Abstract
Gain-of-function studies often require the tedious cloning of transgene cDNA into vectors for overexpression beyond the physiological expression levels. The rapid development of CRISPR/Cas technology presents promising opportunities to address these issues. Here, we report a simple, cloning-free method to induce gene expression at an endogenous locus using CRISPR/Cas9 activators. Our strategy utilizes synthesized sgRNA expression cassettes to direct a nuclease-null Cas9 complex fused with transcriptional activators (VP64, p65, and Rta) for site-specific induction of endogenous genes. This strategy allows rapid initiation of gain-of-function studies in the same day. Using this approach, we tested two CRISPR activation systems, dSpCas9VPR and dSaCas9VPR, for induction of multiple genes in human and rat cells. Our results showed that both CRISPR activators allow efficient induction of six different neural development genes (CRX, RORB, RAX, OTX2, ASCL1, and NEUROD1) in human cells, whereas the rat cells exhibit more variable and less-efficient levels of gene induction, as observed in three different genes (Ascl1, Neurod1, Nrl). Altogether, this study provides a simple method to efficiently activate endogenous gene expression using CRISPR/Cas9 activators, which can be applied as a rapid workflow to initiate gain-of-function studies for a range of molecular- and cell-biology disciplines.
Keywords: CRISPR, cloning free, gene activation, CRISPR activation, SpCas9, SaCas9, multiplex gene activation, transcriptional activator
Introduction
Conventional gene overexpression studies require the need to clone the transgene cDNA into an expression vector and therefore involve DNA ligation, bacterial transformation, screening clones, plasmid purification, and quality check to confirm the vector sequences. This represents a tedious and costly procedure, especially for large-scale genome-wide overexpression studies. Further to this, the use of whole-transgene cDNA imposes a challenge for overexpressing multiple genes simultaneously in cells, due to the large burden of DNA required to be delivered into the cells.
The emergence of CRISPR/Cas technology has revolutionized the field of molecular biology, providing a promising tool for precise gene editing with profound implications for development of gene therapy.1 CRISPR/Cas utilizes an RNA-guided mechanism for site-specific DNA cleavage, which has been used to knock in or knock out genes in vitro2, 3 and in vivo.4, 5 However, the first described Streptococcus pyogenes (Sp)Cas9 is large in size and presents a challenge for packaging into adeno-associated viruses (AAVs) for therapeutic delivery. Subsequently, smaller Cas9 variants have been identified, including Staphylococcus aureus (Sa)Cas9,6, 7 Neisseria meningitidis (Nm)Cas9,8 and compylobacter Jejuni (Cj)Cas9,9 which have higher therapeutic potential due to their smaller sizes. Also, further modifications of Cas9 have expanded the ability of using CRISPR/Cas beyond genome editing, including control of gene regulation, epigenetics, and chromatin imaging.10 For instance, a nuclease-null dead Cas9 can be fused to transcriptional activators to target the regulatory region of a gene to induce its expression.11 Importantly, multiple single-guide RNAs (sgRNAs) targeting different genes can be utilized to induce multiplex gene expression. Since only a short sgRNA is needed to induce expression of a single gene, rather than a whole-transgene cDNA copy, the CRISPR activation approach can potentially reduce the number of viral vectors needed for overexpressing multiple genes. Another advantage of CRISPR activation is it activates gene expression directly at endogenous locus with high specificity, which allows for expression of splice variants as well as endogenous gene regulation via UTR regions.12, 13
Several reports have previously utilized a cloning-free CRISPR approach to knock in or knock out genes. In fission yeasts, Zhang et al.14 showed that the gap-repair mechanism can be used to assemble PCR-amplified sgRNA fragments and linear Cas9 plasmids together. In mammalian cells, Arbab et al.15 have reported a self-cloning CRISPR/Cas9 approach by using palindromic sgRNA, either in expression cassette or short DNA sequences, to allow homologous recombination in target cells to yield a functional site-specific sgRNA plasmid. The authors also showed that co-transfection of the Cas9 expression plasmids and sgRNA expression cassettes synthesized as short 500-bp oligonucleotides allow efficient knockout of target gene in mouse embryonic stem cells. Others have described the use of CRISPR ribonucleoprotein complexes for generation of knockin mice,16, 17 as well as gene editing in mammalian cells in vitro delivered through various methods, such as transfection by cationic lipids, nanoparticles, or cell-penetrating peptides.18, 19, 20, 21, 22 However, a cloning-free approach has not been systematically reviewed for gene activation or repression using CRISPR/Cas9.
