Abstract
Human glioblastoma (GBM) is the most aggressive malignancy of the CNS, with less than 5% survival. Despite great efforts to find effective therapeutics, current options remain very limited. To develop a targeted cancer therapeutic, we selected RNA aptamers against platelet-derived growth factor receptor α (PDGFRα), which is a receptor tyrosine kinase. One RNA aptamer (PDR3) with high affinity (0.25 nM) showed PDGFRα specificity and was internalized in U251-MG cells. Following treatment with the PDR3 aptamer, expression of the transcription factor STAT3 (signal transducer and activator of transcription 3) was inhibited, whereas the expression of the histone demethylase JMJD3 and the tumor suppressor p53 were upregulated. PDR3 also upregulated serine phosphorylation of p53, which subsequently mediated apoptosis through the death receptors: tumor necrosis factor (TNF)-related apoptosis-inducing ligand receptors 1/2 (TRAIL-R1/R2), Fas-associated via death domain (FADD), and Fas. PDR3 significantly decreased cell viability in a dose-dependent manner. Furthermore, translocation of PDR3 into the nucleus induced hypomethylation at the promoters of cyclin D2. To assess the feasibility of targeted delivery, we conjugated PDR3 aptamer with STAT3-siRNA for a chimera. The PDR3-siSTAT3 chimera successfully inhibited the expression of target genes and showed significant inhibition of cell viability. In summary, our results show that well-tailored RNA aptamers targeting the PDGFRα-STAT3 axis have the potential to act as anti-cancer therapeutics in GBM.
Keywords: PDGFRα aptamer, STAT3, p53, apoptosis, GBM
Introduction
In the era of precision medicine, targeted cancer therapies are a critical cornerstone. Practical applications of targeted therapies include targeted delivery or targeting of a specific molecule to increase the therapeutic index. Aptamers are structured nucleic acid ligands that show high affinity and binding specificity for their target molecules.1 These compelling features have driven the search for cancer-specific aptamers that react with the receptors on the surface of cancer cells.2, 3 Recently, cancer-specific aptamers have emerged as targeted delivery vehicles for cargos such as small interfering RNAs (siRNAs),4 small activating RNAs,5 small molecules, and toxins.6, 7 Use of such functionalized aptamers for targeted delivery against cancer cell surface receptors has shown successful outcomes that are promising for cancer therapeutics.8 To take full advantage of the high specificity of aptamers, “molecularly targeted aptamers” may offer another approach for targeted therapies in the cancer field.
Glioblastoma (GBM) is the most common primary malignancy of the CNS, with an incidence of 3.19 per 100,000 people and a 5-year survival rate of less than 5%.9 Despite considerable efforts to develop effective therapeutic options, it still remains incurable. The progression of GBM is characterized by the abnormal activation of receptor tyrosine kinase (RTK) signaling pathways10 that regulate many aspects of tumorigenesis, including cell growth and proliferation.11 Among the RTKs, platelet-derived growth factor receptors (PDGFRs) are overexpressed in a substantial subset of malignant gliomas, and it is widely accepted that the PDGFR pathway is a driver of GBM.12 The various isoforms of platelet-derived growth factor (PDGF) ligands and PDGFRs play diverse important roles in regulating the growth and survival of specific cell types during embryonal development and controlling tissue homeostasis in the adult,13 as well as contributing to tumor growth and metastasis.14 Therefore, PDGFRs are promising targets for therapeutic development in GBM. PDGFRs are composed of two isoforms, PDGFRα and PDGFRβ.15 Biologically, there are fundamental functional differences between PDGFRα and PDGFRβ: PDGFRα impacts the CNS,16 whereas PDGFRβ is essential for development of the vasculature.17 For targeted therapies, RNA aptamers against PDGFRβ have been isolated;18 in particular, a PDGFRβ aptamer-chimera carrying siRNA against the transcription factor STAT3 (signal transducer and activator of transcription 3) showed anti-tumor effects and anti-angiogenesis in GBM.19 This suggests that targeting PDGFRα, the second most commonly amplified gene in GBM20, 21 and widely considered a “bad seed”22 for development of GBM malignancies, could also be effective. Therefore, the development of RNA aptamers against PDGFRα is imperative for the sake of GBM cancer patients.
STAT3 is activated by growth factors and plays a pivotal role in the oncogenesis of many human tumors, including GBM.23, 24 It is involved in GBM stem cell proliferation and multipotency.25 STAT3 also regulates the cellular epigenetic state, causing stable changes in the ability of cells to respond to stimuli in cancer development.26 Depletion of PDGFRα attenuates GBM by modulating STAT3 and multiple oncogenic signaling pathways,27 which suggests the presence of a PDGFRα-STAT3 regulatory axis.
Loss of the histone H3K27me2/3 demethylase JMJD3, also known as KDM6B, enhances aggressiveness in some cancer cells; thus, it is regarded as a tumor suppressor.28 Missense mutations and aberrant methylation of JMJD329 and mutation of H3K2730, 31 have been reported in GBM, indicating that JMJD3 could be involved in GBM malignancy. Furthermore, inhibition of STAT3 upregulates the expression of JMJD3 in GBM stem cells.26 Taken together, these observations suggest that targeting the PDGFRα-STAT3 axis will induce signaling cascades to achieve anti-tumor effects in GBM.
Given that knockdown of STAT3 by RNAi leads to a dramatic decrease of proliferation and neutrosphere formation,25 and the regulatory role of the PDGFRα-STAT3 axis,27 we hypothesized that targeting the PDGFRα-STAT3 axis would induce potent anti-tumor effects in GBM. To test our hypothesis, we isolated an anti-PDGFRα aptamer and assessed its anti-tumor effects in GBM cells. As expected, the anti-PDGFRα aptamer inhibited gene expression of STAT3 over time and upregulated the expression of JMJD3. Therefore, we suggest that aptamers targeting the PDGFRα-STAT3 axis might be useful as cancer therapeutics in the treatment of GBM.
