several reports indicate that prenatal BPA exposure in rodents leads to increased feeding and weight gain in offspring (14). Likewise, adult male and female C57BL/6J mice exposed to BPA for 2 or 4 weeks display increased body weight (15, 16).
The impact of BPA on the brain, especially in the hypothalamus, is only beginning to be explored. Within the ARC, the orexigenic, neuropeptide Y (NPY)/agouti-related peptide (AgRP) and the anorexigenic, pro-opiomelanocortin (POMC) neurons integrate hormonal and metabolic signals to regulate food intake and energy expenditure (17). The few studies investigating the effect of BPA in the ARC have been with either prenatal or perinatal exposure (18–20). Indeed, perinatal BPA exposure upregulates the levels of AgRP and downregulates POMC protein levels in neural progenitor cells of newborn mice (18), illustrating the potential orexigenic effects of BPA in the neonatal hypothalamus. We have found that acute exposure of adult and embryonic hypothalamic neurons to BPA upregulates the expression of Npy,Agrp, and Pomc (21) mRNA, suggesting an overall dysregulation of the hypothalamic control of energy balance at the level of transcription. Whether this dysregulation is linked to circadian rhythm disruption, often tightly associated with metabolic consequences, is unknown.
The expression of the feeding neuropeptides follows a rhythm throughout the day. Pomc gene expression itself exhibits circadian rhythmicity in vitro and in vivo, showing a peak 4 hours after dark phase (ZT 16) and a trough during the day (ZT 4–7) (22). Agrp peaks in vivo in the transition between the light and dark phases, whereas Npy has a diurnal rhythm, peaking once in the dark phase and once in the light phase (23). However, in an isolated population of hypothalamic neurons in vitro, only Npy, not Agrp, had rhythmicity (24).
These rhythms are controlled by the molecular clock, a series of transcriptional-translational feedback loops involving circadian clock genes. The clock genes circadian locomotor output cycles kaput (Clock) and brain and muscle ARNT-like 1 (Bmal1) produce the proteins CLOCK and BMAL1. These transcription factors heterodimerize and bind to E-box promotor elements to upregulate the expression of period (Per1–3) and cryptochrome (Cry1–2). Subsequently, PER and CRY proteins form heterodimers that interact with the BMAL1:CLOCK complex to repress their own transcription. The clock genes cycle in opposite phases and define daily variations in physiological function, which shapes the circadian rhythm (1). The BMAL1:CLOCK heterodimer also activates the transcription of other clock genes, such as reverse erythroblastosis virus (Rev-Erb) α and β, which repress Bmal1 gene expression (25). Approximately 10% of the genome is controlled by these transcription factors, which regulate daily patterns of expression (26, 27). The molecular clock is tightly linked to the control of energy balance as both Clock mutant and Per2 mutant mice develop obesity (28, 29). Furthermore, the saturated fatty acid palmitate, which is elevated in obese states (30), alters circadian clock genes alongside altered hypothalamic neuropeptide expression (31).
Given that circadian dysregulation often underlies metabolic perturbations (1, 32) and that BPA leads to weight gain (15, 16), dysregulates hypothalamic neuropeptide expression, and causes metabolic perturbations (33), we hypothesized that BPA alters the expression of clock genes in hypothalamic NPY/AgRP and POMC neurons. We also hypothesized that changes in neuropeptide gene expression depend on the molecular clock because it is these transcription factors that drive metabolic homeostasis and have led to metabolic consequences when mutated (28, 29). Here, we show that BPA dysregulates gene expression of Bmal1,Per2, and Rev-Erbα in immortalized hypothalamic POMC and NPY/AgRP-expressing neurons. We also describe the generation and characterization of hypothalamic cell lines lacking functional BMAL1 protein and, by using these, show that BMAL1 is necessary for changes in Npy expression. These lines provide tools to examine the role of BMAL1 in a variety of hypothalamic responses to both exogenous and endogenous factors, including chemicals, fatty acids, and hormones.
Methods
Cell culture and reagents
Mouse hypothalamic neurons were immortalized as described previously to generate the POMC-expressing mHypoE-43/5 (34) and mHypoA-POMC/GFP-2 (35) cell lines and the NPY/AgRP-expressing mHypoE-41 (34) and mHypoA-59 (36) cell lines. Neurons were grown in DMEM (MilliporeSigma, Oakville, ON, Canada) containing 4500 mg/L glucose, supplemented with 2% fetal bovine serum (FBS; Gibco, Burlington, ON, Canada) and 1% penicillin/streptomycin (PS; Gibco) and maintained in 5% CO2 at 37°C.
Neurons were split into 60-mm tissue culture plates 24 hours before the experiment. For experiments involving the mHypoE-43/5 and mHypoA-POMC/GFP-2 cells, neurons were cultured to 75% confluency and serum-starved in DMEM modified phenol red–free serum-free medium (Hyclone, Fisher Scientific, Markham, ON, Canada) for 12 hours before treatments, followed by 20 μM forskolin (MilliporeSigma) for 30 minutes, as a standard method used to synchronize neurons in culture. For experiments involving the mHypoE-41 and mHypoA-59 cells, growth medium was replaced with treatment medium 24 hours after plating. BPA (MilliporeSigma) was dissolved in 100% ethyl alcohol (ETOH) to a concentration of 200 mM and diluted 1:1 in sterile H2O to a concentration of 100 mM. A total of 100 mM BPA or vehicle was then diluted 1:1000 in DMEM modified phenol red-free medium, supplemented with 1% charcoal:dextran-stripped FBS (CSFBS; Gemini Bio Products, Burlington, ON, Canada) and 1% PS (Gibco), giving a final concentration of 100 μM BPA or 0.05% ETOH. All experiments were started at the same time of day (~0900 hours), and because medium was changed to treatment medium at time of treatment, all reported treatment times correspond to time since medium change.
For primary culture experiments, cells from the hypothalami of 8-week-old male or female CD-1 mice (Charles River Laboratories, Senneville, QC, Canada) were dispersed by trituration. Cells were cultured in six-well plates in neurobasal A medium (Gibco), supplemented with 10% FBS, 5% horse serum (Gibco), 1% PS (Gibco), 1 × B27 serum-free supplement (Gibco), and 1 × GlutaMAX supplement (Gibco) for 7 to 9 days. Each well received 10 ng/μL ciliary neurotrophic factor (R&D Biosystems, Oakville, ON, Canada) once per day to induce proliferation. After 7 to 9 days, cells were treated with vehicle (0.05% ETOH) or 100 μM BPA for 8 hours in DMEM modified phenol red–free medium, supplemented with 1% CSFBS (Gemini Bio Products) and 1% PS (Gibco). All animal procedures were conducted in accordance with the regulations of the Canadian Council on Animal Care and approved by the University of Toronto’s animal care committee.
Preparation of mHypoA-Bmal1-WT and mHypoA-Bmal1-KO cell lines
Bmal1 heterozygous mice (stock no. 009100; The Jackson Laboratory, Bar Harbor, ME) were purchased and bred by Dr. Patricia Brubaker’s laboratory to obtain Bmal1-knockout (KO) mice and wild-type (WT) littermates. The hypothalami of two Bmal1-KO mice and two WT littermates were then obtained from Dr. Brubaker and neurons were immortalized as previously described (36). Briefly, the hypothalami of an 8- to 9-week-old female Bmal1-KO mouse, male Bmal1-KO mouse, female littermate control, and male littermate control were separately isolated and dispersed into primary culture. Primary cultures were treated with 10 ng/μL ciliary neurotrophic factor for 5 to 7 days to induce neuronal proliferation, followed by viral transformation of a plasmid containing the SV40 T-antigen and a neomycin resistance cassette to induce immortalization. Immortalized cells were selected for with 100 μg/mL G418 (Geneticin, Gibco). This process generated four cell lines representing a mixed population of hypothalamic neurons: mHypoA-Bmal1-KO/F, mHypoA-Bmal1-KO/M, mHypoA-Bmal1-WT/F, and mHypoA-Bmal1-WT/M. Cell lines were screened for Bmal1 expression, circadian clock gene expression, neuropeptide expression, and related markers with real-time quantitative PCR. Cells were grown to 70% to 75% confluency in DMEM containing 4500 mg/L glucose (MilliporeSigma), 2% FBS (Gibco), and 1% PS (Gibco) and treated with vehicle (0.05% ETOH) or 100 μM BPA in DMEM modified phenol red–free medium (1% CSFBS, 1% PS) for 4 or 8 hours. Experiments in mHypoA-Bmal1-KO cell lines were conducted in parallel with mHypoA-Bmal1-WT cells for comparison of effects. All animal procedures were conducted in accordance with the regulations of the Canadian Council on Animal Care and approved by the University of Toronto’s animal care committee.