Here, we present a simple and rapid method to use CRISPR/Cas for gene activation. Our method utilizes commercially synthesized sgRNA expression cassettes, which bypass the need for molecular cloning of site-specific sgRNA plasmids. In this study, we compare the use of synthesized sgRNA expression cassettes with two CRISPR/Cas activators, dSpCas9VPR and dSaCas9VPR, to induce gene expression in human and rat cells. Our strategy vastly simplifies the initiation of gain-of-function studies and as such has implications across many disciplines in cell biology.
Results
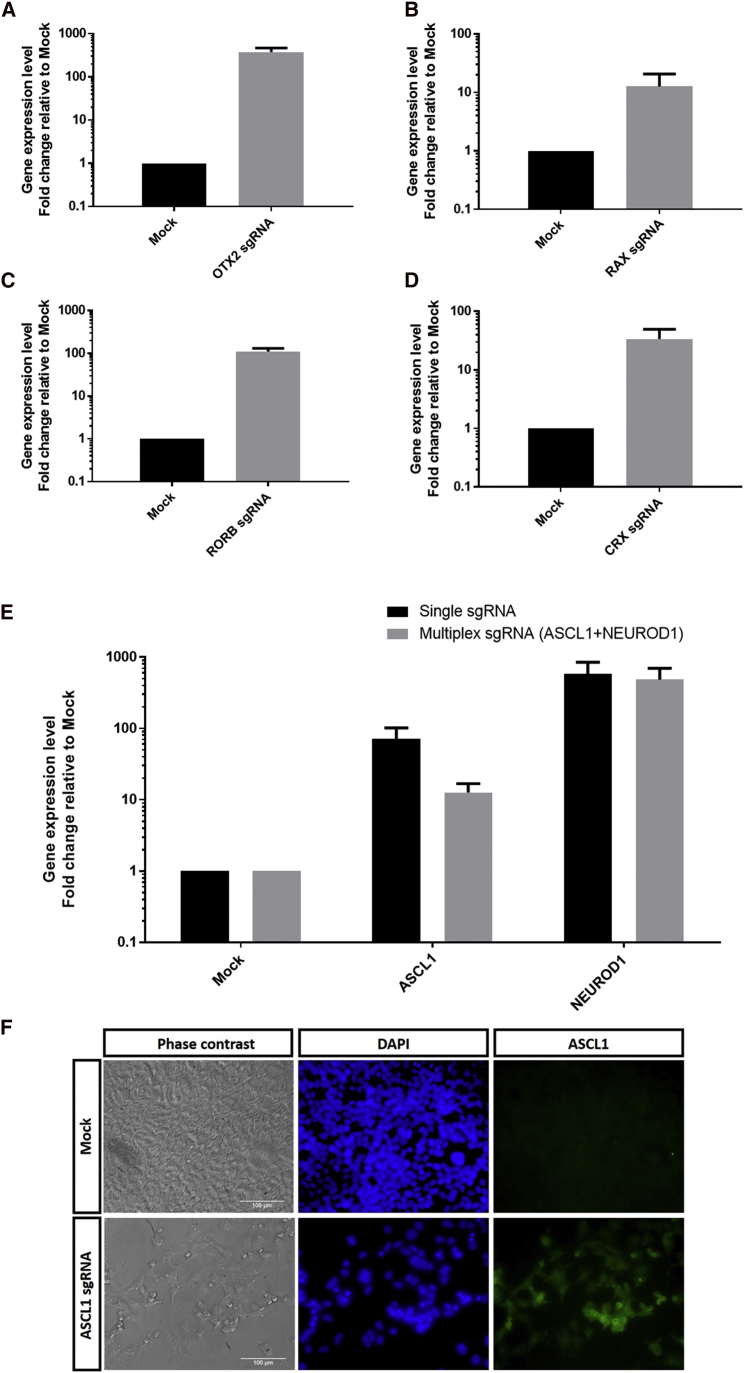
To utilize CRISPR/Cas to activate endogenous genes in mammalian cells, we first tested the dSpCas9VPR system.23 This system utilizes a nuclease-null dead SpCas9 (dSpCas9) coupled with transcription factor activation domains VP64, p65, and Rta (VPR). The sgRNAs were designed using the synergistic activation mediator (SAM) sgRNA design tool. For gene activation, we chose six human genes that encoded for transcription factors that regulate neural and retinal development: RAX, OTX2, RORB, CRX, ASCL1, and NEUROD1 (Table 1). We synthesized sgRNA expression cassettes as linear oligonucleotide fragments containing an upstream U6 promoter, the sgRNA, and the corresponding sgRNA scaffold (Figure S1). The sgRNA expression cassettes and the dSpCas9VPR plasmids were co-transfected into HEK293A cells. By day 3 post-transfection, we can detect robust gene activation in HEK293A cells using dSpCas9VPR, with upregulation of OTX2 (∼375-fold increase; Figure 1A), RAX (∼13-fold increase; Figure 1B), RORB (∼110-fold increase; Figure 1C), and CRX (∼35-fold increase; Figure 1D). Similarly, the dSpCas9VPR system is also efficient in gene activation of ASCL1 and NEUROD1, with ∼70-fold increase and ∼585-fold increase, respectively (Figure 1E). Immunocytochemistry analysis also confirmed efficient expression of ASCL1 protein in HEK293A following dSpCas9VPR gene activation (Figure 1F).
