Abstract
Pseudomonas aeruginosa is one of the three most pathogenic bacteria that frequently cause life-threatening opportunistic human infections, pneumonia, and lower respiratory tract infections in immunocompromised hosts, particularly in the burns ward. The present study aimed to establish a loop-mediated isothermal amplification (LAMP) method for the rapid and sensitive detection of P. aeruginosa-specific gene hypothetical protein (GenBank ID: 882161). The gene was obtained through local and online BLAST, and specific primers were designed for this gene. Reaction conditions were optimized at 65°C for 30 min and 80°C for 2 min, whereas the reaction system contained 5.2 mM Mg2+, 8 U Bst 2.0 DNA polymerase, 1.4 mM deoxyribonucleotide and 0.2 and 1.6 µM of the outer and inner primers, respectively. The LAMP method was evaluated using 150 P. aeruginosa and 170 non-P. aeruginosa strains. Positive reactions were observed on 150 P. aeruginosa strains, whereas all non-P. aeruginosa strains exhibited negative results. Plasmids with the specific gene and mouse blood with P. aeruginosa were used for sensitivity assay. The detection limit of LAMP was 1 bacterium/reaction. Results indicated that the LAMP method targeted to hypothetical protein is a fast, specific, sensitive, inexpensive and suitable method for detection of P. aeruginosa.
Keywords: Pseudomonas aeruginosa, loop-mediated isothermal amplification, polymerase chain reaction, novel specific gene, rapid detection
Introduction
P. aeruginosa is a ubiquitous microorganism and one of the most frequent Gram-negative pathogens that cause human opportunistic infections in burn and other immunocompromised patients (1). These patients are the most susceptible group to infection due to their compromised natural defenses. Intensive care units, respiratory wards and burns units are the main locations where P. aeruginosa infections can spread (2). P. aeruginosa also causes nosocomial pneumonia and cystic fibrosis with high morbidity and mortality (3). Antimicrobial therapy is recommended in the Health Technical Memorandum 04–01 guidelines for the management of pneumonia (4). However, the treatment of pneumonia caused by P. aeruginosa has become increasingly difficult owing to the elevated prevalence of drug resistance and limited therapeutic options (5). Thus, rapid and accurate detection of P. aeruginosa and strict infection control is the most effective strategy in preventing P. aeruginosa infection in burn wards (6).
The main conventional methods for detection of P. aeruginosa include bacterial culture and immunological methods. However, these methods exhibit specific shortcomings, such as being time consuming or inaccurate (7). Biochemical test kits, such as API 20 NE, are commonly used for biochemical identification and exhibit a high oxidase-positive misidentification rate in Gram-negative bacilli, including P. aeruginosa (8). The polymerase chain reaction (PCR) techniques established by Mullis et al (9) specifically amplify the DNA fragment with thermally stable DNA polymerase; these techniques include single, multiplex, and quantitative PCR. PCR is a high-quality, fast, and efficient method that is often utilized to evaluate the specificity, sensitivity, and feasibility of other detection methods (10). However, this method is inconvenient for clinical testing given the requirement of isothermal cyclic amplification (11). The present study aimed to develop an efficient, time-saving, and inexpensive method for the detection of P. aeruginosa.
A novel gene amplification method called loop-mediated isothermal amplification (LAMP) was developed by Notomi et al (12) in 2000, and is possibly a feasible and cost-effective alternate scheme for molecular diagnosis. This technique is based on a set of specific primers with at least four primers to recognize six different regions of the genome and produces a stem-loop DNA with several inverted repeats and cauliflower-like structures with multiple loops (13). The results can be detected by fluorescence signal, turbidity, and the naked eye (14). In the present study, a novel specific gene of P. aeruginosa was obtained through bioinformatics analysis. An efficient and accurate LAMP method was built for the detection of P. aeruginosa and validated via PCR assay.
Materials and methods
Bacterial strains and culturing
All 150 tested P. aeruginosa strains and 170 non-P. aeruginosa strains, including Escherichia coli (ATCC25922), Klebsiella pneumoniae (ATCC700603), Staphylococcus aureus (ATCC25923), Staphylococcus epidermidis (ATCC12228), Enterococcus faecalis (ATCC29212), and Shiga bacillus (ATCC25931) were kindly provided by the First People's Hospital of Yunnan Province from their burns ward (Yunnan, China). The strains were grown in Luria-Bertani (LB) liquid medium (Biotopped Technology Co., Ltd., Taipei, Taiwan) in a shaking incubator (HZQ-F160 full temperature double layer oscillator-cultivating box; Suzhou Pei Ying Experimental Equipment Co., Ltd., Jiangsu, China) at 37°C and 180 rpm for 12 h. The bacterial genome was extracted using the TIANamp genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufacturer's protocol and then stored at −40°C for further experiments (15).