RNA isolation and quantitative RT-PCR
The PureLink RNA isolation kit (Thermo Fisher Scientific, Burlington, ON, Canada) was used according to manufacturer instructions with an on-column DNAse step (PureLink DNAse set) to isolate total RNA. RNA was quantified with the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific); 500 ng to 1 μg of cDNA was synthesized with the SuperScript™ VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific) or the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fischer Scientific). Finally, 12.5 ng of cDNA was amplified with gene-specific primers (Table 1) and Platinum SYBR Green qPCR SuperMix-UDG with ROX by quantitative RT-PCR (qRT-PCR) on an Applied Biosystems Prism 7900HT machine with the following cycling conditions: 2 minutes at 50°C, 2 minutes at 95°C, and 40 cycles of 15 seconds at 95°C and 1 minute at 60°C, followed by melt curve analysis (15 seconds at 95°C, 15 seconds at 60°C, 15 seconds at 95°C). Data were analyzed with the standard curve method for experiments involving the mHypoE-43/5, mHypoA-POMC/GFP-2, mHypoE-41, and mHypoA-59 cell lines and with the ΔΔCT method for primary culture and mHypoA-Bmal1-KO and mHypoA-Bmal1-WT cell line experiments. All genes were normalized to the reference gene, 60S ribosomal protein L7 (Rpl7).
Table 1.
Primer Sequences for qRT-PCR
| Gene Name | Primer Sequence (5′ → 3′) | Amplicon Size |
|---|---|---|
| Bmal1 | F: GGG AGG CCC ACA GTC AGA TT | 78 |
| R: GTA CCA AAG AAG CCA ATT CAT CAA | ||
| Per2 | F: TCA TCA TTG GGA GGC ACA AA | 135 |
| R: GCA TCA GTA GCC GGT GGA TT | ||
| Rev-Erbα | F: TGG AAG ACA GCA GCC GAG TG | 114 |
| R: CAT AGT GGA AGC CTC AGG CCA | ||
| Npy | F: CAG AAA ACG CCC CCA GAA | 77 |
| R: AAA AGT CGG GAG AAC AAG TTT CAT T | ||
| Agrp | F: CGG AGG TCG TAG ATC CAC AGA | 69 |
| R: AGG ACT CGT GCA GCC TTA CAC | ||
| Pomc | F: CCC GCC CAA GGA CAA GCG TT | 112 |
| R: CTG GCC CTT CTT GTG CGC GT | ||
| Clock | F: CAC CGA CAA AGA TCC CTA CTG AT | 151 |
| R: TGA GAC ATC GCT GGC TGT GT | ||
| Cry1 | F: AGA GCT CGG CTT TGA TAC AGA | 120 |
| R: CGT TCA AAG TTT GCC ACC CA | ||
| Nfκb1 | F: GGA TGA CAG AGG CGT GTA TTA G | 114 |
| R: CCT TCT CTC TGT CTG TGA GTT G | ||
| Il6 | F: GTG GCT AAG GAC CAA GAC CA | 85 |
| R: GGT TTG CCG AGT AGA CCT CA | ||
| Esr1 | F: GAG TGC CAG GCT TTG GGG ACT T | 102 |
| R: CCA TGG AGC GCC AGA CGA GA | ||
| Esr2 | F: ATC TGT CCA GCC ACG AAT CAG TGT | 114 |
| R: TCT CCT GGA TCC ACA CTT GAC CAT | ||
| Esrrγ | F: ACT GTT GCA GTT GGA AAG GC | 95 |
| R: TGG AGG GTT CCG TCT TGA TGA | ||
| Pparγ | F: GGT GAC TTT ATG GAG CCT AAG | 110 |
| R: CGG TCT CCA CTG AGA ATA ATG | ||
| Rpl7 | F: TCG CAG AGT TGA AGG TGA AG | 114 |
| R: GCC TGT ACT CCT TGT GAT AGT G | ||
| Npy ChIP | F: TGT GCC TTC CTC CTT ATC AGA | 103 |
| R: GCC ACA AAC ACT GAG CTG TC |
Abbreviations: F, forward; R, reverse.
In silico analysis
Sequences of the 2500 bp region upstream of the transcriptional start site of Npy,Agrp, and Pomc were obtained. The start site of each gene was determined with NCBI GenBank (genome assembly GRCm38.p4, annotation release 106): Npy start chromosome 6, 49822710; Agrp start chromosome 8, 105566695 (reverse strand); and Pomc start chromosome 12, 3954945. BMAL1:CLOCK binding sites (5′CACGTG 3′, 5′CACGNG3′, 5′CACGTT3′, 5′CATG(T/C)G3′, or 5′CANNTG3′) were manually identified. Because of an updated genome annotation, relative positions of binding sites slightly differ from those that were published previously for Npy and Agrp (24). For instance, site 5′CATGTG3′ at –1226 in the Npy promotor in Fick et al. 2010 (24) is listed here as –1207.
Western blotting
Female and male mHypoA-Bmal1-KO and mHypoA-Bmal1-WT cell lines were grown to 90% to 95% confluency. Protein was harvested with 1× lysis buffer (Cell Signaling Technology Inc., New England Biolabs, Whitby, ON, Canada), supplemented with 1 mM phenylmethylsulfonyl fluoride, 1% protease inhibitor cocktail, and 1% phosphatase inhibitor cocktail 2 (MilliporeSigma) and quantified with the BCA protein assay kit (Thermo Fischer Scientific); 25 μg of total protein was separated on 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories Ltd., Mississauga, ON, Canada). Membranes were blocked in 5% skim (nonfat) milk powder dissolved in tris-buffered saline with tween-20 (TBS-T) for 1 hour, before incubation in primary antibody overnight at 4°C. Primary antibodies BMAL1 [D2L7G; Cell Signaling Technology; catalog no. 14020 (37)] and α-tubulin [Cell Signaling Technology; catalog no. 2144 (38)] were diluted 1:1000 in 5% w/v bovine serum albumin in TBS-T. Membranes were then washed with TBS-T, incubated in secondary horseradish peroxidase–linked anti-rabbit antibody [1:7500; Cell Signaling Technology; catalog no. 7074 (39)] for 1 hour, washed with TBS-T, and imaged with the Signal Fire ECL Reagent (Cell Signaling Technology) on the Kodak Image Station 2000R. Membranes were stripped with Restore Plus (Thermo Fisher Scientific) Western blot stripping buffer according to the manufacturer’s instructions before probing for α-tubulin.
Chromatin immunoprecipitation
mHypoA-Bmal1-WT/F cells were grown to 80% to 85% confluence in 10-cm tissue culture dishes and treated with vehicle (0.05% ETOH) or 100 μM BPA for 4 hours as described earlier. Additional dishes of cells were treated with vehicle for immunoprecipitation with positive (histone H3) and negative control (normal rabbit IgG) antibodies. Chromatin immunoprecipitation (ChIP) was performed with the SimpleChIP Enzymatic Chromatin IP kit with magnetic beads (Cell Signaling Technology, Inc.) according to the manufacturer’s instructions. A total of 2 μg of BMAL1 antibody [D2L7G; Cell Signaling Technology (37)] was used for the immunoprecipitation. This was the same antibody used to verify the presence or absence of BMAL1 protein in the cell lines. Purified DNA was analyzed via quantitative PCR on an Applied Biosystem Prism 7900HT machine with cycling conditions as described earlier. Each sample was assayed in duplicate with 2 μL of DNA in a 20-μL reaction with primers specific for the Npy promotor (–1267 to –1165 relative to the transcriptional start site) (Table 1) or the Rpl30 promotor (for positive control H3 provided with the kit). The primer for the Npy promotor was designed previously and the region validated to bind BMAL1 (24). Mean cycle of threshold of each immunoprecipitation sample and its respective 2% input sample were used to calculate the amount of DNA pulled down according to the following equation: percentage input = 2% × 2[C(T) 2% input sample – C(T) IP sample]. Relative binding was calculated by taking the average of the percentage inputs of both vehicle and treated groups per experimental replicate and dividing each value by the respective average.