Table 1.
Information of sgRNAs Used for SpCas9
| Species | Name | TSS Distance | Strand | Sequence | PAM | On-Target Score |
|---|---|---|---|---|---|---|
| Rat | Ascl1 sgRNA 1 | −183 bp | + | 5′-ACGCACTGCAACAACAAACC-3′ | CGG | 46.1 |
| Ascl1 sgRNA 2 | −351 bp | − | 5′-TCCTAGGTAGAAAGTCTGGA-3′ | GGG | 73.4 | |
| Neurod1 sgRNA 1 | −258 bp | + | 5′-TGCGGGTAAAAACAGGTCCG-3′ | CGG | 56.1 | |
| Neurod1 sgRNA 2 | −164 bp | + | 5′-ATACAAATAGGCAGGTCACG-3′ | TGG | 84.1 | |
| Nrl sgRNA 1 | −573 bp | − | 5′-CTTTACCTCTCAAAGCCTTC-3′ | AGG | 27.5 | |
| Nrl sgRNA 2 | −764 bp | + | 5′-CCATCTGCTTAGACTCACCA-3′ | TGG | 77.8 | |
| Human | ASCL1 sgRNA 1 | −181 bp | + | 5′-CGGGAGAAAGGAACGGGAGG-3′ | GGG | 30.9 |
| NEUROD1 sgRNA 1 | −33 bp | + | 5′-AGGGGAGCGGTTGTCGGAGG-3′ | AGG | 30.9 | |
| RAX sgRNA | −99 bp | + | 5′-GAGGGAGGGGCCGAGAGAAG-3′ | GGG | 44.0 | |
| OTX2 sgRNA | −170 bp | + | 5′-AGATTGTAATTGCTTTCTTC-3′ | GGG | 35.2 | |
| CRX sgRNA | −114 bp | − | 5′-AGGGAGGCCCCAGCTCCTGC-3′ | CGG | 51.6 | |
| RORB sgRNA | −172 bp | + | 5′-CCCGGCCACCTCGGACTCCC-3′ | TGG | 41.8 | |
| Mouse | Nkx2.5 sgRNA | −171 bp | + | 5′-GTATTTTCTTTGAGTGTGTC-3′ | TGG | 36.1 |
On-target score is an optimized score for 20-bp sgRNA based on Doench et al.28. Scores range from 0–100. TSS, transcription start site.
Figure 1.
Efficient Gene Activation Using dSpCas9VPR in HEK293A Cells
qPCR analysis of gene activation for (A) OTX2, (B) RAX, (C) RORB, (D) CRX, (E) ASCL1, NEUROD1, or multiplex induction of ASCL1 and NEUROD1. Results are displayed as the mean of three independent biological repeats ± SEM. (F) Immunocytochemistry results showed upregulated protein expression of ASCL1 (green) in HEK293A following gene activation with dSpCas9VPR. Scale bars, 100 μm.
To determine the kinetics of the gene activation, we performed a time-course experiment following ASCL1 gene activation. Our results showed that dSpCas9VPR upregulated ASCL1 expression levels after 2 days and gradually decreased after 3 days, with persistent upregulated levels up to 6 days post-transfection (Figure S3). These results showed that dSpCas9VPR resulted in maximal gene activation level after 2–3 days following transfection.
