Abstract
Xanthomonas vasicola pv. musacearum (Xvm) is a bacterial pathogen responsible for the economically important Xanthomonas wilt disease on banana and enset crops in Sub-Saharan Africa. Given that the symptoms are similar to those of other diseases, molecular diagnosis is essential to unambiguously identify this pathogen and distinguish it from closely related strains not pathogenic on these hosts. Currently, Xvm identification is based on polymerase chain reaction (PCR) with GspDm primers, targeting the gene encoding general secretory protein D. Experimental results and examination of genomic sequences revealed poor specificity of the GspDm PCR. Here, we present and validate five new Xvm-specific primers amplifying only Xvm strains.
Keywords: Bioinformatics, Microbiology, Molecular biology, Plant biology
1. Introduction
Xanthomonas campestris pv. musacearum (Xcm) is a gamma-proteobacterium, causing a devastating bacterial wilt to banana and enset within East and central Africa (ECA) (Nakato et al., 2018). Fingerprinting using rep-PCR, fatty acid methyl ester analysis (FAME) and sequencing of the gyrase B gene suggested that Xcm belongs to the Xanthomonas vasicola species (Aritua et al., 2008). Comparative phylogenomic studies further supported the phylogenetic relatedness of Xcm to Xanthomonas vasicola pv. vasculorum (Xvv) (Studholme et al., 2010; Wasukira et al., 2012). Although the taxonomic reassignment to X. vasicola has yet to be resolved, we opt to refer to the pathogen as Xanthomonas vasicola pv. musacearum (Xvm). The species Xanthomonas vasicola includes pathovars holcicola (Xvh), vasculorum (Xvv), and musacearum (Xvm), respectively pathogenic to sorghum, sugarcane and maize, and Musaceae.
Molecular diagnosis of Xvm has been performed until now using Xvm-specific (GspDm) PCR primers, designed to amplify a 265-bp fragment of the Xvm gspD gene (Adriko et al., 2012) that encodes for the general secretory protein D. The availability of genomic sequences of Xvh, Xvv and Xvm has updated our knowledge of the distribution of genes across pathovar-specific genes. BLASTN searches against the NCBI whole-genome shotgun sequence databases with GspDm revealed hits against three Xvv genomes: two recently published USA isolates collected on maize (isolates 201500744 and 201500181) (Korus et al., 2017; Lang et al., 2017) and Xvv NCPPB 895 (Wasukira et al., 2014). Therefore, the targeted GspD sequence is thus clearly not unique to Xvm. Motivated by the failure of the GspDm PCR assay to unambiguously identify Xvm, we developed new Xvm-specific PCR primers. Such pathovar-specific detection is required for in-planta studies and for in-field detection of the pathogen in symptomatic plants as well as for surveys of alternative host reservoirs (Hodgetts et al., 2015). In this study, we evaluate the effectiveness of five-pathovar-specific primers in Xvm diagnostics. We hypothesize that the five-pathovar specific primers will succinctly and specifically identify Xvm DNA samples.
2. Materials and methods
2.1. Identification of Xvm-specific genes
We aligned all available genomic sequences from 26 closely related genomes against the reference genome of Xvm isolate NCPPB 4379 (Wasukira et al., 2012) and identified 19 genes (Supplementary Table 1) that were present in all the 10 Xvm genomes but absent from the 17 closely related non-Xvm genomes, which included GenBank accessions GCA_002191955.1, GCA_002191965.1, GCA_000159795.2, GCA_000772705.2, GCA_000772775.2, GCA_000772715.1, GCA_000772695.1, GCA_000772785.1, GCA_000772725.1, GCA_000772795.1, GCA_000774005.1, GCA_000770355.1, GCA_000277995.1, GCA_000278015.1, GCA_000278035.1, GCA_000278055.1 and GCA_000278075.1.