Statistical analysis
Data were analyzed for statistical significance with GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA). Groups were compared by t test, one-way ANOVA, or two-way ANOVA, followed by the Bonferroni or Tukey multiple comparison test, where appropriate. Differences were considered statistically significant when P < 0.05. All data are presented as mean ± SEM, and statistical significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Results
Circadian clock gene expression is altered in response to BPA exposure
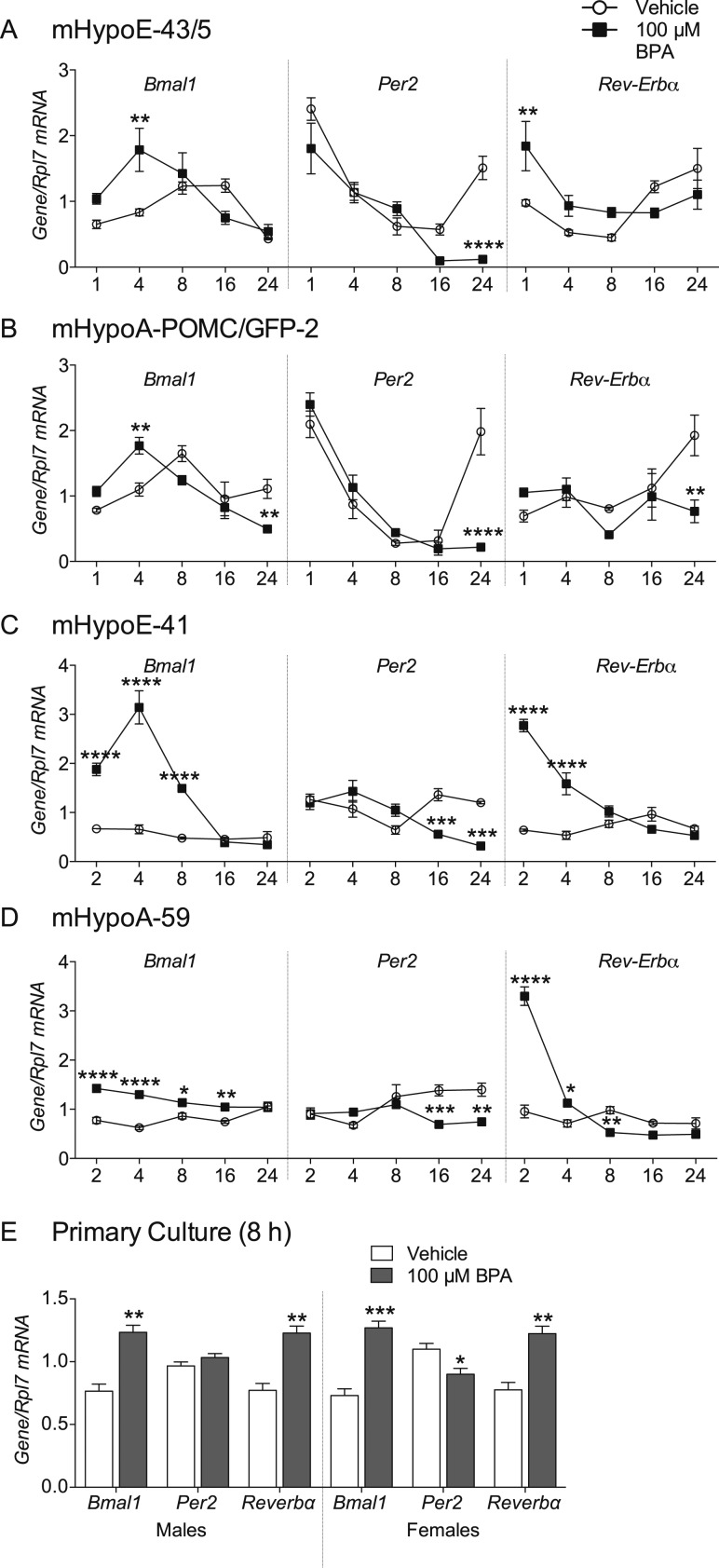
We have previously seen that exposure to the obesogenic endocrine disrupting chemical BPA upregulates Pomc expression in the mHypoE-43/5 and mHypoA-POMC/GFP-2 cell lines (21) and Npy and Agrp expression in the mHypoE-41 and mHypoA-59 cell lines (data not shown). Because disruption of the hypothalamic circadian system is associated with changes in energy regulation (7, 31), we investigated whether BPA alters the expression of three central clock genes in cell lines where neuropeptide expression is dysregulated. A total of 100 μM BPA increased Bmal1 expression at 4 hours in both mHypoA-POMC/GFP-2 [F(4, 24)interaction = 9.097, P = 0.0001; F(4, 24)time = 12.26, P = 0.0001; F(1, 24)treatment = 0.2295, P = 0.6362] and mHypoE-43/5 cell lines [F(4, 29)interaction = 5.131, P = 0.0030; F(4, 29)time = 8.620, P = 0.0001; F(1, 29)treatment = 4.867, P = 0.0355] and from 2 to 8 or 16 hours in mHypoE-41 [F(4, 22)interaction = 37.40, P < 0.0001; F(4, 22)time = 50.29, P < 0.0001; F(1, 24)treatment = 138.4, P < 0.0001] and mHypoA-59 [F(4, 30)interaction = 10.99, P < 0.0001; F(4, 30)time = 3.270, P < 0.0244; F(1, 30)treatment = 95.83, P < 0.0001] cell lines, respectively (Fig. 1A–1D). Furthermore, Bmal1 mRNA levels were decreased at 24 hours in the mHypoA-POMC/GFP-2 cell line (Fig. 1B). BPA downregulated Per2 expression at 24 hours in both POMC-expressing cell lines and at 16 and 24 hours [mHypoA-POMC/GFP-2: F(4, 24)interaction = 11.08, P < 0.0001; F(4, 24)time = 28.84, P < 0.0001; F(1, 24)treatment = 3.417, P = 0.0769; mHypoE-43/5: F(4, 27)interaction = 8.079, P = 0.0002; F(4, 27)time = 37.22, P < 0.0001; F(1, 27)treatment = 20.51, P = 0.0001] in NPY/AgRP-expressing lines [mHypoA-59: F(4, 30)interaction = 6.959, P = 0.0004; F(4, 30)time = 3.309, P = 0.0233; F(1, 30)treatment = 12.14, P = 0.0015; mHypoE-41: F(4, 22)interaction = 12.92, P < 0.0001; F(4, 22)time = 6.569, P = 0.0012; F(1, 22)treatment = 6.627, P = 0.0173]. Rev-Erbα expression was significantly increased at early time points (1 to 4 hours) in mHypoE-43/5 [F(4, 29)interaction = 5.343, P = 0.0024; F(4, 29)time = 8.074, P = 0.0002; F(1, 29)treatment = 2.619, P = 0.1164], mHypoE-41 [F(4, 26)interaction = 33.23, P < 0.0001; F(4, 26)time = 21.84, P < 0.0001; F(1, 26)treatment = 63.27, P < 0.0001], and mHypoA-59 [F(4, 30)interaction = 77.86, P < 0.0001; F(4, 30)time = 97.55, P < 0.0001; F(1, 30)treatment = 40.00, P < 0.0001] cells, and significantly decreased at later time points in the mHypoA-POMC/GFP-2 [F(4, 22)interaction = 4.218, P = 0.0110; F(4, 22)time = 3.441, P = 0.0250; F(1, 22)treatment = 3.214, P = 0.0868], and mHypoA-59 lines (24 and 8 hours, respectively) (Fig. 1A–1D). POMC-expressing cell lines were synchronized with forskolin, therefore exhibiting a rhythm in Bmal1,Per2, and Rev-Erbα basal expression (Fig. 1A and 1B). This is not seen in the unsynchronized NPY/AgRP-expressing cells (Fig. 1C and 1D). Although slight differences exist in the timing and magnitude of response to BPA between each cell line, there is an overall increase in Bmal1, a decrease in Per2, and an upregulation in Rev-Erbα, illustrating a dysregulation in the normal expression of these circadian rhythm–associated genes. These effects are replicated in primary culture from CD-1 mice, where 8 hours of BPA treatment upregulated Bmal1 and Rev-Erbα in both male-derived (Bmal1, t6 = 5.901, P = 0.00105; Rev-Erbα, t6 = 5.788, P = 0.00116), and female-derived (Bmal1, t6 = 7.106, P = 0.000390; Rev-Erbα, t6 = 5.350, P = 0.00175) cultures and downregulated Per2 (t6 = 3.032, P = 0.0230) in female-derived cultures (Fig. 1E).
Figure 1.