Results
PDR3 Binds to PDGFRα with High Affinity
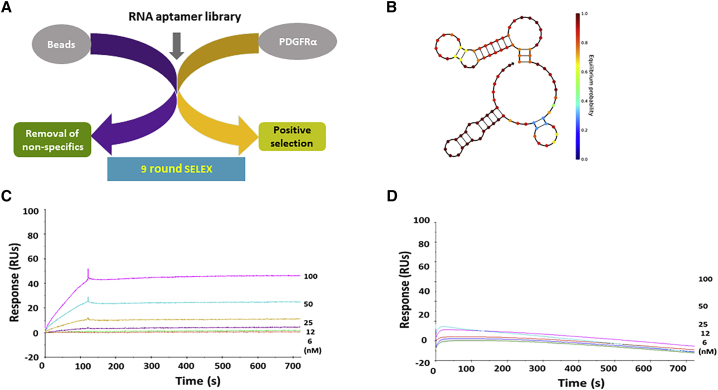
We performed RNA aptamer selection against the ectodomain of recombinant human PDGFRα protein. To remove any non-specific binders, we used beads without target protein for negative selection. After removal of non-specific binders, we incubated the aptamer library with the target protein for positive selection (Figure 1A). After nine rounds of selection, we identified an anti-PDGFRα aptamer that we called PDR3. We used NUPACK32 to predict its computational-based secondary structure and showed multiple stem loop structures (Figure 1B). To perform label-free kinetic analysis of aptamer-PDGFRα interactions in real time, we used a biosensor assay. We measured the equilibrium dissociation constant (KD) of PDR3 for PDGFRα as 0.25 nM, with 6.78 × 103 M−1S−1 and 1.68 × 10−6 S−1 as the corresponding association (KA) and dissociation (KD) rate constants (Figure 1C). The binding affinity against PDGFRβ was unmeasurable by biosensor assay (Figure 1D), suggesting that PDR3 is a PDGFRα-specific aptamer.
Figure 1.
Secondary Structure and Biosensor Assays
(A) A protein-based SELEX method was used to isolate anti-PDGFRα RNA aptamers from a randomized 40-nt RNA library. After nine rounds of SELEX, the amplified aptamers were cloned, and individual clones were identified by DNA sequencing. (B) The secondary structure of the anti-PDGFRα aptamer, PDR3, was predicted using NUPACK software. (C) Surface plasmon resonance (SPR) sensorgrams of binding between PDR3 and recombinant PDGFRα. To assess binding, we used a Biacore T100 to monitor label-free interactions and to measure the increase in response units (RUs) from baseline. BIAevaluation software was used to calculate binding affinity (KD). (D) The binding of PDR3 to recombinant PDGFRβ was measured using SPR. Binding affinity was not measurable.
PDR3 Aptamer Efficiently Binds to Human GBM Cells
To confirm that the aptamer binds to GBM cells, we used flow cytometry and confocal microscopy to assess live cell binding of PDR3 to the human GBM cell line U251-MG, which overexpresses PDGFRα.33 We incubated U251-MG cells with a Cy3-labeled initial non-selected aptamer library as a negative control (NC) or with the PDR3 aptamer. Flow cytometry analyses confirmed enriched cell surface binding of PDR3 to U251-MG cells, compared with NC (Figure 2A). Live cell confocal imaging showed punctate cytoplasmic staining in U251-MG cells after incubation with Cy3-labeled PDR3 aptamer, but no staining was observed with NC (Figure 2B). The punctate pattern of cytoplasmic staining suggests that the PDR3 aptamer is endocytosed into U251-MG cells. We quantified the extent of internalization using mean fluorescence intensity in confocal images, which showed significantly increased intensity following incubation with Cy3-labeled PDR3 (Figure 2C).
Figure 2.
Internalization Assays of PDR3 in Live Cells and Relative Target Gene Expression Analysis
(A) Cy3-labeled PDR3 aptamer and aptamer library as negative control (NC) were assessed for binding efficiency by flow cytometry in U251-MG cells. The data show measurements of positively stained cells and are representative of duplicates. (B) Cy3-labeled PDR3 aptamer was assessed for intracellular uptake in live cells. Human GBM U251-MG cells were treated with 200 nM Cy3-labeled PDR3 aptamer or aptamer library for 2 hr, and internalization was visualized using confocal microscopy. Cells show punctate regions of Cy3 intracellular labeling. Red: Cy3-labeled RNAs; blue: Hoechst 33342 for nuclear staining. Scale bars: 5 μm. NC, Cy3-labeled initial aptamer library; PDR3, Cy3-labeled anti-PDGFRα aptamer. (C) Quantification of internalization. Images were quantified to yield mean intensity per cell of the red fluorophore (Cy3) using Zen Blue (v2.3 Carl Zeiss Microimaging). Intensity level (12-bit) is presented on the y axis. Data are presented as the mean ± SD. NC, Cy3 labeled initial aptamer library; PDR3, Cy3 labeled anti-PDGFRα aptamers. (D) Relative expression of STAT3 mRNA transcripts was quantified using real-time PCR (qPCR) in U251-MG cells treated with 200 nM PDR3 or irrelevant aptamer (IRRE) over time. Data are presented as the mean ± SD. (E) Protein expression of STAT3 after treatment was measured by western blot. Cells were treated with PDR3 (200 nM) at 24-hr intervals, and total protein was extracted at 72 hr. For loading control, GAPDH was used. Each group is shown in duplicate. (F) Pixel intensity was quantified using Image-pro premier. Data are presented as the mean ± SD. (G) Relative expression of JMJD3 mRNA transcripts was quantified using qPCR in U251-MG cells treated with 200 nM PDR3 or IRRE for 48 hr. Data are presented as the mean ± SD. (H) Relative expression of p53 mRNA transcripts was quantified using qPCR on U251-MG cells treated with 200 nM PDR3 or IRRE for 48 hr. Data are presented as the mean ± SD. One-way ANOVA was used to assess statistical significance; *p ≤ 0.05; **p ≤ 0.01. CC, untreated control cells; PDR3, anti-PDGFRα aptamer.
PDR3 Suppresses Expression of STAT3 and Upregulates Gene Expression of JMJD3 and p53 in the Downstream Pathway
In previous studies, depletion of PDGFRα reduced expression of the transcription factor STAT3 in the PDGFRα-STAT3 regulatory axis.27 Therefore, we used qPCR to assess STAT3 gene expression after PDR3 treatment. We observed downregulation of STAT3 for up to 120 hr (Figure 2D). We assessed effects on STAT3 protein level using western blot, which confirmed significant downregulation of STAT3 protein expression following PDR3 treatment (Figures 2E and 2F). In the downstream pathway, PDR3-induced STAT3 inhibition induced upregulation of the histone H3K27me2/3 demethylase JMJD3, suggesting that JMJD3 is a direct target of STAT3.26 Therefore, we used qPCR to measure the expression level of JMJD3. We observed the significant upregulation of JMJD3 following PDR3 treatment (Figure 2G). In the downstream biological pathway, JMJD3 interacts with the tumor suppressor protein p53.34, 35 Therefore, we used qPCR to determine p53 gene expression. As expected, p53 gene expression was increased in the PDR3 treatment group (Figure 2H).