From the 19 candidates we chose five genes, encoding two predicted avirulence proteins (KFA14425.1 and KFA05711.1), FIS transcriptional regulator, histone-like nucleoid structuring protein, and the XRE (Xenobiotic response element) family transcriptional regulator (Table 1). These were prioritized because they belong to phylogenetically informative COGs (Comas et al., 2006). BLASTP searches against RefSeq (NCBI) and PKGDB (Genoscope) revealed that KFA14425.1 is an ortholog of the XopJ5 Type III Effector (T3E) described in X. citri pv. fuscans (Genbank ID: ATB59468.1) (Bansal et al., 2017; (Bansal et al., 2017; Kremer et al., 2017), while KFA05711.1 matched the C-terminal part of the T3E AvrXrV, also named XopJ3, with homologues detectable at the amino-acid sequence level in several Xanthomonas but not X. vasicola.
Table 1.
Polymerase chain reaction primers used to distinguish Xanthomonas campestris pv. musacearum isolates.
| Gene | Primer name | Sequence | Target sequence RefSeq Accession number and coordinates | Expected amplicon size (bp) |
|---|---|---|---|---|
| KFA14425.1 Avirulence protein | AvP1 - F | ACGTCGTATGCCGGAAGAAGCT |
KB372850.1: 46840–47631 |
500 |
| AvP1 - R | TCACATCCACCCCACTCTCGAG | |||
| KFA05711.1 Avirulence protein | AvP2 - F | TCAGGATTCTAAGGCGTGACGGA |
KB372883.1: 21500–21994 |
495 |
| AvP2 - R | ATGCCTGGTTTCGTGAAAATGAGAGAA | |||
| KFA11440.1 FIS family transcriptional regulator | FTR - F | TGCCCGTCCACGTTTCTTGG |
KB372863.1: 7776–8144 |
280 |
| FTR - R | TCAATTGCCTCCGCCAAAGCC | |||
| KFA10484.1 Histone-like nucleoid-structuring protein | HNS - F | TCGGCTGGCCTCATCAAGCA |
KB372866.1: 3030–3434 |
364 |
| HNS - R | GGGCAGGAAGGAAACCGAGGAA | |||
| KFA11135.1 XRE family transcriptional regulator | XFTR - F | TGTGGGACGCGATCGAAGAGA |
KB372863.1: 134541–134822 |
252 |
| XFTR - R | CCGCTTCCAGCACTCGCATT |
2.2. DNA extraction
For all the bacterial strains used in this study, total DNA was extracted using the protocol described by Mahuku (2004). Briefly, a loopful of 3-day-old bacterial cells were harvested and washed twice in 500 μL of 1M NaCl in Eppendorf tubes to reduce and separate the bacterial cells from the polysaccharide xanthan gum. The bacterial cells were washed twice with sterile distilled water to reduce salt concentration. The bacterial cell pellets were suspended in 500 μL of pre-warmed (55 °C) TES extraction buffer (0.2 M Tris-HCl, pH 8; 10 mM EDTA, pH 8; 0.5 M NaCl; 1% SDS) containing proteinase K (50 μgmL−1); vortexed for 30 s and incubated at 65 °C for 15 min. One-half volume (250 μL) of 7.5M ammonium acetate was added, gently mixed and the samples left to stand for 10 min at room temperature. Tubes were centrifuged at 13 000 rpm for 15 min and 500 μL of the supernatant transferred to a fresh tube. The DNA was precipitated by adding an equal volume (500 μL) of ice-cold isopropanol, gently mixing and incubating in a refrigerator at -20 °C overnight. Tubes were centrifuged at 15 000 rpm and 4 °C for 10 min and the DNA pellet was washed with 800 μL of cold 70% ethanol. The DNA pellet was dried by inverting tubes on clean paper towels for 30 min at room temperature. The DNA pellet was re-suspended in 100 μL of nuclease-free water. Integrity of DNA was determined using the NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific Inc., Pittsburgh, PA), adjusted to 10 ngμL−1, and stored at -20 °C until use.