Selection of specific genes of P. aeruginosa
The formatted non-redundant nucleic acid database was downloaded from National Center for Biotechnology Information (NCBI; ftp://ftp.ncbi.nih.gov/blast/db) as the local database. Local BLAST sequencing in BLAST software (version 2.7.1+; http://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/) was performed using the P. aeruginosa genome (KI518973.1) as query sequence against this database. Potential specific genes were further analyzed by online BLAST (version 2.7.1+; http://blast.ncbi.nlm.nih.gov/Blast.cgi) with the database excluding or including the sequences of P. aeruginosa. The specific gene of P. aeruginosa was required to be highly conserved among the P. aeruginosa strains and show no significant similarity to other species for inclusion.
Initially, the genomic sequence of P. aeruginosa and the non-redundant nucleic acid database of NCBI were downloaded to the local server. Then, the potential specific genes of P. aeruginosa were screened by sequence similarity alignment. Online BLAST was used to further identify the screened potential specific genes due to the slow update of the local database. Two-step strategies using interspecies-specific and intraspecies commonality were used to identify specific genes by online BLAST. The first step excluded P. aeruginosa during the alignment; the gene may be considered a possible target gene if the alignment result is different. The second included P. aeruginosa during alignment, with the highly conserved genes considered as possible target genes. The retrieval range was limited to the species with known sequences except for P. aeruginosa. The specific gene of P. aeruginosa can only be considered when the alignment differs from those of other species, is similar to those of a few species or features a very low similarity.
Primer design and reaction
Four oligonucleotide primers targeting the specific gene identified using BLAST, hypothetical protein gene (GenBank ID 882161), were designed using Primer Explorer version 5 software (http://primerexplorer.jp/lampv5/index.html) for the LAMP assay. The outer primers F3 and B3 were used as the forward and reverse primer in the PCR assays, respectively. Table I presents all the primers used in the present study. PCR was performed using 2X Tsingke Master Mix, which was purchased from Tsingke Biotech Co., Ltd (Kunming, China). According to the manufacturer's protocol, the PCR reaction system containing 12.5 µl 2X Tsingke Master Mix, 1.0 µl primers (10 µM) and 1.0 µg DNA template from each strain was added with nuclease-free water up to 25 µl volume. The reactions were performed in a GeneAmp PCR System 9700 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with the following amplification conditions: Pre-denaturation at 95°C for 5 min, followed by 32 cycles of denaturation at 95°C for 30 sec, annealing at 57°C for 30 sec, extension at 72°C for 30 sec and a final extension at 72°C for 7 min. The PCR products were verified using gel electrophoresis on a 2% agarose gel and stained with GelStain (Beijing Transgen Biotech Co., Ltd., Beijing, China). LAMP assay reactions were optimized at 65°C for 30 min and 80°C for 2 min. The system contained 2.5 µl 10X Bio Labs buffer, 8.0 U Bst 2.0 DNA polymerase [both New England Biolabs (Beijing) Ltd., Beijing, China], 5.2 mM Mg2+, 1.4 mM deoxyribonucleotide, 0.2 µM of the outer primer and 1.6 µM of the inner primer, with nuclease-free water up to 25 µl volume. SYBR-Green I (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), a nuclear dye, was added into the LAMP reaction tubes for fluorescence visualization of the LAMP products.
Table I.
Loop-mediated isothermal amplification primers for hypothetical protein gene.