PDR3 Induces Apoptosis via Tumor Suppressor p53
To investigate the induction of apoptosis by the PDR3 aptamer, we assessed early apoptosis using Annexin V staining. The results of confocal microscopy (Figure 3A) and flow cytometry (Figure 3B) showed a significant increase in apoptosis following treatment of U251-MG cells with PDR3. To better understand the apoptotic mechanism, we used an antibody-based apoptosis proteome array to capture specific apoptotic signaling molecules. Compared with control, we observed significant upregulation of phospho-p53 (serine 15), phospho-p53 (serine 46), and phospho-p53 (serine 392) following PDR3 treatment (Figures 3C, top left, and 3D). In addition, we observed upregulation of the death receptors: tumor necrosis factor (TNF)-related apoptosis-inducing ligand receptor 1/death receptor 4 (TRAIL-R1/DR4), TNF-related apoptosis-inducing ligand receptor 2/death receptor 5 (TRAIL-R2/DR5), Fas-associated via death domain (FADD), and Fas in the PDR3 treatment group (Figures 3C, top right, and 3E). In contrast, we observed no difference in the expression of Bad, Bax, pro-caspase 3, or cleaved caspase 3 (Figure 3C).
Figure 3.
Apoptosis Assays after PDR3 Treatment
(A) U251-MG cells were treated with 500 nM PDR3 aptamer for 24 hr. The cells were stained with Annexin V-Alexa Fluor 488, and fluorescent images were taken using confocal microscopy. Green: Annexin V-Alexa Fluor 488; blue: Hoechst 33342. Scale bars: 10 μm. (B) Cells were stained with Annexin V and analyzed using flow cytometry after treatment with PDR3 for 24 hr. Propidium iodide (PI) was used to discriminate dead cells. (C) Apoptosis-related molecules were determined using protein array. Total proteins (200 μg) after treatment with PDR3 aptamer for 72 hr were used for apoptosis assays. Upregulated serine phosphorylation of p53 and death receptors TRAIL-R1, TRAIL-R2, FADD, and Fas were observed. (D and E) Pixel intensity of phospho-p53 (D) and death receptors (E) were quantified using ImageJ. Data are presented as the mean ± SD. (F) The EC50 of PDR3 was calculated, using PDGFRα in vitro kinase assay, to be 55 ng. (G) Cell viability was measured in U251-MG cells treated twice with various concentrations of PDR3 or IRRE at 24-hr intervals and harvested at a final incubation time of 48 hr. Cell viability was measured using MTS assay. Data were normalized to untreated control cells. Data are presented as the mean ± SD. (H) Inhibition of cell proliferation after PDGFRα activation with PDGF-AA ligands was measured using MTS assay. After pretreatment with PDR3 aptamer, PDGF-AA ligands were incubated with cells. Cell viability was measured using MTS assay. Cell proliferation was normalized to untreated control cells. Data are presented as the mean ± SD. One-way ANOVA was used to assess statistical significance; *p ≤ 0.05; **p ≤ 0.01. CC, untreated control cells; PDR3, anti-PDGFRα aptamer.
PDR3 Reduces Cell Viability
Our discovery that the PDR3 aptamer itself inhibited STAT3 expression and induced p53-mediated apoptosis was surprising. To measure the direct inhibition of kinase activity by PDR3, we used a luminance kinase assay to calculate the half maximal effective concentration (EC50 ) as 55.3 ng (Figure 3F). In addition, we determined cell viability; PDR3 significantly inhibited proliferation of U251-MG cells in a dose-dependent manner (Figure 3G). To determine whether PDR3 inhibits tumor cell growth by blocking PDGFRα activation, we pretreated U215-MG cells with PDR3, followed by incubation with PDGF-AA ligands. Our results showed that PDR3 did not block ligand-mediated activation of PDGFRα (Figure 3H).
PDR3 Affects the Regulation of Nuclear DNA Methylation
Because some cells showed nuclear translocation of PDR3 (Figure 4A), we assessed whole genome nuclear DNA hypomethylation after treatment of U215-MG cells with PDR3. We summarize the results for differentially methylated regions (DMRs) in promoters, gene bodies, and intergenic regions in Figure 4B. A complete list of DMR genes is shown in Table S1. We clustered the DMRs using a hierarchical heatmap (Figure 4C) and observed variance between samples. Chromosomal views of methylation differences in specific genes, Cyclin D2 (CCDN2), zinc-finger protein (ZNF)286A, ZNF607, and ZNF876P, are shown in Figure 4D and Figure S1. Based on all of these data, we developed a working model of the intracellular cascade resulting from PDR3 treatment (Figure 4E).
Figure 4.
DNA Methylation by PDR3 Aptamer
(A) Nuclear translocation of PDR3 was observed in live U215-MG cells using confocal microscopy. The nuclear region is indicated with a line. Nuclear translocation of PDR3 is indicated with an arrow. Red: Cy3-labeled RNAs; blue: Hoechst 33342 for nuclear staining. Scale bars: 10 μm. (B) The diagram summarizes the relative percentages of hypomethylated regions. Changes in DNA methylation were measured using the bisulfite method. (C) Hierarchical heatmap of hypomethylation changes induced by PDR3 treatment. Heatmap shows promoter methylation levels. Each group is shown in duplicate. (D) Chromosomal views of methylation differences in the CCND2 gene between control and PDR3 treatment. (E) Schematic working model of biological pathways affected by PDR3. Inhibition of STAT3 by binding of PDR3 to PDGFRα upregulates the expression of JMJD3 and its downstream effector p53. Activated p53 induces apoptosis-related genes such as TRAIL R1/R2, FADD, and Fas to promote cell death. Along with apoptosis, translocation of PDR3 into the nucleus induces methylation changes. NC, Cy3-labeled initial aptamer library; PDR3, anti-PDGFRα aptamer; CC, untreated cell control.