Subsequently, we tested the feasibility of using dSpCas9VPR for multiplex gene activation. We co-transfected sgRNAs for ASCL1 and NEUROD1 into HEK293A cells. Notably, both genes can be upregulated simultaneously and efficiently using the dSpCas9VPR, with ∼13-fold induction of ASCL1 expression and ∼485-fold induction of NEUROD1 expression (Figure 1E). However, multiplexing resulted in a lower level of gene induction compared to using single sgRNA, an effect more prominent in induction of ASCL1 (Figure 1E). Taken together, our results demonstrated that dSpCas9VPR can be used to efficiently activate gene expression in human cells.
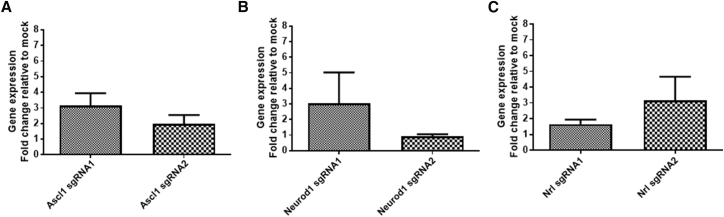
Furthermore, we assessed the feasibility of using dSpCas9VPR for gene activation in rat cells. As the SAM sgRNA design tool does not support design for the rat genome, we utilized the gene-activator sgRNA design tool in Benchling (Table 1). We analyzed dSpCas9VPR-mediated activation of three genes, Ascl1, Neurod1, and Nrl, in a rat Müller glial cell line rMC1, which can be transfected efficiently (Figure S5). Our results showed that Ascl1 can be activated to modest levels using two sgRNAs, resulting in an ∼3- and ∼2-fold increase, respectively (Figure 2A). Similarly, two sgRNAs were tested for Neurod1 gene activation. While one sgRNA resulted in a modest increase in Neurod1 expression levels (∼3-fold increase), another sgRNA failed to activate Neurod1 (Figure 2B). For Nrl, the two designed sgRNAs resulted in an ∼3- and ∼1.6-fold induction in gene expression, respectively (Figure 2C). We also tested gene activation with dSpCas9VPR in another rat fibroblast cell line R12. However, we did not observe significant gene activation in Ascl1, Neurod1, nor Nrl (Figure S4). This is unlikely due to problems with transfection, as R12 can be efficiently transfected in this context (Figure S5). Moreover, we showed that the dSpCas9VPR can efficiently activate gene expression in mouse embryonic fibroblasts (∼19-fold increase in Nkx2.5; Figure S6), suggesting that the inefficient CRISPR activation seen is specific to rat cells and not other rodent cells. Collectively, these results demonstrated that dSpCas9VPR can be used to activate genes in rat cells, albeit only limited to modest levels of upregulation in certain rat cells and is not as efficient as in human cells.
Figure 2.
Assessment of Multiple sgRNAs for dSpCas9VPR to Induce Gene Activation in Rat Müller Glial Cell rMC1
qPCR analysis of gene activation using two sgRNAs, respectively, for (A) Ascl1, (B) Neurod1, and (C) Nrl. Results are displayed as the mean of four independent biological repeats ± SEM.