2.3. Primer design and PCR amplification
PCR primers, targeting 250–500bp within each of these five gene sequences were designed using Geneious 10 (Kearse et al., 2012). Each PCR mix contained 1 μL of 2mM MgCl2, 5 μL 5X Go Taq buffer, 0.5 μL of 10 mM (each) dNTPs, 0.25 μL of 5U Go Taq, 1.5 μL of 10X primer Mix (corresponding to 3 pmol of each primer), 2μL DNA template (10 ng μL-1), in a total of 15 μL per reaction. The simplex PCR cycles consisted of (i) initial denaturation step at 95 °C for 10 min; (ii) 25 cycles of denaturation at 94 °C for 30s, annealing at 60 °C for 90s, elongation at 72 °C for 90 s; then (iii) a final extension step at 60 °C for 30 min. The amplicons were separated by electrophoresis in a 2% agarose gel in 0.5X TBE buffer at 100 V for 45 min. Gels stained with ethidium bromide were visualized and images captured with the Vilber UV Transilluminator (www.vilber.com).
2.4. Primer specificity
The specificity of these five primer pairs was tested on a collection containing 20 reference Xvm strains (20 NCPPB-referenced Xvm strains), other pathovars of X. vasicola (Xanthomonas vasicola pv. holcicola (Xvh), Xanthomonas vasicola pv. vasculorum (Xvh)), and closely related Xanthomonas species, Xanthomonas campestris pv. cannabis (Xcc), and Xanthomonas oryzae pv. oryzae (Xoo). We further tested the five new Xvm-specific primers on a collection of 142 bacterial isolates phenotypically similar to Xvm on YPGA growth media (Mwangi et al., 2007) and Wilbrink medium (Sands et al., 1986).
3. Results
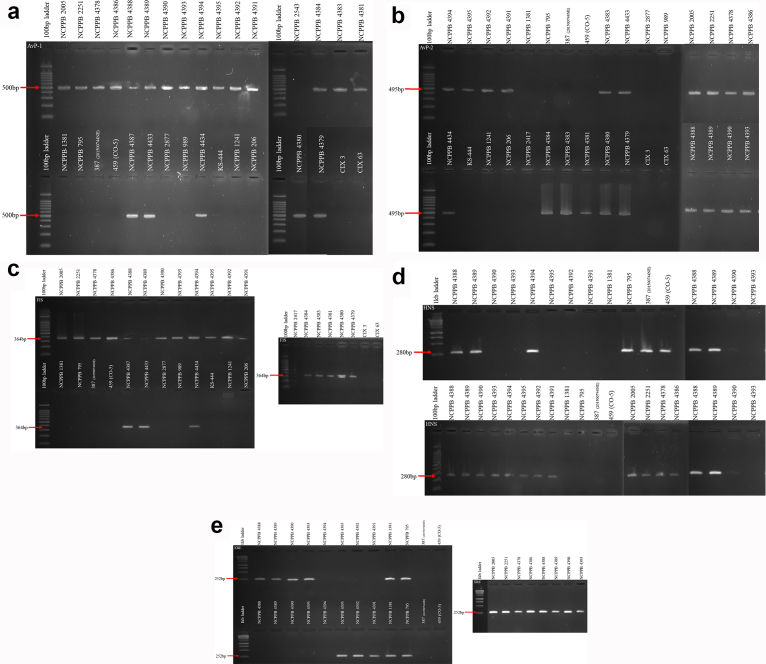
Fragments of the expected size were successfully amplified from all 20 reference Xvm strains, while no amplification was obtained from any of the 12 related Xanthomonas species DNAs (Table 2) (Fig. 1a–e). Considering the second collection, all 142 strains tested positive with GspDm primers. However, using the new primers 107 strains were confirmed to be Xvm (positive with all five primer pairs) while strain BCC280 had positive amplification for only four of the primers and none for the primers amplifying the FIS family transcriptional regulator gene (Supplementary Table 2). BCC280 was further confirmed to be Xvm using whole-genome sequencing. The remaining 34 bacterial strains were negative for all five primer pairs (Supplementary Table 2).
Table 2.