Chimeric PDR3 Inhibits the Expression of STAT3
Because PDR3 showed endocytosis into cells in binding assays, we constructed chimeras combining the PDR3 aptamer with siRNAs to investigate the feasibility of targeted delivery. In this study, we created chimeras by conjugating PDR3 to siRNA against the transcription factor STAT3 (Figure 5A). The sequences are depicted in Table 1. We compared in vitro internalization of PDR3 with chimeric constructs of PDR3 conjugated to scrambled siRNA control (PDR3-Scr) or siRNA to STAT3 (siSTAT3; PDR3-siSTAT3), or with irrelevant aptamer (IRRE) conjugated to siSTAT3 (IRRE-siSTAT3). Live confocal microscopy images, taken 2 hr after incubation, demonstrated the typical pattern of punctate cytoplasmic staining of Cy3-labeled PDR3 chimeras (Figure 5B). To confirm the ability of the chimeras to inhibit STAT3 gene expression in vitro, we treated U251-MG cells with PDR3-Scr, PDR3-siSTAT3, or IRRE-siSTAT3 chimeras. Cells treated with chimeric PDR3-siSTAT3 and PDR3-Scr showed significantly reduced expression of STAT3 mRNA for up to 120 hr, compared with control (Figure 5C). Given that PDR3 itself showed the suppression of STAT3, this may explain STAT3 suppression following treatment with PDR3-Scr. The PDR3-siSTAT3 chimeras showed slightly greater downregulation of STAT3, compared with PDR3-Scr. However, the difference was only statistically significant at the 72-hr time point. Both PDR3 chimeras decreased cell viability in a dose-dependent manner (Figure 5D).
Figure 5.
The Inhibition of STAT3 Expression by PDR3-siSTAT3 chimeras
(A) Schematic of cell surface receptor-mediated siRNA delivery. An RNA aptamer targeting PDGFRα, which is overexpressed on GBM, allows specific delivery of therapeutic siSTAT3 into target cells. (B) Intracellular uptake of chimeras. U251-MG cells were treated with 200 nM Cy3-labeled chimeric aptamer-siRNAs and analyzed using confocal microscopy, which shows punctate regions of Cy3 labeling in the cytoplasm. Red: Cy3-labeled RNA; blue: Hoechst 33342. Scale bars: 10 μm. (C) Relative expression of STAT3 mRNA transcripts was quantified using qPCR over time in U251-MG cells treated with 200 nM PDR3 chimeras. Data are presented as the mean ± SD. (D) U251-MG cells were treated with various concentrations of chimeras twice at 24-hr intervals and harvested at a final incubation time of 48 hr. Cell viability was measured using MTS assay, normalized to untreated control cells. One-way ANOVA was used to assess statistical significance; *p ≤ 0.05; **p ≤ 0.01. IRRE-STAT3, irrelevant aptamer with siSTAT3 sequence; PDR3-scr, PDR3 aptamer conjugated to scrambled siRNA control; PDR3-STAT3, PDR3 aptamer conjugated to siSTAT3 sequence.
Table 1.
Sequences of PDR3 and Sense and Antisense Strands of STAT3 siRNA
| Name | Sequences |
|---|---|
| PDR3 | 5′-GGGAGAGCGGAAGCGUGCUGGGCCUGCUCUUUAAUAAACCCACUUUCGAACAUCAGCGUAUGUCCAUAACCCAGAGGUGAUGGAUCCCCC-3′ |
| STAT3 sense | 5′-GCUGCAGAAAGAUACGACUUU-3′ |
| STAT3 antisense | 5′-AGUCGUAUCUUUCUGCAGCUU-3′ |
Discussion
To increase the therapeutic index, aptamers that target cancer cell-specific surface receptors to achieve targeted delivery have become popular as cancer therapeutics.5, 6, 8 Aptamers have been well characterized for targeted delivery of cargoes. However, the molecular function of aptamers themselves has not yet been investigated. In this study, for the first time, we show that an anti-PDGFRα aptamer has gene regulatory functions. We prove that the anti-PDGFRα aptamer PDR3 inhibits expression of STAT3 in the regulatory axis of PDGFRα-STAT3. The histone demethylase JMJD3 is a direct downstream target of STAT3, which binds to the JMJD3 promoter.26 Therefore, inhibition of STAT3 could lead to upregulation of JMJD3. As expected, we showed that STAT3 suppression by PDR3 induced the upregulation of JMJD3. Further downstream, JMJD3 is recruited to p53-bound promoters34 and induces p53 stabilization by direct interaction between JMJD3 and p53.35 Therefore, we determined the gene expression level of p53 following PDR3 treatment and showed that p53 was activated via upregulated expression of JMJD3.
p53 is one of the most intensively studied tumor suppressors over the past 30 years as a potential therapeutic target. In particular, apoptotic cell death is closely associated with activation of p53 in human cancers.36 p53 targets multiple elements involved in apoptotic pathways: it acts as a transcription factor to induce pro-apoptotic genes,37 and it translocates to the mitochondria to interact with BCL-2 family members and induce apoptosis.38 Two major apoptotic mechanisms exist: the “intrinsic” pathway is activated by caspases and mitochondrial disruption, and the “extrinsic” pathway is induced by ligand binding to cell surface death receptors such TRAIL-R1/DR4, TRAIL-R2/DR5, FADD, and Fas.39 In the extrinsic apoptosis pathway, p53 activation increases expression of Fas, redistributes it from the Golgi complex to the cell surface, induces the binding of Fas and FADD, and sensitizes cells to Fas-induced apoptosis.40 Activated p53 is also involved in upregulating the TRAIL receptors DR441 and DR542 as cancer-specific apoptotic inducers;43 both TRAIL receptors bind FADD to induce apoptosis.44 Additionally, the transcriptional activity of p53 is regulated and stabilized by post-translational modifications such as phosphorylation.45 Phosphorylation at specific residues affects p53 transcriptional activity and its selectivity for genes that induce specific cellular responses. Phosphorylation of p53-serine 15 (pS15), p53-serine 46 (pS46), and p53-serine 392 (pS392) has been well studied in apoptosis. For example, p53-pS15 stimulates transactivation at p53 promoters.46 Activated p53-pS15 upregulates the pro-apoptotic gene Fas but does not upregulate the pro-apoptotic protein Bax.47 p53-pS46 regulates transcriptional activation of apoptosis-related genes such as p53AIP1.48 p53-pS392 induces non-BCL-2 family-mediated apoptosis.49 In this study, we observed upregulation of p53-pS15, p53-pS46, and p53-pS392 following activation of TRAIL DR4/5, FADD, and Fas to induce apoptosis after PDR3 treatment. However, we did not observe changes to the BCL-family, including Bax, after PDR3 treatment. Taken together, our results suggest that PDR3 mediates a biological molecular cascade in which downregulated expression of STAT3 via the PDGFRα-STAT3 axis activates expression of JMJD3. In turn, JMJD3 upregulation reverses polycomb complex-mediated repression of genes, which induce upregulation of p53 in GBM. p53 upregulation induces differential serine phosphorylation of p53, followed by activation of the death receptor-mediated extrinsic apoptotic pathway.