| Primer | Sequence (5′-3′) | Size (bp) |
|---|---|---|
| F3 | CAAGCGCAAGATAGTCGCC | 19 |
| B3 | TCCGCTTGAACAGGCTGGTG | 20 |
| FIP | GAAGATATCCGGCTGGTTGCTTTTCAAGAGGGAATGCCGCAGT | 43 |
| BIP | AACGGATCATCGGCATCCTGGTTTTCATCGCCGTCCACAGGTAGA | 45 |
Construction and verification of positive plasmids
The positive plasmids were constructed via the following steps. The DNA fragment of target genes was obtained by PCR and the genome DNA of P. aeruginosa as a template. The target fragment was then inserted into the pMD 19-T simple vector and transformed into E. coli JM109 cells using the E. coli JM109 Competent Cell kit [cat. no. 9052; both Takara Biomedical Technology (Beijing) Co., Ltd., Beijing, China] at 0°C for 30 min, 42°C for 45 sec and 0°C for 2 min, according to the manufacturer's protocol. The 37°C preheated blank LB medium was added into the transform system culture at 37°C and 180 rpm for 1 h. Positive clones were selected from the cultured bacterial solution in the LB solid medium (Biotopped Technology Co., Ltd., Taipei, Taiwan) by spreading the bacteria around the plate, which was cultured at 37°C for overnight. Then, following an overnight culture at 37°C, the plasmid were extracted from the cells using TIANprep Mini Plasmid kit (Tiangen Biotech Co., Ltd.), according to the manufacturer's protocol and verified by sequencing. Finally, the number of copies of recombinant plasmids was calculated by the deduced polynomial model as follows:
where C denotes the copy of the recombinant plasmid, X and Y represent the concentration of the recombinant plasmid and the number of base pairs in the target fragment, respectively, NA is Avogadro's constant, 2692 is the number of base pairs in the vector, and 660 is the mean molecular weight of 1 base pair.
As described above, we constructed positive plasmids with the hypothetical protein gene. The plasmids were transformed into JM109 competent cells. The positive clones were used for bacterial liquid PCR. The sequence alignment of the result was analyzed in NCBI following sequencing and the sequence correctness was verified. The positive clones were then cultured in LB liquid medium at 37°C and 180 rpm for 12 h prior to the extraction of positive plasmids by TIANprep Mini Plasmid kit (Tiangen Biotech Co., Ltd.), according to the manufacturer's protocol. The plasmids were verified by gel electrophoresis on 2% agarose gels and stained with GelStain, and then stored at −20°C for further use.
Specificity of PCR and LAMP reactions
A total of 150 clinical P. aeruginosa strains and 170 non-P. aeruginosa strains, including 24 E. coli, 32 K. pneumonia, 37 S. aureus, 15 S. epidermidis, 17 E. faecalis, and 45 S. bacillus, were tested in this study to evaluate the specificity of PCR and LAMP reactions. The PCR test was used as the gold standard in preliminary experiments for the specificity test prior to the LAMP test. All experiments were repeated twice.
Sensitivity of PCR and LAMP reactions
The sensitivity of PCR and LAMP reactions were evaluated using two different templates, the serially diluted 10-fold positive plasmids (108−10° copies) and blood sample mimicking infection. The counted P. aeruginosa strain was serially diluted by 10-fold in PBS and then mixed with blood from 3 healthy specific pathogen free female Kunming mice (Laboratory Animal Center of Kunming Medical University, Kunming, Yunnan; weight, 22–25 g; age, 5 weeks) at 1:1 proportion to mimic infection. The mice were housed in the animal experiment center of Kunming University of Science and Technology (Kunming, China) at 25°C with a 12/12 h light/dark cycle and access to food and water ad libitum. The blood sample was lysed using a solution containing 125 mM NaOH, 1 mM EDTA and 0.1% Tween-20, and then a solution containing 125 mM HCl and 10 mM Tris-HCl, and the suspension was used as the template for PCR and LAMP assays. All experiments were repeated six times.
Results
Screening the specific gene and primer design
Following the preliminary screening of possible specific genes by local BLAST, 1,599 potential specific genes were obtained. These genes were then used in the second screening by online BLAST. Conclusively, 5 potential specific genes were attained, and they met the criterion regarding interspecific specificity and intraspecies universality. In the primer design step, four primers were designed for the six regions of the target gene in LAMP, and the primers were confirmed by primer BLAST and PCR assay. Considering the above selection criteria, the hypothetical protein gene was finally identified as the only one that can be used in the detection of P. aeruginosa.