PCR amplification of the Xanthomonas species amplified by Xanthomonas vasicola pv. musacearum specific primers.
| Isolate name | Host | Country of origin | Species and pathovar | Sourcea | Amplification byb |
||||
|---|---|---|---|---|---|---|---|---|---|
| AvP-1 | AvP-2 | FTR | HNS | XFTR | |||||
| NCPPB 2005 | Enset | Ethiopia | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 2251 | Banana | Ethiopia | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4378 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4386 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4388 | Banana | DRC | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4389 | Banana | Rwanda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4390 | Banana | Rwanda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4393 | Banana | Tanzania | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4394 | Banana | Tanzania | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4395 | Banana | Tanzania | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4392 | Banana | Tanzania | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4391 | Banana | Rwanda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 1381 | Sugarcane | Zimbabwe | Xanthomonas vasicola pv. vasculorum | D. Studholme, Exeter | − | − | − | − | − |
| CFBP 5830 | Sugarcane | Madagascar | Xanthomonas vasicola pv. vasculorum | CFBP | − | − | − | − | − |
| 201500744NE | Maize | USA | Xanthomonas vasicola pv. vasculorum | J. Lang, CSU | − | − | − | − | − |
| CO-5 | Maize | USA | Xanthomonas vasicola pv. vasculorum | J. Lang, CSU | − | − | − | − | − |
| NCPPB 4387 | Banana | DRC | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4433 | Banana | Burundi | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 2877 | Cannabis | Romania | Xanthomonas campestris pv. cannabis | NCPPB | − | − | − | − | − |
| NCPPB 989 | Holcus sp | USA | Xanthomonas vasicola pv. holcicola | NCPPB | − | − | − | − | − |
| NCPPB 4434 | Banana | Kenya | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| KS-444 | Maize | USA | Xanthomonas vasicola pv. vasculorum | J. Lang, CSU | − | − | − | − | − |
| NCPPB 1241 | Sorghum | Australia | Xanthomonas vasicola pv. holcicola | D.Studholme, Exeter | − | − | − | − | − |
| NCPPB 206 | Maize | South Africa | Xanthomonas vasicola pv. vasculorum | NCPPB | − | − | − | − | − |
| NCPPB 2417 | Sorghum | Newzealand | Xanthomonas vasicola pv. holcicola | NCPPB | − | − | − | − | − |
| NCPPB 4384 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4383 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4381 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4380 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| NCPPB 4397 | Banana | Uganda | Xanthomonas vasicola pv. musacearum | NCPPB | + | + | + | + | + |
| CIX 3 = CFBP2532T | Rice | India | Xanthomonas oryzae pv. oryzae | IPME | − | − | − | − | − |
| CIX 63 = MAI55 | Rice | Mali | Xanthomonas oryzae pv. oryzae | IPME | − | − | − | − | − |
NCPPB: National Collection of Plant Pathogenic Bacteria; CFBP: Collection Franҫaise de Bactéries Phytopathogènes.
AvP - Avirulence protein; FTR - FIS transcriptional regulator; HNS - Histone-like nucleoid structuring protein; XFTR - XRE family transcriptional regulator.
Fig. 1.
a). Gel documentation results for the Avirulence protein (KFA14425.1) - Xvm specific primers on 32 Xanthomonas species to include Xanthomonas vasicola pv. musacearum (NCPPB 2005, 2251, 4378, 4379, 4380, 4381, 4383, 4384, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4433 and 4434); Xanthomonas vasicola pv. vasculorum (NCPPB (206, 795 and1381), 201500744NE, CO-5, KS-444); Xanthomonas vasicola pv. holcicola (NCPPB 989, 1241 and 2417); Xanthomonas oryzae pv. oryzae (CIX3 and 63) and Xanthomonas campestris pv. cannabis (NCPPB 2877). (b). Gel documentation results for the Avirulence protein (KFA05711.1) - Xvm specific primers on 32 Xanthomonas species to include Xanthomonas vasicola pv. musacearum (NCPPB 2005, 2251, 4378, 4379, 4380, 4381, 4383, 4384, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4433 and 4434); Xanthomonas vasicola pv. vasculorum (NCPPB (206, 795 and1381), 201500744NE, CO-5, KS-444); Xanthomonas vasicola pv. holcicola (NCPPB 989, 1241 and 2417); Xanthomonas oryzae pv. oryzae (CIX3 and 63) and Xanthomonas campestris pv. cannabis (NCPPB 2877). (c). Gel documentation results for the FIS transcriptional regulator - Xvm specific primers on 32 Xanthomonas species to include Xanthomonas vasicola pv. musacearum (NCPPB 2005, 2251, 4378, 4379, 4380, 4381, 4383, 4384, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4433 and 4434); Xanthomonas vasicola pv. vasculorum (NCPPB (206, 795 and1381), 201500744NE, CO-5, KS-444); Xanthomonas vasicola pv. holcicola (NCPPB 989, 1241 and 2417); Xanthomonas oryzae pv. oryzae (CIX3 and 63) and Xanthomonas campestris pv. cannabis (NCPPB 2877). (d). Gel documentation results for the histone-like nucleoid structuring protein - Xvm specific primers on 32 Xanthomonas species to include Xanthomonas vasicola pv. musacearum (NCPPB 2005, 2251, 4378, 4379, 4380, 4381, 4383, 4384, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4433 and 4434); Xanthomonas vasicola pv. vasculorum (NCPPB (206, 795 and1381), 201500744NE, CO-5, KS-444); Xanthomonas vasicola pv. holcicola (NCPPB 989, 1241 and 2417); Xanthomonas oryzae pv. oryzae (CIX3 and 63) and Xanthomonas campestris pv. cannabis (NCPPB 2877). (e). Gel documentation results for the Xenobiotic response element - Xvm specific primers on 32 Xanthomonas species to include Xanthomonas vasicola pv. musacearum (NCPPB 2005, 2251, 4378, 4379, 4380, 4381, 4383, 4384, 4386, 4387, 4388, 4389, 4390, 4391, 4392, 4393, 4394, 4395, 4433 and 4434); Xanthomonas vasicola pv. vasculorum (NCPPB (206, 795 and1381), 201500744NE, CO-5, KS-444); Xanthomonas vasicola pv. holcicola (NCPPB 989, 1241 and 2417); Xanthomonas oryzae pv. oryzae (CIX3 and 63) and Xanthomonas campestris pv. cannabis (NCPPB 2877).
4. Discussion
Effective management of plant diseases requires the use of reliable and specific diagnostic tools to detect the pathogens causing biotic stress to the plants. Previous tools that were developed to identify Xvm were discovered to amplify other X. vasicola species, and thus were not specific to Xvm. Based on X. vasicola genomic resources, we developed five new markers and we demonstrated their high specificity within the X. vasicola species. Our results further highlight the potential to use these new markers in Xvm diagnostic studies and during field collection of isolates. Although only amplified by four of the five primers, strain BCC280 was confirmed to be Xvm using whole genome sequencing. The inability for the FIS primers to amplify strain BCC280 could possibly imply presence of a SNP within the FIS region of this strain. In fact, these markers can be multiplexed based on the product expected band size. In conclusion, these five new primers provide a tool that will precisely identify Xvm and can be used to compliment visual observation in the field and colonies that seemingly look like Xvm. These specific genes can easily be used in future development of Xvm detection methods in plant tissues, soil, water, or insect tissues.
Declarations
Author contribution statement
Gloria Valentine Nakato: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Emmanuel Wicker, David J. Studholme: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Teresa A. Coutinho: Wrote the paper.
George Mahuku: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was a part of the BAXEPI project, supported by Agropolis Fondation under the reference ID 1605-025 through the « Investissements d'avenir » program (Labex Agro: ANR-10-LABX-0001-01), under the frame of I-SITE MUSE (ANR-16-IDEX-0006). Additional financial support came from the CRP-Roots Tubers and Banana.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Emmanuel Wicker, Email: wicker@cirad.fr.