We also observed nuclear translocation of PDR3 in some cells. Previous studies have shown that PDGFRα and other RTKs are localized in the nucleus.50, 51, 52 Therefore, our observation of PDR3 nuclear localization suggests that it is mediated by the PDGFRα receptor. In some aspects of molecular biology, the nuclear localization of RTKs acts like a transcription factor.53 Thus, it is highly possible that gene regulation induced by the PDR3 aptamer could follow three possible mechanisms: (1) PDR3 directly inhibits the transcriptional role of PDGFRα in the nucleus; (2) PDR3 inhibits the binding of PDGFRα to specific DNA sequences for transcriptional gene regulation, because its transactivation activity is evidently dependent on DNA binding; or (3) additional associated molecules might be involved. To validate these possibilities, further studies are planned in the near future.
Furthermore, PDGF and PDGFRα are reported to form complexes with chromatin.54 Aberrant promoter methylation of tumor suppressor genes is a common feature of glioma cancer cells.55 Thus, to investigate PDR3-induced chromatin changes, we studied whole genome methylation. After PDR3 treatment, we identified DMRs at the promoters of CCND2, ZNF286A, ZNF607, and ZNF876P. CCND2 plays a pivotal role in cell-cycle regulation, differentiation, and malignant transformation,56 and loss of CCND2 is closely associated with aberrant promoter methylation.57 Our results suggest that hypomethylation of CCND2 might promote the transcriptional activation of tumor suppressors. ZNFs are the most common transcription factors in mammals.58 Some ZNFs such as ZAC, ST18, ZNF282, and ZNF331 are reported tumor suppressors.59, 60, 61, 62 In our study, we detected hypomethylation at several ZNF promoters (ZNF286A, ZNF607, and ZNF876P) following PDR3 treatment; we will investigate the biological function of these ZNFs in GBM in further studies. Additionally, we will investigate key remaining questions, such as how DNA methylation is orchestrated by PDR3 and whether additional players are involved.
Structured single-strand RNAs (ssRNAs) such as aptamers are reported to elicit innate immune activation and induction of the caspase-mediated intrinsic apoptotic pathway.63 Our PDR3 aptamer induced a different mechanism of cancer cell death, i.e., the death receptor-mediated extrinsic apoptotic pathway. Because cancer cells are more sensitive to death receptor-mediated apoptosis than normal cells, PDR3 aptamers might increase the therapeutic index and be less harmful to normal cells. Given that ssRNAs activate the innate immune response,63 we will investigate the activation of the innate immune response such as expression of interferon-β and pro-inflammatory cytokines by PDR3.
In conclusion, we showed that PDR3, an RNA aptamer against the cell surface RTK PDGFRα, has therapeutic potential, based on downregulation of STAT3 and its induction of apoptosis via p53-mediated death receptors in GBM. Furthermore, translocation of the PDR3 aptamer to the nucleus might have potential for transcriptionally regulated anti-cancer therapeutics in GBM.
Materials and Methods
Cell Lines
The U251-MG cell line (human glioblastoma, 0906300) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and cultured according to the supplier’s instructions.
Chemicals
The DuraScribe T7 transcription kit (Lucigen, Middleton, WI, USA) was used to incorporate 2F′-modified pyrimidines into aptamers. Micro Bio-spin P30 columns (Bio-Rad, Hercules, CA, USA) were used to remove unincorporated nucleotide triphosphates (NTPs).
Recombinant Target Protein
Human PDGFRα protein (10556-H08H) was purchased from Sino Biological (Beijing, PR China). The recombinant protein was produced in human cells and expressed as the extracellular domain of human PDGFRα (NP_006197.1; Met 1–Glu 524) with a polyhistidine tag at the C terminus.
Protein SELEX
In vitro selection was carried out essentially as described previously,64 with a few modifications. The 2F′-RNA aptamers were selected from 40-nt randomized sequences constructed by in vitro transcription of synthetic DNA templates with NTPs (2′F UTP, 2′F CTP, GTP, ATP; Epicenter Biotechnologies) and T7 RNA polymerase. To remove RNAs that bind nonspecifically to agarose beads, 1.44 μM of the RNA library was pre-incubated with 20 μL of Ni-NTA agarose beads in 100 μL of binding buffer (30 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol, and 1% BSA) for 30 min at room temperature with shaking, precipitated by centrifugation, and discarded. The precleared supernatant was transferred to a new tube and incubated with 333 nM his-tagged human PDGFRα (hPDGFRα) for 30 min at room temperature. RNAs that bound to hPDGFRα were recovered, amplified by RT-PCR and in vitro transcription, and used in subsequent selection rounds. In subsequent rounds, hPDGFRα concentration was reduced by 2-fold at every third round for more stringent conditions. After nine rounds of SELEX, the resulting cDNA was amplified. The amplified DNA was cloned, and individual clones were identified by DNA sequencing. Structures of aptamers were predicted using NUPACK.32
Flow Cytometry-Based Binding Assays
Aptamer binding was - assessed using flow cytometry. For this assay, U251-MG cells were detached using a non-enzymatic cell dissociation solution, washed with PBS, and suspended in binding buffer. Next, Cy3-labeled aptamers at 500 nM were added to target cells for 30 min at 0°C. Cells were washed with binding buffer and immediately analyzed using a Fortessa flow cytometer (BD, San Jose, CA, USA). For the exclusion of dead cells, DAPI (1 μg/mL) was used. The data were analyzed with FlowJo software.
Aptamer Internalization Studies
1 × 105 U251-MG cells were seeded in 35-mm glass-bottom dishes (MatTek, Ashland, MA, USA) and grown in appropriate media for 24 hr. Aptamer RNA was labeled with Cy3 fluorescent dye using the Cy3 Silencer siRNA labeling kit (Thermo Fisher Scientific, Waltham, MA, USA). Cy3-labeled aptamers were added to the cells at 200 nM, incubated for 2 hr, and washed for imaging. The images were taken using a Zeiss LSM 880 confocal laser scanning microscope with Airyscan using a C-Apo 63×/1.3NA water immersion objective. Internalization was quantified to yield mean intensity per cell of the red fluorophore (Cy3) using Zen Blue (v2.3 Carl Zeiss Microimaging).