BPA dysregulates circadian gene expression in hypothalamic POMC and NPY/AgRP-expressing neurons. POMC-expressing (A) mHypoE-43/5 and (B) mHypoA-POMC/GFP-2 cells lines and NPY/AgRP-expressing (C) mHypoE-41 and (D) mHypoA-59 cell lines were treated with 100 µM BPA or vehicle (0.05% ETOH) for 1 or 2, 4, 8, 16, and 24 hours and mRNA expression of Bmal1,Per2, and Rev-Erbα was measured by qRT-PCR (n = 3 to 4). (E) Hypothalamic primary cultures from CD-1 male and female mice were also treated with 100 µM BPA for 8 hours and mRNA expression measured (n = 4). Data are expressed as mean ± SEM, with open circles and bars representing vehicle (0.05% ETOH) and shaded circles and bars representing 100 µM BPA treatment. Statistical significance was determined with a (A–D) two-way ANOVA, followed by the Bonferroni post hoc test or (E) a multiple t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
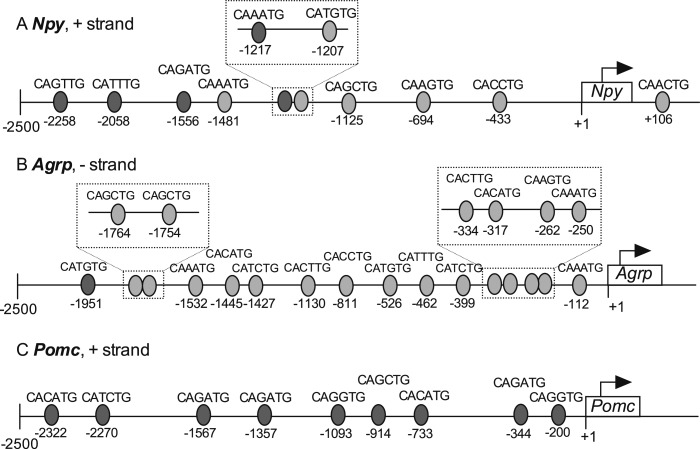
Putative binding sites for BMAL1-CLOCK heterodimers exist in the regulatory regions of Npy, Agrp, and Pomc
Considering that BPA-induced alteration of Bmal1 expression precedes, or occurs simultaneously with, changes in Npy,Agrp, or Pomc, we questioned whether BMAL1 may be a mechanistic link between BPA and the changes seen in neuropeptide gene expression. To address this question, we first determined potential BMAL1:CLOCK heterodimer binding sites in the 2500-bp 5′ regulatory regions of neuropeptide genes. BMAL1:CLOCK heterodimer canonically binds to E-boxes 5′CACGTG3′ (40) in DNA; however, it can also bind to other sequences including 5′CACGNG3′, 5′CACGTT3′, 5′CATG(T/C)G3′ (41), and 5′CANNTG3′ (42). Several positions containing the 5′CANNTG3′ sequence are illustrated in Fig. 2. Binding sites in the Npy and Agrp promotor have been previously reported (24). We identified four additional sites in the Npy and one additional site in the Agrp promotor. These newly identified sites were located between the −2500 bp and −1500 bp regions of the promotors, except for the 5′CAAATG3′ site at −1217 in the Npy promotor. In total, the regulatory region of Npy contained 10 sites, Agrp contained 16 sites (Fig. 2B), and Pomc contained 9 (Fig. 2C). The presence of E-box elements in these regulatory regions suggests the potential action of BMAL1 as a transcriptional regulator of Npy, Agrp, and Pomc gene expression.
Figure 2.
Potential BMAL1:CLOCK binding sites in the Npy,Agrp, and Pomc promotors. Putative binding sites for BMAL1:CLOCK are represented by gray ovals in the 2500-bp upstream (5′) region of the transcriptional start site (TSS, +1) of the (A) Npy, (B) Agrp, and (C) Pomc genes. The sequence of each binding site is written above the oval with the location relative to the TSS listed below. Where two or more sites are close together, the area is expanded and detailed in the inserts. Lighter ovals represent sites identified previously by Fick et al. (24), and darker ovals represent newly identified binding sites. All sequences are listed 5′ to 3′.
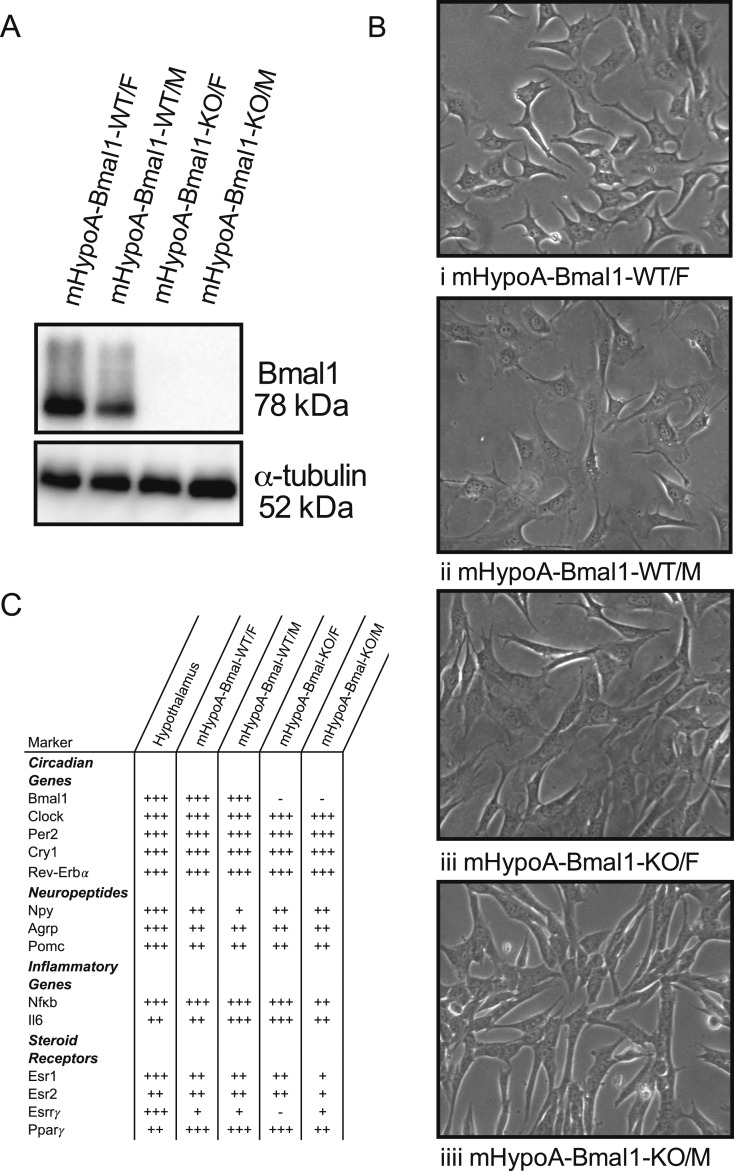
Characterization of mHypoA-Bmal1-KO cell lines
To elucidate the role of BMAL1 on BPA-induced changes in neuropeptide expression, a neuronal model lacking functional BMAL1 protein expression was needed. Having previously encountered difficulty with small interfering RNA knockdown of Bmal1, our laboratory generated immortalized hypothalamic cell lines from female and male whole-body Bmal1-KO mice, alongside WT littermate controls. These cell lines are nonclonal, exhibit neuronal morphology (Fig. 3B), and express Npy,Agrp, and Pomc (Fig. 3C). The absence of BMAL1 protein and mRNA was verified by Western blot (Fig. 3A) and qRT-PCR (Fig. 3C). Although the KO models lack Bmal1 expression, basal expression of other genes in the circadian feedback loop (Clock, Per2, Cry, Rev-Erbα) were similar to WT cells (Fig. 3C). The expression of inflammatory markers and receptors, including the estrogen receptors, estrogen-related receptor-γ, and peroxisome proliferator–activated receptor-γ, were verified with qRT-PCR. Besides the lack of estrogen-related receptor-γ expression in the mHypoA-Bmal1-KO/M cells, the expression profiles of the cell lines are similar. These receptors have been linked to the mechanism of BPA action in peripheral tissues and neuronal models (21, 43), suggesting the cells are likely to respond to BPA exposure.
Figure 3.