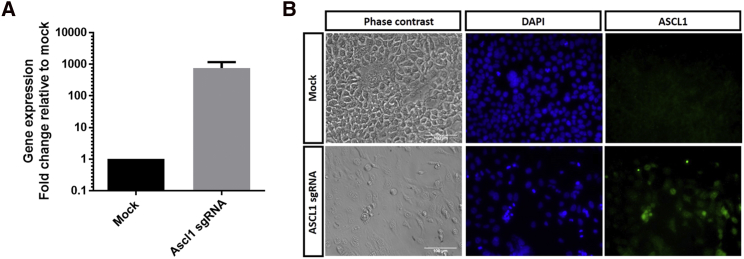
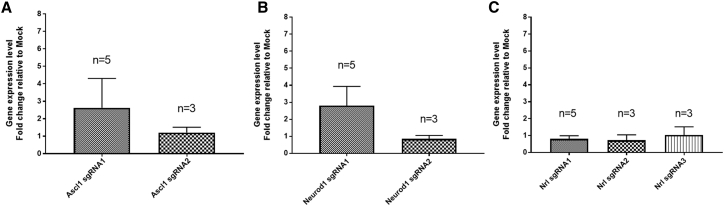
Next, we assessed the efficiency of gene activation using another CRISPR/Cas activation system, dSaCas9VPR, which utilizes a nuclease-null SaCas9 coupled with VPR. Notably, the dSaCas9VPR is much smaller in size than dSpCas9VPR, which has important implications for packaging into viral vectors for gene therapy. We designed sgRNAs using Benchling for both human and rat genes and selected two to three sgRNAs against each gene for evaluation (Table 2). The sgRNA expression cassette for dSaCas9VPR contained a 5′ Myc tag, upstream U6 promoter, sgRNA, sgRNA scaffold, and a 3′ HA tag (Figure S2). In HEK293A cells, we showed that dSaCas9VPR can activate the expression levels of endogenous ASCL1 efficiently (∼760-fold increase; Figure 3A). Also, we detected upregulation of the ASCL1 protein levels following dSaCas9VPR gene activation (Figure 3B). However, dSaCas9VPR exhibited variable efficiency in gene activation in rMC1 cells (Figure 4), a result similar to those observed in dSpCas9VPR. Two sgRNAs were tested for activation of Ascl1 or Neurod1, with one sgRNA inducing modest levels of gene upregulation and the other failing to do so (Ascl1, ∼2.6, ∼1 fold changes; Neurod1, ∼2.8, ∼1 fold changes; Figures 4A and 4B). However, for Nrl activation, all three sgRNAs tested failed to upregulate Nrl expression levels (Figure 4C). In summary, we showed that dSaCas9VPR can be used to efficiently activate gene expression in human cells; however, its effect in rat cells is variable and remained inefficient.
Table 2.
Information of sgRNAs Used for SaCas9
| Species | Name | TSS Distance | Strand | Sequence | PAM | On-Target Score | On-Target Score Ranking |
|---|---|---|---|---|---|---|---|
| Rat | Ascl1 sgRNA 1 | −180 bp | + | 5′-GCACTGCAACAACAAACCCGG-3′ | CTGAAT | 70.9 | 1 |
| Ascl1 sgRNA 2 | −223 bp | − | 5′-TGGCGCGTGCCGGACTCCCGG-3′ | CTGAAT | 64.2 | 5 | |
| Neurod1 sgRNA 1 | −258 bp | + | 5′-CTGCGGGTAAAAACAGGTCCG-3′ | CGGAGT | 56.1 | 2 | |
| Neurod1 sgRNA 2 | −205 bp | + | 5′-TTCTTCTGGCCACAAAGGGGC-3′ | CGGAAT | 38.5 | 5 | |
| Nrl sgRNA 1 | −575 bp | − | 5′-TTTACCTCTCAAAGCCTTCAG-3′ | GAGAGT | 79.3 | 1 | |
| Nrl sgRNA 2 | −145 bp | + | 5′-TTCAGGGCTGCTTCATTACTC-3′ | CGGAAT | 45.1 | 9 | |
| Nrl sgRNA 3 | −760 bp | − | 5′-TTTAACTTAGCACCTGCCATG-3′ | GTGAGT | 72.7 | 2 | |
| Human | ASCL1 sgRNA | −239 bp | + | 5′-GCACTGCAACAACAAACCCAG-3′ | CTGAAT | 74.2 | 2 |
On-target score is an optimized score for 20-bp sgRNA based on Doench et al.28. The on-target scores are ranked for sgRNAs targeting 2,000-bp upstream of transcription start site of genes. Scores range from 0–100. TSS, Transcription start site.
Figure 3.
Efficient Gene Activation Using dSaCas9VPR in HEK293A Cells
(A) qPCR analysis of gene activation for ASCL1 in HEK293A cells. Results are displayed as mean of three independent biological repeats ± SEM. (B) Immunocytochemistry results showed upregulated protein expression of ASCL1 (green) in HEK293A following gene activation with dSaCas9VPR. Scale bars, 100 μm.
Figure 4.
Comparison of Multiple sgRNAs to Induce Gene Activation in Rat Müller Glial Cell rMC1 Using dSaCas9VPR
qPCR analysis of gene activation for (A) Ascl1 using two sgRNAs, (B) Neurod1 using two sgRNAs, and (C) Nrl using three sgRNAs. Results are displayed as the mean of three to five independent biological repeats ± SEM.