Biosensor Assays
A Biacore T100 (GE Healthcare, Uppsala, Sweden) was used to perform label-free monitoring of aptamer-PDGFRα interactions in real time. Biotinylated aptamers were coupled to a streptavidin-coated Biacore chip (SensorChip SA, BR-1003-98; General Electric Company) by an injection at a concentration of 25 μg/mL in binding buffer (30 mM Tris-HCl [pH 7.5]; 150 mM NaCl; 5 mM MgCl2) at 10 μL/min. The RNA was refolded by heating to 65°C, followed by cooling to 37°C before immobilization. To measure binding kinetics, five concentrations of purified PDGFRα or PDGFRβ receptor proteins were injected at a flow rate of 10 μL/min. After binding, the surface was regenerated by injecting 50 mM NaOH at a flow rate of 15 μL/min for 20 s. Data from the control surface were subtracted. BIAevaluation software (GE Healthcare) was used for analysis. The binding data were fit to a 1:1 binding with a mass transfer model to calculate kinetics parameters as previously described.65, 66
Relative Gene Expression Analysis by qPCR
1 × 105 cells were seeded in six-well plates 1 day before treatment. Cells were treated with 200 nM IRRE or PDR3 twice at 24-hr intervals and harvested at various final incubation times. For IRRE, gp120 aptamers67 were used in the following assays. Total RNA was extracted and converted to cDNA using iScript reverse transcriptase (Bio-Rad), and the target genes were amplified using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). The comparative threshold cycle (CT) method (ΔΔCT method)68 was used to determine the expression level of targets. The housekeeping genes, hypoxanthine phosphoribosyltransferase (HPRT) or 18S, were used as endogenous controls to normalize the data.
Protein Expression by Western Blot Analysis
For analyzing protein expression, U251-MG cells were seeded into six-well plates at a density of 1 × 105 cells/well. PDR3 was incubated with the cells, in duplicate reactions, at a concentration of 200 nM. The treatment was repeated 24 hr later, and the cells were harvested at a final incubation time of 72 hr. Total protein was extracted using a conventional radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 150 mM sodium chloride, 1.0% Igepal, 0.5% sodium deoxycholate, and 0.1% SDS) with protease inhibitors. Total protein content was then quantitated using a Bradford assay, following the manufacturer’s instructions (Bio-Rad Bradford Assay). Total protein extracts were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes, then probed with antibodies against STAT3 (Santa Cruz) or GAPDH (Santa Cruz). The proteins of interest were detected with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000) and visualized with Bio-Rad Western Sure ECL substrate, according to the manufacturer’s protocol. Image-Pro Premier 9 was used to quantify pixel intensity.
Apoptosis Analysis
For early apoptosis analysis, Annexin V conjugated with Alexa Fluor 488 (A13201; Thermo Fisher) was used. 1 × 105 cells were seeded in six-well plates 1 day before treatment with IRRE or PDR3 at 500 nM. After staining with Annexin V-Alexa Fluor 488, confocal microscopy or flow cytometry was used to assess apoptosis. To discriminate dead cells, we used propidium iodide (PI) to stain cells. For determining apoptotic pathways, a human apoptosis array kit (ARY009; R&D Systems) was used following the manufacturer’s instruction. Image J was used to quantify the pixel intensity.
Cell Viability Assays
To assess inhibition of cell proliferation, 5 × 103 U251-MG cells/well were seeded in 96-well plates and grown in appropriate media 1 day before treatment. Cells were treated with aptamers at various concentrations twice at 24-hr intervals. Cell viability was measured using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)) assay at a final incubation time of 48 hr (Promega, Madison, WI, USA). To activate PDGFRα receptors, PDGF-AA ligands (221-AA-010; R&D Systems) were incubated at 200 ng/mL with U251-MG cells that were pretreated with aptamers at various concentrations. As a NC for peptides, cells were treated with 0.1% BSA. Cell viability was measured at 48 hr using MTS assay.
Tyrosine Kinase Assays
Tyrosine kinase was assayed to determine EC50 using a PDGFRα kinase enzyme system (V4487; Promega). Kinase or ATPase activity was detected using a luminescence system, according to the manufacturer’s instructions.
DNA Methylation Assays
1 × 105 U251-MG cells/well were seeded in six-well plates 1 day before treatment with PDR3 aptamer. Cells were treated with 500 nM PDR3 twice at 24-hr intervals and harvested at a final incubation time of 48 hr to analyze DNA methylation. Reduced-representation bisulfite sequencing was performed by the City of Hope Integrative Genomics Core as previously described.69 DNA sequencing was performed using a HiSeq 2500 Illumina sequencer. Sequences were aligned to human genome assembly hg19 using Bismark v0.8.3. Subsequent analysis was implemented using R statistical language. Methylation levels were calculated for CpG sites covered by at least five or more reads. DMRs were identified using the following steps: CpG sites were first filtered to select the ones with methylation levels ≥0.5 as one group and <0.5 as the other group, with a difference between them ≥0.25. Filtered CpG sites within 50 bp of each other were merged into regions as candidate DMRs. Linear regression was then applied to identify DMRs that showed significant differences between PFD3 and untreated control cell (CC) groups. False discovery rate (FDR) was calculated to adjust for multiple comparisons. The location of significant DMRs was annotated relative to Ref seq genes as promoters (transcriptional start site [TSS] ± 1 kb), gene bodies, or intergenic regions.
Statistical Analysis
Data were analyzed using a one-way ANOVA test to assess statistical significance; *p ≤ 0.05; **p ≤ 0.01. Bar graphs represent the mean and SD across multiple independent experimental repeats.
Author Contributions
S.Y., N.H., and J.J.R. developed the concept. S.Y. designed and performed the experiments, including RNA aptamer selection, cell internalization, measurements of cell binding affinity, qPCR, data organization, and statistical analyses. S.Y. and J.J.R. prepared the manuscript. X.W. analyzed whole genome DNA methylation. B.A. was involved in confocal imaging of live cells and quantified aptamer internalization.
Conflicts of Interest
J.J.R. and N.H. are co-founders of privately owned Apterna Therapeutics.