Characterization of the mHypoA-Bmal1-WT and -KO cell models. mHypoA-Bmal1-KO/F and mHypoA-Bmal1-KO/M cell lines do not express BMAL1. (A) Expression or absence of BMAL1 protein in the WT vs KO cell lines was verified with Western blotting with a BMAL1 antibody and alpha-tubulin used as a loading control. (B) Cell lines were imaged with an Olympus CKX41 microscope (10× lens objective) with the Tucsen 10.0 MP IS1000 USB camera and exhibit neuronal morphology. (C) Summary of circadian, neuropeptide, inflammatory marker and steroid receptor mRNA expression in hypothalamus tissue, mHypoA-Bmal1-WT/F, mHypoA-Bmal1-WT/M, mHypoA-Bmal1-KO/F, and mHypoA-Bmal1-KO/M cell lines. RNA was isolated from untreated hypothalamic tissue or cells before cDNA synthesis and analysis by qRT-PCR. Relative expression is denoted by + or −, where − (not expressed) cycle at threshold (CT) ≤ 35, + CT = 30 to 34.9, ++ CT = 25 to 29.9, +++ (highly expressed) CT = 20 to 24.9. Esr1, estrogen receptor α;Esr2, estrogen receptor β;Esrrγ, estrogen-related receptor γ;Nfkb, nuclear factor κb.
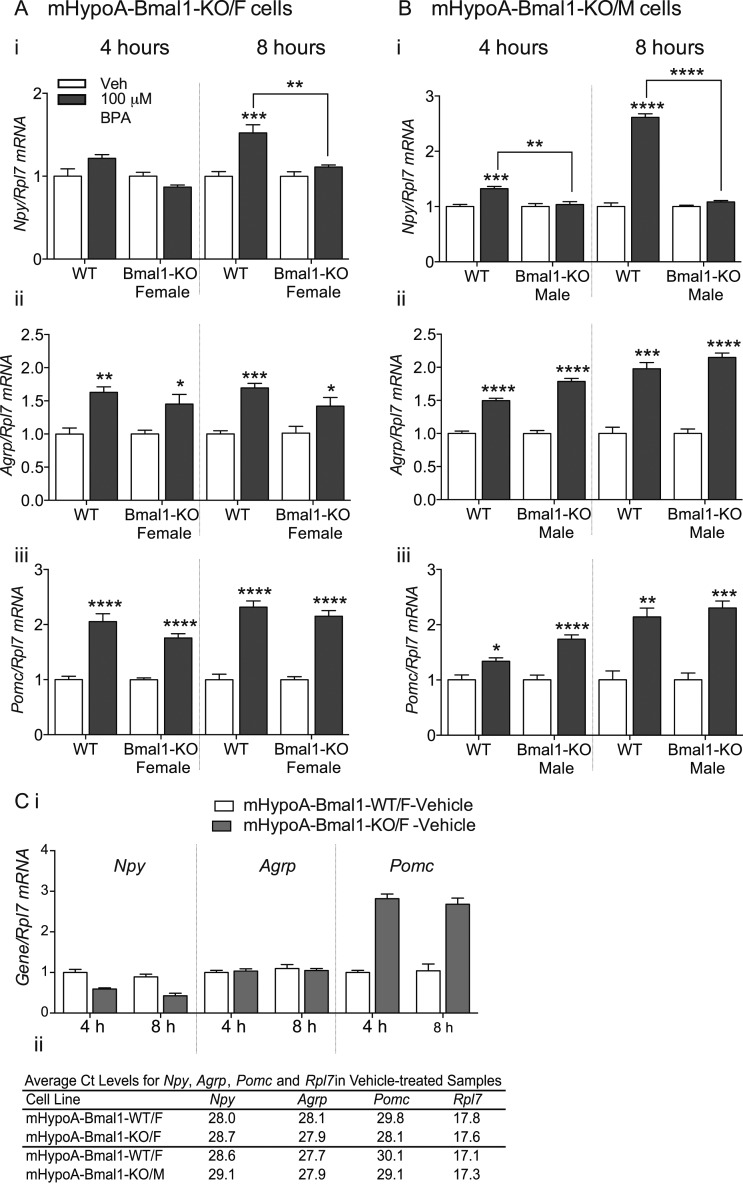
The effect of BPA on Npy expression, but not Agrp or Pomc, depends on Bmal1
Using the mHypoA-Bmal1-KO and WT littermate control cell lines, we investigated whether BMAL1 was involved in BPA-mediated upregulation of the neuropeptide gene expression. The mHypoA-Bmal1-KO/F (Fig. 4A) and mHypoA-Bmal1-KO/M (Fig. 4B) cell lines were treated with 100 μM BPA for 8 hours alongside mHypoA-Bmal1-WT/F controls. Agrp and Pomc expression were increased at 4 and 8 hours in both WT and KO cell lines [Fig. 4A, ii 4 hours, F(1, 16)treatment = 29.49, P < 0.0001; 8 hours, F(1, 14)treatment = 39.73, P < 0.0001; Fig. 4A, iii 4 hours, F(1, 16)treatment = 106.1, P < 0.0001; 8 hours, F(1, 16)treatment = 171.6, P < 0.0001; and Fig. 4B, ii 4 hours, F(1, 20)treatment = 250.4, P < 0.0001; 8 hours, F(1, 8)treatment = 169.1, P < 0.0001; Fig. 4B, iii 4 hours, F(1, 20)treatment = 45.40, P < 0.0001; 8 hours, F(1, 8)treatment = 71.62, P < 0.0001)]. However, the increase in Npy expression seen in the mHypoA-Bmal1-WT/F cell line at 8 hours was not present in male mHypoA-Bmal1-KO/F [Fig. 4A, i 8 hours, F(1, 16)interaction = 10.30, P = 0.0055; F(1, 16)cell-type = 10.30, P = 0.0055; F(1, 22)treatment = 24.43, P = 0.0001] or mHypoA-Bmal1-KO/M [Fig. 4B, 4 hours, F(1, 20)interaction = 9.326, P = 0.00063; F(1, 20)cell-type = 9.326, P = 0.00063; F(1, 20)treatment = 14.72, P = 0.0001; 8 hours, F(1, 8)interaction = 238.0, P < 0.0001; F(1, 8)cell-type = 238.0, P < 0.0001; F(1, 8)treatment = 293.5, P < 0.0001] cell model, implicating the involvement of BMAL1 in the upregulation of Npy but not Agrp or Pomc in response to BPA. Interestingly, basal mRNA levels of both Npy and Pomc were altered in mHypoA-Bmal1-KO cells compared with mHypoA-Bmal1-WT/F cells (Fig. 4C).
Figure 4.