David J. Studholme, Email: D.J.Studholme@exeter.ac.uk.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Adriko J., Aritua V., Mortensen C.N., Tushemereirwe W.K., Kubiriba J., Lund O.S. Multiplex PCR for specific and robust detection of Xanthomonas campestris pv. musacearum in pure culture and infected plant material. Plant Pathol. 2012;61:489–497. [Google Scholar]
- Aritua V., Parkinson N., Thwaites R., Heeney J.V., Jones D.R., Tushemereirwe W., Crozier J., Reeder R., Stead D.E., Smith J. Characterization of the Xanthomonas sp causing wilt of enset and banana and its proposed reclassification as a strain of X-vasicola. Plant Pathol. 2008;57:170–177. [Google Scholar]
- Bansal K., Midha S., Kumar S., Patil P.B. Ecological and evolutionary insights into Xanthomonas citri pathovar diversity. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.02993-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I., Moya A., Azad R.K., Lawrence J.G., Gonzalez-Candelas F. The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol. Biol. Evol. 2006;23:2049–2057. doi: 10.1093/molbev/msl075. [DOI] [PubMed] [Google Scholar]
- Hodgetts J., Hall J., Karamura G., Grant M., Studholme D.J., Boonham N., Karamura E., Smith J.J. Rapid, specific, simple, in-field detection of Xanthomonas campestris pathovar musacearum by loop-mediated isothermal amplification. J. Appl. Microbial. 2015;119:1651–1658. doi: 10.1111/jam.12959. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korus K., Lang J.M., Adesemoye A.O., Block C.C., Pal N., Leach J.E., Jackson-Ziems T.A. First report of Xanthomonas vasicola causing bacterial leaf streak on corn in the United States. Plant Dis. 2017;101:1030–1031. [Google Scholar]
- Kremer F.S., de Souza I.T., Guimaraes A.M., Moura A.B., Pinto L.S. In: Genome Sequence of a New Brazilian Strain of X. Fuscans. GenBank, editor. 2017. https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP023294.1?report=genbank [Google Scholar]
- Lang J.M., DuCharme E., Ibarra Caballero J., Luna E., Hartman T., Ortiz-Castro M., Korus K., Rascoe J., Jackson-Ziems T.A., Broders K., Leach J.E. Detection and characterization of Xanthomonas vasicola pv. vasculorum (Cobb 1894) comb. Nov. Causing bacterial leaf streak of corn in the United States. Phytopathology. 2017;107:1312–1321. doi: 10.1094/PHYTO-05-17-0168-R. [DOI] [PubMed] [Google Scholar]
- Mahuku G. A simple extraction method suitable for PCR based analysis of plant, fungal, and bacterial DNA. Plant Mol. Biol. Rep. 2004;22:71–81. [Google Scholar]
- Mwangi M., Mwebaze M., Bandyopadhyay R. Development of a semi-selective medium for the isolation of Xanthomonas campestris pv. musacearum from insect vectors, infected plant material and soil. Plant Pathol. 2007;56:383–390. [Google Scholar]
- Nakato V., Mahuku G., Coutinho T. Xanthomonas campestris pv. musacearum: a major constraint to banana, plantain and enset production in central and east Africa over the past decade. Mol. Plant Pathol. 2018;19:525–536. doi: 10.1111/mpp.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands D.C., Mizrak G., Hall V.N., Kim H.K., Bockelman H.E., Golden M.J. Seed transmitted bacterial diseases of cereals: epidemiology and control. Arab J. Plant Prot. 1986;4:117–125. https://asplantprotection.org/wp-content/uploads/2018/07/V4-2_Art10.pdf [Google Scholar]
- Studholme D.J., Kemen E., MacLean D., Schornack S., Aritua V., Thwaites R., Grant M., Smith J., Jones J.D. Genome-wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 2010;310:182–192. doi: 10.1111/j.1574-6968.2010.02065.x. [DOI] [PubMed] [Google Scholar]
- Wasukira A., Tayebwa J., Thwaites R., Paszkiewicz K., Aritua V., Kubiriba J., Smith J., Grant M., Studholme D.J. Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pathovar musacearum. Genes. 2012;3:361–377. doi: 10.3390/genes3030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasukira A., Coulter M., Al-Sowayeh N., Thwaites R., Paszkiewicz K., Kubiriba J., Smith J., Grant M., Studholme D.J. Genome Sequencing of Xanthomonas vasicola pathovar vasculorum reveals variation in plasmids and genes encoding lipopolysaccharide synthesis, Type-IV pilus and Type-III Secretion effectors. Pathogens. 2014;3:211–237. doi: 10.3390/pathogens3010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.