Acknowledgments
Research reported in this publication includes work performed in the City of Hope Integrative Genomics, Analytical Cytometry, and X-ray Crystallography Cores supported by the National Cancer Institute of the NIH under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would also like to thank the Light Microscopy Digital Imaging Core at the Beckman Research Institute of City of Hope for their technical assistance and City of Hope scientific writer, Sarah T. Wilkinson, PhD. This work is funded by NIH grant AI42552 to J.J.R.
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.012.
Supplemental Information
References
- 1.Tuerk C. Using the SELEX combinatorial chemistry process to find high affinity nucleic acid ligands to target molecules. Methods Mol. Biol. 1997;67:219–230. doi: 10.1385/0-89603-483-6:219. [DOI] [PubMed] [Google Scholar]
- 2.Yoon S., Armstrong B., Habib N., Rossi J.J. Blind SELEX approach identifies RNA aptamers that regulate EMT and inhibit metastasis. Mol. Cancer Res. 2017;15:811–820. doi: 10.1158/1541-7786.MCR-16-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia W., Ren C., Wang L., Zhu B., Jia W., Gao M., Zeng F., Zeng L., Xia X., Zhang X. CD109 is identified as a potential nasopharyngeal carcinoma biomarker using aptamer selected by cell-SELEX. Oncotarget. 2016;7:55328–55342. doi: 10.18632/oncotarget.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruspe S., Giangrande P.H. Aptamer-siRNA chimeras: discovery, progress, and future prospects. Biomedicines. 2017;5:E45. doi: 10.3390/biomedicines5030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon S., Huang K.W., Reebye V., Mintz P., Tien Y.W., Lai H.S., Sætrom P., Reccia I., Swiderski P., Armstrong B. Targeted delivery of C/EBPalpha -saRNA by pancreatic ductal adenocarcinoma-specific RNA aptamers inhibits tumor growth in vivo. Mol. Ther. 2016;24:1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon S., Huang K.W., Reebye V., Spalding D., Przytycka T.M., Wang Y., Swiderski P., Li L., Armstrong B., Reccia I. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. Mol. Ther. Nucleic Acids. 2017;6:80–88. doi: 10.1016/j.omtn.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kratschmer C., Levy M. Targeted delivery of auristatin-modified toxins to pancreatic cancer using aptamers. Mol. Ther. Nucleic Acids. 2018;10:227–236. doi: 10.1016/j.omtn.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29:366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostrom Q.T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N.E., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro-oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wade A., Robinson A.E., Engler J.R., Petritsch C., James C.D., Phillips J.J. Proteoglycans and their roles in brain cancer. FEBS J. 2013;280:2399–2417. doi: 10.1111/febs.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regad T. Targeting RTK Signaling Pathways in Cancer. Cancers (Basel) 2015;7:1758–1784. doi: 10.3390/cancers7030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westermark B. Platelet-derived growth factor in glioblastoma-driver or biomarker? Ups. J. Med. Sci. 2014;119:298–305. doi: 10.3109/03009734.2014.970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldin C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 2013;11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y. Multifarious functions of PDGFs and PDGFRs in tumor growth and metastasis. Trends Mol. Med. 2013;19:460–473. doi: 10.1016/j.molmed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Heldin C.H., Westermark B. Platelet-derived growth factor: three isoforms and two receptor types. Trends Genet. 1989;5:108–111. doi: 10.1016/0168-9525(89)90040-1. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 17.Song S., Ewald A.J., Stallcup W., Werb Z., Bergers G. PDGFRbeta+ perivascular progenitor cells in tumours regulate pericyte differentiation and vascular survival. Nat. Cell Biol. 2005;7:870–879. doi: 10.1038/ncb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camorani S., Esposito C.L., Rienzo A., Catuogno S., Iaboni M., Condorelli G., de Franciscis V., Cerchia L. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol. Ther. 2014;22:828–841. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esposito C.L., Nuzzo S., Catuogno S., Romano S., de Nigris F., de Franciscis V. STAT3 gene silencing by aptamer-siRNA chimera as selective therapeutic for glioblastoma. Mol. Ther. Nucleic Acids. 2018;10:398–411. doi: 10.1016/j.omtn.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozawa T., Brennan C.W., Wang L., Squatrito M., Sasayama T., Nakada M., Huse J.T., Pedraza A., Utsuki S., Yasui Y. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K.W., Hu B., Cheng S.Y. Platelet-derived growth factor receptor alpha in glioma: a bad seed. Chin. J. Cancer. 2011;30:590–602. doi: 10.5732/cjc.011.10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell J.E. Validating Stat3 in cancer therapy. Nat. Med. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 24.Yu H., Jove R. The STATs of cancer—new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 25.Sherry M.M., Reeves A., Wu J.K., Cochran B.H. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherry-Lynes M.M., Sengupta S., Kulkarni S., Cochran B.H. Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS ONE. 2017;12:e0174775. doi: 10.1371/journal.pone.0174775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cenciarelli C., Marei H.E., Felsani A., Casalbore P., Sica G., Puglisi M.A., Cameron A.J., Olivi A., Mangiola A. PDGFRα depletion attenuates glioblastoma stem cells features by modulation of STAT3, RB1 and multiple oncogenic signals. Oncotarget. 2016;7:53047–53063. doi: 10.18632/oncotarget.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto K., Tateishi K., Kudo Y., Sato T., Yamamoto S., Miyabayashi K., Matsusaka K., Asaoka Y., Ijichi H., Hirata Y. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPα. Carcinogenesis. 2014;35:2404–2414. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- 29.Ene C.I., Edwards L., Riddick G., Baysan M., Woolard K., Kotliarova S., Lai C., Belova G., Cam M., Walling J. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS ONE. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan M.A., Shilatifard A. Medicine. (Poly)combing the pediatric cancer genome for answers. Science. 2013;340:823–824. doi: 10.1126/science.1239223. [DOI] [PubMed] [Google Scholar]
- 31.Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 33.Nistér M., Claesson-Welsh L., Eriksson A., Heldin C.H., Westermark B. Differential expression of platelet-derived growth factor receptors in human malignant glioma cell lines. J. Biol. Chem. 1991;266:16755–16763. [PubMed] [Google Scholar]