BPA upregulates Npy,Agrp, and Pomc in mHypoA-Bmal1-WT/F cells, whereas the upregulation in Npy is absent in mHypoA-Bmal1-KO/F and mHypoA-Bmal1-KO/M cells. (A) mHypoA-Bmal1-KO/F (n = 4 to 5) and (B) mHypoA-Bmal1-KO/M (n = 6 for 4 hours, n = 3 for 8 hours) cells were treated with 100 µM BPA or vehicle (0.05% ETOH) for 4 or 8 hours alongside mHypoA-Bmal1-WT/F. (A i and B i) Npy, (A ii and B ii) Agrp, and (A iii and B iii) Pomc mRNA expression was quantified with qRT-PCR. Data are expressed as mean ± SEM. Statistical significance were determined with a two-way ANOVA, followed by the Tukey multiple comparison test. (C i) Npy,Agrp, and Pomc expression (n = 5) in mHypoA-Bmal1-KO/F cells compared with mHypoA-Bmal1-WT/F cells treated with vehicle (0.05% ETOH) for 4 or 8 hours. All groups are expressed relative to the 4-hour vehicle-treated mHypoA-Bmal1-WT/F group, which was normalized to 1. Data are expressed as mean ± SEM. (C ii) Mean cycle at threshold (CT) levels of Npy, Agrp, Pomc, and Rpl7 as measured by qRT-PCR in vehicle-treated mHypoA-Bmal1-KO/F or mHypoA-Bmal1-KO/M cells compared with mHypoA-Bmal1-WT/F cells that were treated in parallel. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
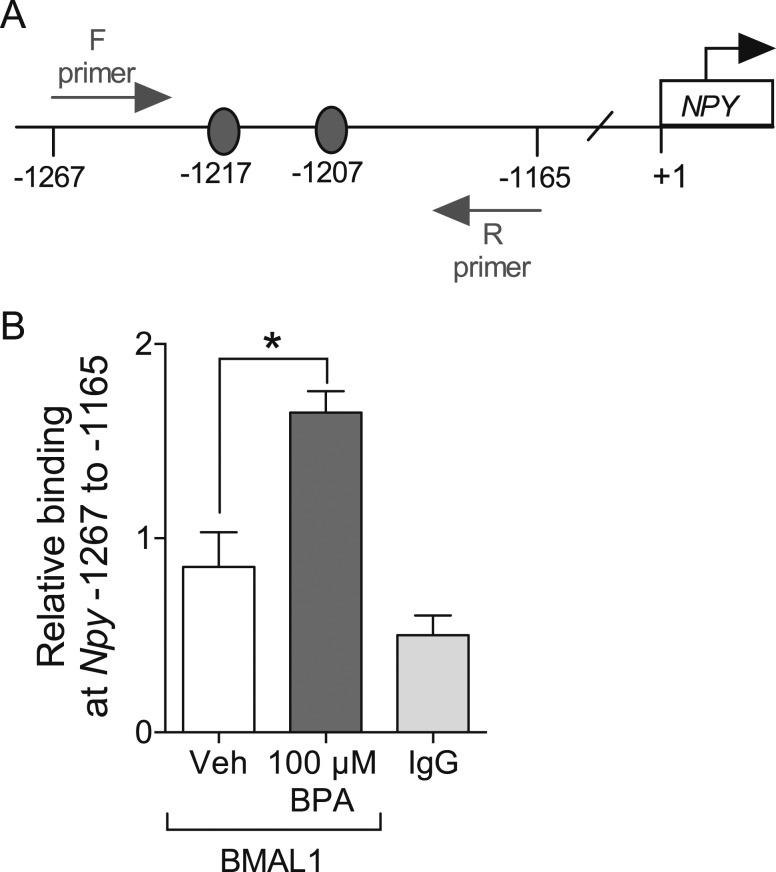
BPA treatment increases BMAL1 binding to the Npy promotor
To elucidate whether BMAL1 is directly involved in regulating the BPA-mediated induction of Npy by binding to the Npy promotor, we performed ChIP using a BMAL1 antibody in the mHypoA-Bmal1-WT/F cells. Compared with negative control IgG, binding was slightly greater in the chromatin immunoprecipitated with BMAL1, suggesting a basal degree of BMAL1 binding to this region of the Npy promotor at 4 hours. Treatment with 100 μM BPA significantly increased the amount BMAL1 protein binding to the −1267 to −1165 region of the Npy promotor compared with vehicle treatment after 4 hours (Fig. 5B, F(2, 6) = 18.97, P = 0.0025). Two putative binding sites exist for the BMAL1:CLOCK heterodimer in this region of the promotor, including 5′CAAATG3′ at −1217 and 5′CATGTG3′ at −1207 (Fig. 5A, Fig. 2A). This finding suggests that BMAL1 may be involved in regulating Npy at the transcriptional level upon BPA exposure.
Figure 5.
BPA increases BMAL1 binding to the Npy promotor. (A) Schematic representing the 103-bp region of the Npy promotor, containing two potential BMAL1 binding sites (gray ovals), amplified for the ChIP assay with qRT-PCR. (B) mHypoA-Bmal1-WT/F cells were treated with 100 µM BPA or vehicle (0.05% ETOH) for 4 hours, and changes in BMAL1 binding to the −1267 to −1165 region of the Npy promotor were measured with a ChIP assay (n = 3). A vehicle-treated chromatin sample was incubated with normal IgG antibody as a negative control. Data are expressed as mean ± SEM. Statistical significance was determined with a one-way ANOVA, followed by the Bonferroni multiple comparison test. *P < 0.05. F, forward; R, reverse.
Discussion
Under normal conditions, the SCN synchronizes the feeding centers of the hypothalamus, allowing neurons in these regions to respond appropriately to timed signals from the rest of the body (1, 6). However, metabolic factors may dysregulate the expression of clock genes, leading to a loss of synchrony (1). Such factors include high glucose and dietary fatty acids, which can alter the expression patterns of Bmal1,Per2, and Rev-Erbα by changing the peak amplitude and period of the 24-hour expression profile (31, 44). In turn, this change alters expression profiles of genes that are rhythmically controlled by these clock-associated transcription factors (26, 27, 31). Here, we describe changes in hypothalamic circadian gene expression induced by BPA, a chemical known to lead to metabolic perturbations. In immortalized hypothalamic cell lines and primary culture, we show that exposure to BPA increased Bmal1 expression within 2 to 4 hours and decreased Per2 expression at 16 and 24 hours. This effect was observed across four hypothalamic cell lines, representing both adult and embryonic cells as well as anorexigenic POMC and orexigenic NPY/AgRP-expressing cells, illustrating the robust nature of these effects across heterogeneous populations of neurons. Rev-Erbα was also altered, although the pattern was more heterogeneous across the cell lines.
Although there are no studies to date investigating circadian dysregulation by BPA in the adult mammalian hypothalamus, Sen and Sellix (45) have reported altered clock gene expression in reproductive tract tissues. Furthermore, Choi et al. (46) showed that Per2 and Cry1 levels were downregulated with increasing BPA concentration and exposure time in goldfish brain and liver. Circulating concentrations of melatonin, a hormone involved in entrainment of circadian rhythms, and the expression of the melatonin receptor were also decreased with BPA exposure (46). The impact of endocrine disrupting chemicals besides BPA on circadian systems, particularly pertaining to reproductive tissues, has been described (45). Interestingly, prenatal exposure of Sprague-Dawley rats to a weakly estrogenic polychlorinated biphenyl mixture (Aroclor 1221) altered Bmal1 and Per2 expression in the anteroventral periventricular nucleus of the hypothalamus in female pups (47). These studies, along with our work, illustrate that the circadian clock system is vulnerable to dysregulation by endocrine disrupting chemicals, including BPA.
A dysregulation in clock gene expression has broad implications for cellular function, such as cell migration (48) and gene transcription (26). Changes in core clock genes by BPA may disrupt the transcription-translation feedback loop that exists within these hypothalamic neurons, thereby disrupting rhythmicity. Circadian clock genes not only act to maintain a 24-hour rhythmicity within the body, they bind to promotor regions of ∼10% of genes and can alter expression of these several E-box domain containing genes (26, 27). We and others have described the presence of potential BMAL1:CLOCK heterodimer binding sites on the Npy,Agrp (24), and Pomc gene promotors, suggesting that BPA-induced disruption of components of the molecular clock may be responsible for the changes observed in Npy, Agrp, and Pomc expression.
To experimentally determine the role of clock genes in the regulation of feeding neuropeptides by BPA, we chose a Bmal1-KO model. BMAL1 is critical for the circadian regulation of energy homeostasis as whole-body Bmal1-KO mice exhibit disruptions in the daily rhythm of glucose and triglyceride levels (49). Unlike Cry and Per-KO mice, Bmal1-KO mice lack functional redundancy because BMAL1 controls the transcription of the paralogous gene Bmal2, leading to the complete absence of BMAL function (50, 51). Using previously established techniques (36), we immortalized hypothalamic neurons from Bmal1-KO animals and their WT control littermates. With the generation of these cell models, we showed that BPA-mediated upregulation of Npy, but not Agrp or Pomc, requires functional BMAL protein expression. This finding may imply that the inability of BPA to change BMAL1 expression, synthesis, or binding protects the mHypoA-Bmal1-KO hypothalamic cells from BPA-induced dysregulation of Npy expression. Although both Agrp and Pomc contain several putative BMAL1 binding sites and may be basally regulated by rhythmic BMAL1 binding, the BMAL1 dependency of the BPA-induced changes is specific to Npy. However, the fact that the Agrp effect is not altered in mHypoA-Bmal1-KO lines coincides with the finding that Agrp mRNA was previously found to lack rhythmicity in one population of hypothalamic neurons (24). The role of BMAL1 in basal Npy and Pomc, but potentially not Agrp, expression is further illustrated in the differences in basal Npy and Pomc mRNA levels in the mHypoA-Bmal1-KO cell lines compared with the mHypoA-Bmal1-WT/F cell line (Fig. 4C). As BMAL1 rhythmically binds to the Npy promotor (24), the lack of BMAL1 expression in the mHypoA-Bmal1-KO cells may explain the slightly lower expression of Npy. Interestingly, the difference in basal expression of Pomc between the mHypoA-Bmal1-WT/F and the mHypoA-Bmal1-KO cell lines does not alter the magnitude of the BPA-induced Pomc response, again emphasizing the specificity of the BMAL1-dependent effect of BPA on Npy.