Discussion
A key limiting factor in the conventional design of overexpression studies is the requirement of tedious plasmid construction for each transgene, which involves molecular cloning steps that take more than 1 week. Here, we present a simplified method for gene activation using CRISPR/Cas9 activators in mammalian cells that is feasible in 1 day. Notably, our strategy to transfect sgRNA as a synthesized linear oligonucleotide fragment with U6 promoter allows sgRNA expression in the cells and eliminates the need to clone sgRNA into a designated vector prior to transfection. Our strategy provides a rapid way to initiate gene-overexpression studies using the CRISPR activation system.
This study compared two CRISPR activation systems, dSpCas9VPR and dSaCas9VPR, in human and rat cells. Our results demonstrated that both dSpCas9VPR and dSaCas9VPR can efficiently induce gene expression in human cells. We showed the use of dSpCas9VPR for multiplex activation of two genes in human cells, as well as its high efficiency for gene activation in mouse cells. Furthermore, in rat cells we have assessed activation of three genes using six sgRNAs for dSpCas9VPR and seven sgRNAs for dSaCas9VPR in two different rat cell lines. However, our results showed only modest levels and, in some cases, negligible levels of gene activation in rat Müller glial cells and fibroblasts. This is unlikely due to issues with transfection efficiency, as both rat cell types can be efficiently transfected with robust GFP expression. We speculate that this could be due to inadequate support of the current sgRNA design algorithm for the rat genome. For instance, many of the sgRNAs used for rat genes have high ranking of on-target scores (Table 2), but they are mostly inefficient in inducing expression of rat genes using the CRISPR activation systems. This highlights the need to improve the design and accuracy of predicting functional sgRNAs for the rat genome. Furthermore, it is also possible that the differences in gene activation we observed are due to variations in chromatic state and accessibility in different rat and human cells. Therefore, alternative CRISPR activation systems may be more effective at enhancing gene activation in rat cells. In addition to the dCas9-VPR system from the Church laboratory, other CRISPR activation systems have been described, such as the SAM system, the SunTag activator system, and dCas9-VP128 system.24, 25, 26 These alternative CRISPR activation systems utilize distinct transcriptional activators to enhance gene activity. Chavez et al.11 undertook a comprehensive head-to-head comparison of various CRISPR activation systems and found that different systems showed varying activities at different gene loci as well as in different cell types tested.
Notably, the simplified method for gene activation described here can also be adopted for other CRISPR systems for gene editing or repression, which allows rapid testing of effective sgRNA sequences that can be selected for further experiments in different delivery systems. Also, our sgRNA expression cassette design can be applied to different Cas9 activator systems, since the same sgRNA design can be used for different Cas9 activator systems (such as SpCas9VPR, SpCas9-Suntag, SpCas9-VP64).
A potential challenge of using CRISPR/Cas systems is that not all sgRNAs are efficient in Cas9 targeting; thus, multiple sgRNAs are often screened to identify the functional sgRNAs. In our experience, the sgRNA design tools for predicting functional sgRNAs are generally very accurate for human cells. For dSpCas9VPR, the first sgRNA designed is functional for gene activation in 13 out of 15 genes tested (∼87%; Table S1). Similarly, although we only tested gene activation for a single gene using dSaCas9VPR, the first sgRNA designed is highly efficient for gene activation (Table S1). Therefore, we recommend designing two sgRNAs within 300 bp upstream of the transcription start site to be tested for gene activation in human cells. It should be noted that sgRNAs that target different proximities of the transcriptional start site can result in different efficiencies of gene activation,18 and this can be further optimized for individual target genes of interest. Additionally, it is also possible to combine several sgRNAs targeting different promoter regions to improve the levels of gene activation using CRISPRa.
In summary, this study outlines a simple and robust workflow to efficiently activate endogenous gene expression in mammalian cells using CRISPR/Cas activators, which can be applied as a rapid workflow to initiate gain-of-function studies for a range of molecular- and cell-biology subjects.
Materials and Methods
Institutional Approval
This study was approved by the Institutional Biosafety Committee at Monash University (#10969) in compliance with the regulations by the Office of the Gene Technology Regulator in Australia.
sgRNA Design and Preparation of Expression Cassette
For SpCas9, sgRNAs with NGG protospacer adjacent motifs (PAMs) were designed using the SAM sgRNA design tool (http://sam.genome-engineering.org/database/) for human genes and Benchling (https://benchling.com/) for rat genes. The SpCas9 sgRNA expression cassette contains an upstream U6 promoter, sgRNA, and sgRNA scaffold with stem extension and stem loop (Figure S1).