- 34.Williams K., Christensen J., Rappsilber J., Nielsen A.L., Johansen J.V., Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS ONE. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solá S., Xavier J.M., Santos D.M., Aranha M.M., Morgado A.L., Jepsen K., Rodrigues C.M. p53 interaction with JMJD3 results in its nuclear distribution during mouse neural stem cell differentiation. PLoS ONE. 2011;6:e18421. doi: 10.1371/journal.pone.0018421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer J.H., Helfand S.L. New tricks of an old molecule: lifespan regulation by p53. Aging Cell. 2006;5:437–440. doi: 10.1111/j.1474-9726.2006.00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aubrey B.J., Kelly G.L., Janic A., Herold M.J., Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuler M., Bossy-Wetzel E., Goldstein J.C., Fitzgerald P., Green D.R. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 39.Kumar R., Herbert P.E., Warrens A.N. An introduction to death receptors in apoptosis. Int. J. Surg. 2005;3:268–277. doi: 10.1016/j.ijsu.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Bennett M., Macdonald K., Chan S.W., Luzio J.P., Simari R., Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 41.Guan B., Yue P., Clayman G.L., Sun S.Y. Evidence that the death receptor DR4 is a DNA damage-inducible, p53-regulated gene. J. Cell. Physiol. 2001;188:98–105. doi: 10.1002/jcp.1101. [DOI] [PubMed] [Google Scholar]
- 42.Wu G.S., Burns T.F., McDonald E.R., 3rd, Jiang W., Meng R., Krantz I.D., Kao G., Gan D.D., Zhou J.Y., Muschel R. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 43.de Miguel D., Lemke J., Anel A., Walczak H., Martinez-Lostao L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23:733–747. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider P., Thome M., Burns K., Bodmer J.L., Hofmann K., Kataoka T., Holler N., Tschopp J. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity. 1997;7:831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 45.Ashcroft M., Kubbutat M.H., Vousden K.H. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loughery J., Cox M., Smith L.M., Meek D.W. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amano T., Nakamizo A., Mishra S.K., Gumin J., Shinojima N., Sawaya R., Lang F.F. Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. J. Neurooncol. 2009;92:357–371. doi: 10.1007/s11060-009-9844-1. [DOI] [PubMed] [Google Scholar]
- 48.Oda K., Arakawa H., Tanaka T., Matsuda K., Tanikawa C., Mori T., Nishimori H., Tamai K., Tokino T., Nakamura Y., Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 49.Castrogiovanni C., Waterschoot B., De Backer O., Dumont P. Serine 392 phosphorylation modulates p53 mitochondrial translocation and transcription-independent apoptosis. Cell Death Differ. 2018;25:190–203. doi: 10.1038/cdd.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aslam M.I., Hettmer S., Abraham J., Latocha D., Soundararajan A., Huang E.T., Goros M.W., Michalek J.E., Wang S., Mansoor A. Dynamic and nuclear expression of PDGFRα and IGF-1R in alveolar Rhabdomyosarcoma. Mol. Cancer Res. 2013;11:1303–1313. doi: 10.1158/1541-7786.MCR-12-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouveia L., Betsholtz C., Andrae J. Expression analysis of platelet-derived growth factor receptor alpha and its ligands in the developing mouse lung. Physiol. Rep. 2017;5:e13092. doi: 10.14814/phy2.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr. Opin. Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 53.Lin S.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y., Bourguignon L., Hung M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 54.Rakowicz-Szulczynska E.M., Rodeck U., Herlyn M., Koprowski H. Chromatin binding of epidermal growth factor, nerve growth factor, and platelet-derived growth factor in cells bearing the appropriate surface receptors. Proc. Natl. Acad. Sci. USA. 1986;83:3728–3732. doi: 10.1073/pnas.83.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q., Sakamoto K., Wagner K.U. D-type Cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol. Cell. Endocrinol. 2014;382:583–592. doi: 10.1016/j.mce.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Cui Y., Zhang L., Sheng J., Yang Y., Kuang G., Fan Y., Zhang Q., Jin J. The silencing of CCND2 by promoter aberrant methylation in renal cell cancer and analysis of the correlation between CCND2 methylation status and clinical features. PLoS ONE. 2016;11:e0161859. doi: 10.1371/journal.pone.0161859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamikihara T., Arima T., Kato K., Matsuda T., Kato H., Douchi T., Nagata Y., Nakao M., Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int. J. Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 60.Jandrig B., Seitz S., Hinzmann B., Arnold W., Micheel B., Koelble K., Siebert R., Schwartz A., Ruecker K., Schlag P.M. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene. 2004;23:9295–9302. doi: 10.1038/sj.onc.1208131. [DOI] [PubMed] [Google Scholar]
- 61.Cheng Y., Geng H., Cheng S.H., Liang P., Bai Y., Li J., Srivastava G., Ng M.H., Fukagawa T., Wu X. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res. 2010;70:6516–6526. doi: 10.1158/0008-5472.CAN-09-4566. [DOI] [PubMed] [Google Scholar]
- 62.Yu J., Liang Q.Y., Wang J., Cheng Y., Wang S., Poon T.C., Go M.Y., Tao Q., Chang Z., Sung J.J. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–317. doi: 10.1038/onc.2012.54. [DOI] [PubMed] [Google Scholar]
- 63.Lee J., Lee Y., Xu L., White R., Sullenger B.A. Differential induction of immunogenic cell death and interferon expression in cancer cells by structured ssRNAs. Mol. Ther. 2017;25:1295–1305. doi: 10.1016/j.ymthe.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon S., Lee G., Han D., Song J.Y., Kang K.S., Lee Y.S. Neutralization of infectivity of porcine circovirus type 2 (PCV2) by capsid-binding 2'F-RNA aptamers. Antiviral Res. 2010;88:19–24. doi: 10.1016/j.antiviral.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 65.Hernandez F.J., Kalra N., Wengel J., Vester B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg. Med. Chem. Lett. 2009;19:6585–6587. doi: 10.1016/j.bmcl.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 66.Soontornworajit B., Zhou J., Snipes M.P., Battig M.R., Wang Y. Affinity hydrogels for controlled protein release using nucleic acid aptamers and complementary oligonucleotides. Biomaterials. 2011;32:6839–6849. doi: 10.1016/j.biomaterials.2011.05.074. [DOI] [PubMed] [Google Scholar]
- 67.Zhou J., Swiderski P., Li H., Zhang J., Neff C.P., Akkina R., Rossi J.J. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Hahn M.A., Li A.X., Wu X., Pfeifer G.P. Single base resolution analysis of 5-methylcytosine and 5-hydroxymethylcytosine by RRBS and TAB-RRBS. Methods Mol. Biol. 2015;1238:273–287. doi: 10.1007/978-1-4939-1804-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.