Others have alluded to the relationship between Bmal1 and Npy. For example, a low-protein diet in pregnant dams ablated the circadian rhythm of Bmal1 expression in the hypothalamus of 17-day-old offspring, with a concurrent shift in the diurnal rhythm of Npy expression (52). Furthermore, in a clonal Npy-expressing cell line, palmitate increased Bmal1 expression and decreased its amplitude of oscillation along with an increase in Npy mRNA levels (31). However, in these studies it remained unclear whether BMAL1 has direct transcriptional control of Npy expression. Fick et al. (24) demonstrated a rhythmic pattern of BMAL1 binding to the Npy promotor across a 24-hour time course in the mHypoE-44 hypothalamic cell line, suggesting this rhythmic binding may contribute to the rhythmic expression of Npy mRNA. Specifically, using ChIP analysis, we have shown here that BPA increases the relative amount of BMAL1 binding to the Npy promotor before the changes seen in Npy expression, suggesting transcriptional upregulation of Npy. In line with our findings, nervous system–specific Bmal1-KO mice have decreased body weight and food intake (53). Because increased BMAL1 binding may contribute to increased expression of Npy, downregulation of Npy may partially underlie this anorexigenic phenotype of Bmal1-KO mice.
Whether factors other than BMAL1 lead to the lack of BPA-induced Npy expression in the mHypoA-Bmal1-KO cells warrants investigation. The mHypoA-Bmal1-KO and mHypoA-Bmal1-WT cell lines are similar in terms of basal expression of neuropeptides, circadian rhythm genes, and inflammatory genes. The lack of Esrrγ expression in the mHypoA-Bmal1-KO/F cells probably does not contribute to the ablated Npy response because the mHypoA-Bmal1-KO/M cells express Esrrγ at the same level as mHypoA-Bmal1-WT cells. Although basal expression levels of these genes are similar across both KO and WT cell lines, their rhythmic expression over 24 hours may be affected by the absence of BMAL1. For instance, rhythmic expression of circadian genes Rev-Erbα and d-box binding PAR bZIP transcription factor is abolished in adrenal cortex Bmal1-KO animals (54). Future investigations may include whether changes in rhythmic expression profiles of certain genes contribute to the abolished Npy response. Regardless, ablation of a crucial circadian gene is not a feasible or desirable method to protect hypothalamic cells from BPA-mediated neuropeptide dysregulation. Elucidating the mechanism by which this dysregulation in circadian gene expression or binding occurs may provide beneficial information to block the effects.
The mechanisms by which BPA alters clock gene expression in feeding-related hypothalamic neurons remains unknown. BPA has been thought to act primarily through nuclear and membrane-bound estrogen receptors and other nuclear receptors, including peroxisome proliferator activated receptor-γ (PPARγ), estrogen-related receptor-γ, aryl hydrocarbon receptor, androgen receptor, and the glucocorticoid receptor (43, 55). These receptors can bind to response elements in promotors and act as transcription factors to regulate gene expression. In fact, Rhee et al. (56) described the presence of xenobiotic response elements, including aryl hydrocarbon receptor response elements and estrogen receptor response elements, in the promotor regions of killfish Clock, Bmal1, Per2, Cry1, and Cry2. Furthermore, PPARγ has been shown to act as a transcriptional regulator of Bmal1. We have recently identified PPARγ as a mediator of BPA-induced upregulation of Pomc in the mHypoA-POMC/GFP-2 cell line (21). In addition to steroid and nuclear receptor activation, BPA can induce inflammatory signaling (16, 21, 57), oxidative stress (15), endoplasmic reticulum stress, and MAP kinase signaling (16, 57, 58). Whether these receptors and pathways are involved in BPA-mediated Bmal1,Per2, or Rev-Erbα dysregulation in hypothalamic feeding-related neurons remains to be determined.
In conclusion, this study explored the direct effects of BPA on circadian clock gene expression in POMC and NPY/AgRP mammalian hypothalamic cell models, demonstrating altered expression of circadian rhythm genes. We also described the development, characterization, and experimental use of Bmal1-KO hypothalamic cell lines that can be used to define the role of BMAL1 in hypothalamic responses to various hormonal and metabolic signals. Using these cell lines, we showed the involvement of BMAL1 in BPA-mediated Npy dysregulation, implicating hypothalamic circadian rhythm alterations in the metabolic consequences of BPA exposure. These findings in turn shed light on the fundamental connection between metabolism and the circadian molecular clock.
Acknowledgments
We thank Dr. Patricia Brubaker, Alex Martchenko, and Sarah Wheeler for breeding and providing the Bmal1-KO and WT mice. We also thank Emma McIlwraith, Matthew Clemenzi, Chantel Kowalchuk, and Andy Tran for helpful discussions.
Financial Support: We acknowledge funding from the Canadian Institutes for Health Research and Canada Foundation for Innovation and Canada Research Chairs Program (to D.D.B.). N.L. was supported by the Natural Sciences and Engineering Research Council and the Banting & Best Diabetes Centre. A.S. was supported by the Ontario Graduate Studentship.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- AgRP
agouti-related peptide
- ARC
arcuate nucleus
- Bmal1
brain and muscle ARNT-like 1
- BPA
bisphenol A
- ChIP
chromatin immunoprecipitation
- CSFBS
charcoal:dextran-stripped fetal bovine serum
- ETOH
ethyl alcohol
- FBS
fetal bovine serum
- KO
knockout
- NPY
neuropeptide Y
- POMC
pro-opiomelanocortin
- PPARγ
peroxisome proliferator activated receptor-γ
- PS
penicillin/streptomycin
- qRT-PCR
quantitative RT-PCR
- Rev-Erb
reverse erythroblastosis virus
- SCN
suprachiasmatic nucleus
- TBS-T
tris-buffered saline with tween-20
- WT
wild-type
References
- 1. Oosterman JE, Kalsbeek A, la Fleur SE, Belsham DD. Impact of nutrients on circadian rhythmicity. Am J Physiol Regul Integr Comp Physiol. 2015;308(5):R337–R350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagai K, Nishio T, Nakagawa H, Nakamura S, Fukuda Y. Effect of bilateral lesions of the suprachiasmatic nuclei on the circadian rhythm of food-intake. Brain Res. 1978;142(2):384–389. [DOI] [PubMed] [Google Scholar]
- 3. Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring). 2009;17(11):2100–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di Lorenzo L, De Pergola G, Zocchetti C, L’Abbate N, Basso A, Pannacciulli N, Cignarelli M, Giorgino R, Soleo L. Effect of shift work on body mass index: results of a study performed in 319 glucose-tolerant men working in a southern Italian industry. Int J Obes Relat Metab Disord. 2003;27(11):1353–1358. [DOI] [PubMed] [Google Scholar]
- 5. Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58(11):747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buijs FN, Guzmán-Ruiz M, León-Mercado L, Basualdo MC, Escobar C, Kalsbeek A, Buijs RM. Suprachiasmatic nucleus interaction with the arcuate nucleus; essential for organizing physiological rhythms. eNeuro. 2017;4(2):ENEURO.0028-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. [DOI] [PubMed] [Google Scholar]
- 8. Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev. 2009;30(4):293–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Statistics Canada. Bisphenol A concentrations in Canadians, 2012 and 2013. Available at: www150.statcan.gc.ca/n1/pub/82-625-x/2015001/article/14208-eng.htm. Accessed 15 July 2015.