For SaCas9, sgRNAs with NNGRRT PAMs were designed using Benchling (https://benchling.com/). Predicted sgRNAs with long stretches of repeating nucleotides are excluded from selection. The SaCas9 sgRNA expression cassette contains a Myc tag, U6 promoter, sgRNA, sgRNA scaffold, and HA tag (Figure S2).
Both SpCas9 and SaCas9 sgRNA expression cassettes (<500 bp) were synthesized as gBLOCK gene fragments (Integrated DNA Technologies). The sgRNA expression cassettes were amplified by PCR using the following primers: SpCas9 forward primer, 5′-TGAGTATTACGGCATGTGAGGGC-3′; SpCas9 reverse primer, 5′-TCAATGTATCTTATCATGTCTGCTCGA-3′; SaCas9 forward primer, 5′-GAACAAAAACTCATCTCAGAAGAGGATCTG-3′; SaCas9 reverse primer, 5′-TACCCATACGATGTTCCAGATTACGCT-3′. PCR was performed using KOD Hot Start DNA polymerase (Merck Millipore) with the following thermal profile: 95°C for 2 min; 30 cycles of 95°C for 20 s, 66°C (SpCas9 sgRNA) or 64°C (SaCas9 sgRNA) for 10 s, 70°C for 8 s; 70°C for 5 min. The PCR amplicons were separated by gel electrophoresis, and the sgRNA expression cassettes were extracted using the Wizard SV gel and PCR cleanup kit (Promega). The amplified sgRNA expression cassettes were checked with Nanodrop to confirm good DNA quality.
Cell Culture
Rat Müller glial cells rMC1, rat fibroblasts R12, mouse embryonic fibroblasts, and HEK293A cells were maintained in DMEM high-glucose media supplemented with 10% fetal calf serum, 2 mM L-glutamine, and 0.5% penicillin/streptomycin (all from Thermo Fisher). All cells were passaged using 0.25% trypsin before the culture become confluent and maintained in incubators at 37°C with 5% CO2 level.
Transfection Efficiency Assay
rMC1 and R12 cells were transfected with the pmaxGFP construct (Lonza) using Lipofectamine 3000 overnight, following the manufacturer’s instruction. GFP expression is determined 1 day after transfection using a fluorescence microscope (Olympus CKX53).
Gene Activation Using CRISPR/Cas Activation
dSaCas9-VPR and dSpCas9-VPR plasmids were gifts from George Church (Addgene #68495 and #63798, respectively). rMC1, R12, mouse embryonic fibroblasts and HEK293A cells were transfected using Lipofectamine 3000. In brief, cells were plated down on a 12-well plate at day 0 (6 × 104/well). At day 1, the cells were transfected with 360 ng sgRNA expression cassettes and 800 ng dSaCas9-VPR or dSpCas9-VPR plasmids using Lipofectamine 3000 overnight. In some experiments, the cells and DNA were upscale proportionally to obtain more RNA. Mock control (no DNA transfected) was utilized as a negative control. At day 4, the samples were harvested for RNA to assess gene expression levels or fixed for immunocytochemistry analysis. In some experiments, RNA was harvested at different time points (days 2, 3, 4, 5, and 6) to determine kinetics of CRISPR gene activation.
qPCR Analysis
Total RNA was extracted using the RNeasy kit (QIAGEN) or the Illustra RNAspin kit (GE Healthcare) followed by DNase treatment. For mouse embryonic fibroblasts, RNA was extracted from cells using TriReagent (Thermo Fisher) followed by RNA precipitation with chloroform and isopropanol (Sigma-Aldrich). cDNA synthesis was performed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher) following the manufacturer’s instructions. Taqman gene-expression assay (Thermo Fisher) was performed using the Taqman Fast Advanced Master Mix with the following probes: human RAX (Hs00429459_m1), human OTX2 (Hs00222238_m1), human ASCL1 (Hs00269932_m1), human NEUROD1 (Hs00159598_m1), human CRX (Hs00230899_m1), human RORB (Hs00199445_m1), human ACTB (Hs99999903_m1), rat Ascl1 (Rn00574345_m1), rat Neurod1 (Rn00824571_s1), rat Nrl (Rn01481925_m1), rat Gapdh (Rn01775763_g1), mouse Nkx2.5 (Mm00657783_m1), and mouse Gapdh (Mm99999915_g1). qPCR was processed using the ABI Step One Plus system, the ABI 7500 system, or the QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems). The delta delta Ct method was used to calculate relative gene expression compared to control. Gene expression for each transfected condition was normalized to its corresponding mock control. The housekeeping genes ACTB or Gapdh were used for normalizing gene expression in human and rat cells, respectively.