- 10. Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect. 2005;113(4):391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rancière F, Lyons JG, Loh VHY, Botton J, Galloway T, Wang T, Shaw JE, Magliano DJ. Bisphenol A and the risk of cardiometabolic disorders: a systematic review with meta-analysis of the epidemiological evidence. Environ Health. 2015;14:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong SH, Sung Y-A, Hong YS, Ha E, Jeong K, Chung H, Lee H. Urinary bisphenol A is associated with insulin resistance and obesity in reproductive-aged women. Clin Endocrinol (Oxf). 2017;86(4):506–512. [DOI] [PubMed] [Google Scholar]
- 13. Provvisiero DP, Pivonello C, Muscogiuri G, Negri M, de Angelis C, Simeoli C, Pivonello R, Colao A. Influence of bisphenol A on type 2 diabetes mellitus. Int J Environ Res Public Health. 2016;13(10):E989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alonso-Magdalena P, Quesada I, Nadal Á. Prenatal exposure to BPA and offspring outcomes: the diabesogenic behavior of BPA. Dose Response. 2015;13(2):1559325815590395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moghaddam HS, Samarghandian S, Farkhondeh T. Effect of bisphenol A on blood glucose, lipid profile and oxidative stress indices in adult male mice. Toxicol Mech Methods. 2015;25(7):507–513. [DOI] [PubMed] [Google Scholar]
- 16. Yang M, Chen M, Wang J, Xu M, Sun J, Ding L, Lv X, Ma Q, Bi Y, Liu R, Hong J, Ning G. Bisphenol A promotes adiposity and inflammation in a nonmonotonic dose-response way in 5-week-old male and female C57BL/6J mice fed a low-calorie diet. Endocrinology. 2016;157(6):2333–2345. [DOI] [PubMed] [Google Scholar]
- 17. Arora S, Anubhuti. Role of neuropeptides in appetite regulation and obesity: a review. Neuropeptides. 2006;40(6):375–401. [DOI] [PubMed] [Google Scholar]
- 18. Desai M, Ferrini MG, Han G, Jellyman JK, Ross MG. In vivo maternal and in vitro BPA exposure effects on hypothalamic neurogenesis and appetite regulators. Environ Res. 2018;164:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. MacKay H, Patterson ZR, Abizaid A. Perinatal exposure to low-dose bisphenol-A disrupts the structural and functional development of the hypothalamic feeding circuitry. Endocrinology. 2017;158(4):768–777. [DOI] [PubMed] [Google Scholar]
- 20. Mackay H, Patterson ZR, Khazall R, Patel S, Tsirlin D, Abizaid A. Organizational effects of perinatal exposure to bisphenol-A and diethylstilbestrol on arcuate nucleus circuitry controlling food intake and energy expenditure in male and female CD-1 mice. Endocrinology. 2013;154(4):1465–1475. [DOI] [PubMed] [Google Scholar]
- 21. Salehi A, Loganathan N, Belsham DD. Bisphenol A induces Pomc gene expression through neuroinflammatory and PPARgamma nuclear receptor-mediated mechanisms in POMC-expressing hypothalamic neuronal models. Mol Cell Endocrinol. 2019;479:12–19. [DOI] [PubMed] [Google Scholar]
- 22. Chalmers JA, Lin SY, Martino TA, Arab S, Liu P, Husain M, Sole MJ, Belsham DD. Diurnal profiling of neuroendocrine genes in murine heart, and shift in proopiomelanocortin gene expression with pressure-overload cardiac hypertrophy. J Mol Endocrinol. 2008;41(3):117–124. [DOI] [PubMed] [Google Scholar]
- 23. Stütz AM, Staszkiewicz J, Ptitsyn A, Argyropoulos G. Circadian expression of genes regulating food intake. Obesity (Silver Spring). 2007;15(3):607–615. [DOI] [PubMed] [Google Scholar]
- 24. Fick LJ, Fick GH, Belsham DD. Rhythmic clock and neuropeptide gene expression in hypothalamic mHypoE-44 neurons. Mol Cell Endocrinol. 2010;323(2):298–306. [DOI] [PubMed] [Google Scholar]
- 25. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. [DOI] [PubMed] [Google Scholar]
- 26. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. [DOI] [PubMed] [Google Scholar]
- 27. Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5(1):407–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology. 2009;150(5):2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boden G. Interaction between free fatty acids and glucose metabolism. Curr Opin Clin Nutr Metab Care. 2002;5(5):545–549. [DOI] [PubMed] [Google Scholar]
- 31. Fick LJ, Fick GH, Belsham DD. Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem Biophys Res Commun. 2011;413(3):414–419. [DOI] [PubMed] [Google Scholar]
- 32. Angelousi A, Kassi E, Nasiri-Ansari N, Weickert MO, Randeva H, Kaltsas G. Clock genes alterations and endocrine disorders. Eur J Clin Invest. 2018;48(6):e12927. [DOI] [PubMed] [Google Scholar]
- 33. Chevalier N, Fénichel P. Bisphenol A: targeting metabolic tissues. Rev Endocr Metab Disord. 2015;16(4):299–309. [DOI] [PubMed] [Google Scholar]
- 34. Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology. 2004;145(1):393–400. [DOI] [PubMed] [Google Scholar]
- 35. Nazarians-Armavil A, Chalmers JA, Lee CB, Ye W, Belsham DD. Cellular insulin resistance disrupts hypothalamic mHypoA-POMC/GFP neuronal signaling pathways. J Endocrinol. 2013;220(1):13–24. [DOI] [PubMed] [Google Scholar]
- 36. Belsham DD, Fick LJ, Dalvi PS, Centeno ML, Chalmers JA, Lee PK, Wang Y, Drucker DJ, Koletar MM. Ciliary neurotrophic factor recruitment of glucagon-like peptide-1 mediates neurogenesis, allowing immortalization of adult murine hypothalamic neurons. FASEB J. 2009;23(12):4256–4265. [DOI] [PubMed] [Google Scholar]
- 37. RRID:AB_2728705.
- 38. RRID:AB_2210548.
- 39. RRID:AB_2099233.
- 40. Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17(7):3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoshitane H, Ozaki H, Terajima H, Du NH, Suzuki Y, Fujimori T, Kosaka N, Shimba S, Sugano S, Takagi T, Iwasaki W, Fukada Y. CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol Cell Biol. 2014;34(10):1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14(6):679–689. [PMC free article] [PubMed] [Google Scholar]
- 43. MacKay H, Abizaid A. A plurality of molecular targets: the receptor ecosystem for bisphenol-A (BPA). Horm Behav. 2018;101:59–67. [DOI] [PubMed] [Google Scholar]
- 44. Oosterman JE, Belsham DD. Glucose alters Per2 rhythmicity independent of AMPK, whereas AMPK inhibitor compound C causes Profound repression of clock genes and AgRP in mHypoE-37 hypothalamic neurons. PLoS One. 2016;11(1):e0146969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sen A, Sellix MT. The circadian timing system and environmental circadian disruption: from follicles to fertility. Endocrinology. 2016;157(9):3366–3373. [DOI] [PubMed] [Google Scholar]
- 46. Choi JY, Choe JR, Lee TH, Choi CY. Effects of bisphenol A and light conditions on the circadian rhythm of the goldfish Carassius auratus. Biol Rhythm Res. 2018;49(4):502–514. [Google Scholar]
- 47. Walker DM, Goetz BM, Gore AC. Dynamic postnatal developmental and sex-specific neuroendocrine effects of prenatal polychlorinated biphenyls in rats. Mol Endocrinol. 2014;28(1):99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ali AAH, Schwarz-Herzke B, Mir S, Sahlender B, Victor M, Görg B, Schmuck M, Dach K, Fritsche E, Kremer A, von Gall C. Deficiency of the clock gene Bmal1 affects neural progenitor cell migration [published online ahead of print 19 October 2018] Brain Struct Funct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010;20(4):316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Birky TL, Bray MS. Understanding circadian gene function: animal models of tissue-specific circadian disruption. IUBMB Life. 2014;66(1):34–41. [DOI] [PubMed] [Google Scholar]
- 52. Orozco-Solís R, Matos RJ, Lopes de Souza S, Grit I, Kaeffer B, Manhães de Castro R, Bolaños-Jiménez F. Perinatal nutrient restriction induces long-lasting alterations in the circadian expression pattern of genes regulating food intake and energy metabolism. Int J Obes. 2011;35(7):990–1000. [DOI] [PubMed] [Google Scholar]
- 53. Mieda M, Sakurai T. Bmal1 in the nervous system is essential for normal adaptation of circadian locomotor activity and food intake to periodic feeding. J Neurosci. 2011;31(43):15391–15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dumbell R, Leliavski A, Matveeva O, Blaum C, Tsang AH, Oster H. Dissociation of molecular and endocrine circadian rhythms in male mice lacking Bmal1 in the adrenal cortex. Endocrinology. 2016;157(11):4222–4233. [DOI] [PubMed] [Google Scholar]
- 55. Acconcia F, Pallottini V, Marino M. Molecular mechanisms of action of BPA. Dose Response. 2015;13(4):1559325815610582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rhee JS, Kim BM, Lee BY, Hwang UK, Lee YS, Lee JS. Cloning of circadian rhythmic pathway genes and perturbation of oscillation patterns in endocrine disrupting chemicals (EDCs)-exposed mangrove killifish Kryptolebias marmoratus. Comp Biochem Physiol C Toxicol Pharmacol. 2014;164:11–20. [DOI] [PubMed] [Google Scholar]
- 57. Zhu J, Jiang L, Liu Y, Qian W, Liu J, Zhou J, Gao R, Xiao H, Wang J. MAPK and NF-κB pathways are involved in bisphenol A–induced TNF-α and IL-6 production in BV2 microglial cells. Inflammation. 2015;38(2):637–648. [DOI] [PubMed] [Google Scholar]
- 58. Dong S, Terasaka S, Kiyama R.. Bisphenol A induces a rapid activation of Erk1/2 through GPR30 in human breast cancer cells. Environ Pollut. 2011;159(1):212–218. [DOI] [PubMed] [Google Scholar]