Immunocytochemistry Analysis
Standard immunocytochemistry procedures were carried out as previously described.27 In brief, samples were fixed in methanol, followed by blocking and permeabilization (0.1% Tween 20). Subsequently, the samples were immunostained with antibodies against ASCL1 (Abcam, #ab74065, 5 μg/mL), the appropriate Alexa Fluor 488 secondary antibodies (Abcam), and nuclear counterstain with DAPI (Sigma). Samples were imaged using a Nikon Eclipse TE2000-U fluorescent microscope.
Author Contributions
S.S.C.H., A.W.H., and R.C.B.W. designed the experiments; L.F., J.Y., L.E.W., T.N., S.K., S.Y.L., and S.S.C.H. performed the experiments; L.F., J.Y., L.E.W., S.S.C.H., S.Y.L., A.W.H., and R.C.B.W. analyzed the data; R.C.B.W. wrote the manuscript; all authors approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Chris Karelas for technical support. rMC1 is a generous gift from Mark Gillies’s laboratory, and R12 is a generous gift from Peter Van Wijngaarden. This work was supported by funding from the Ophthalmic Research Institute of Australia (R.C.B.W.), the University of Melbourne De Brettville Trust (R.C.B.W.), and the Kel and Rosie Day Foundation (R.C.B.W.). The Centre for Eye Research Australia receives operational infrastructure support from the Victorian government.
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.008.
Supplemental Information
References
- 1.Hung S.S.C., Chrysostomou V., Li F., Lim J.K.H., Wang J.-H., Powell J.E., Tu L., Daniszewski M., Lo C., Wong R.C. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest. Ophthalmol. Vis. Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 2.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung S.S.C., McCaughey T., Swann O., Pébay A., Hewitt A.W. Genome engineering in ophthalmology: Application of CRISPR/Cas to the treatment of eye disease. Prog. Retin. Eye Res. 2016;53:1–20. doi: 10.1016/j.preteyeres.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.-F., Sontheimer E.J., Thomson J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.-Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 13.La Russa M.F., Qi L.S. The New State of the Art: Cas9 for Gene Activation and Repression. Mol. Cell. Biol. 2015;35:3800–3809. doi: 10.1128/MCB.00512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X.-R., He J.-B., Wang Y.-Z., Du L.-L. A Cloning-Free Method for CRISPR/Cas9-Mediated Genome Editing in Fission Yeast. G3 (Bethesda) 2018;8:2067–2077. doi: 10.1534/g3.118.200164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arbab M., Srinivasan S., Hashimoto T., Geijsen N., Sherwood R.I. Cloning-free CRISPR. Stem Cell Reports. 2015;5:908–917. doi: 10.1016/j.stemcr.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aida T., Chiyo K., Usami T., Ishikubo H., Imahashi R., Wada Y., Tanaka K.F., Sakuma T., Yamamoto T., Tanaka K. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 2015;16:87. doi: 10.1186/s13059-015-0653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X., Chen C., Veevers J., Zhou X., Ross R.S., Feng W., Chen J. CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci. Rep. 2017;7:42244. doi: 10.1038/srep42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Kim S., Kim D., Cho S.W., Kim J., Kim J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z., Zhang D., Xiong X., Yan B., Xie W., Sheen J., Li J.F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants. 2017;3:930–936. doi: 10.1038/s41477-017-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanenbaum M.E., Gilbert L.A., Qi L.S., Weissman J.S., Vale R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung S.S.C., Wong R.C.B., Sharov A.A., Nakatake Y., Yu H., Ko M.S.H. Repression of global protein synthesis by Eif1a-like genes that are expressed specifically in the two-cell embryos and the transient Zscan4-positive state of embryonic stem cells. DNA Res. 2013;20:391–402. doi: 10.1093/dnares/dst018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.