Specificity of PCR and LAMP reactions
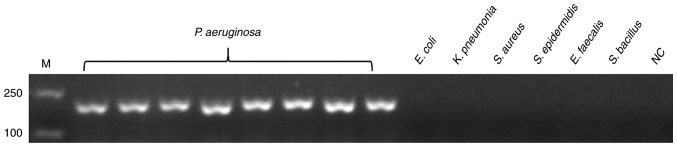
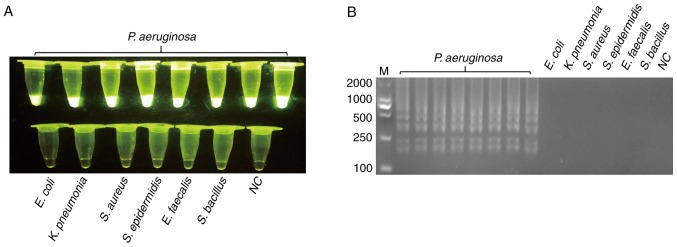
The genomic DNA of P. aeruginosa and non-P. aeruginosa were extracted as templates for the specificity test of PCR and LAMP, as detailed above. Prior to LAMP reaction, the PCR test was used for the specificity test, and all 170 non-P. aeruginosa strains were negative. Fig. 1 presents the representative results. Following validation by PCR, LAMP was used to test specificity. SYBR-Green I was added to the LAMP reaction tubes for fluorescence visualization. Fig. 2 presented the results of LAMP; the findings of which were consistent with PCR.
Figure 1.
Specificity of the PCR assay for detecting the target gene of hypothetical protein gene. Genomic DNA of P. aeruginosa was used as the template for PCR in lanes 1–8, the template in lanes 9–14 were ordinal of control strains, E. coli, K. pneumonia, S. aureus, S. epidermidis, E. faecalis and S. bacillus, whereas lane 15 is NC. All experiments were repeated twice. PCR, polymerase chain reaction; NC, negative control; M, marker.
Figure 2.
Specificity of the loop-mediated isothermal amplification assay for detecting the target gene of hypothetical protein gene. (A) The template of tubes 1–8 were genomic DNA of P. aeruginosa; tubes 9–14 were ordinal of control strains, E. coli, K. pneumonia, S. aureus, S. epidermidis, E. faecalis and S. bacillus, whereas 15 is NC. (B) The agarose gel electrophoresis figure of (A). All experiments were repeated twice. NC, negative control; M, marker.
Sensitivity of PCR and LAMP reactions
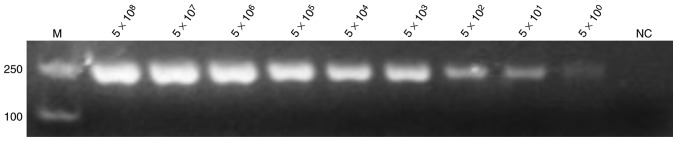
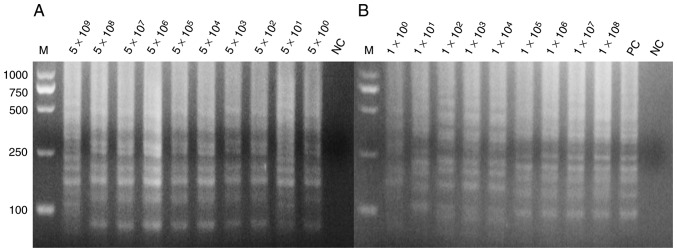
The positive plasmids and bacterial solution were serially diluted 10-fold based on the calculated copy number and plate counting, respectively. The positive plasmids were directly used as templates for PCR and LAMP assay. Figs. 3 and 4 demonstrate the extreme sensitivities, which can reach up to 10° copies per reaction, of PCR and LAMP assay. The bacterial solutions were mixed with the blood of mice with a volume ratio of 1:1. These mixtures were lysed by direct extraction reagent and then used for the LAMP assay. Fig. 4 indicates that the LAMP assay exhibits a high sensitivity that may reach up to 10° copies per reaction. SYBR-Green I into was also added to the LAMP reaction tubes for fluorescence visualization detection (Fig. 5).
Figure 3.
Sensitivity of the polymerase chain reaction assay for detection of the hypothetical protein gene. The positive plasmid was used as a template at concentrations of 5×108, 5×107, 5×106, 5×105, 5×104, 5×103, 5×102, 5×101 and 5×10° per reaction, with nuclease-free water used as template for NC. All experiments were repeated six times. NC, negative control; M, marker.
Figure 4.
Sensitivity of the loop-mediated isothermal amplification assay for detection of the hypothetical protein gene. (A) The positive plasmids as template at concentrations of 5×109, 5×108, 5×107, 5×106, 5×105, 5×104, 5×103, 5×102, 5×101 and 5×10° per reaction, with nuclease-free water as template in lane 1 for NC. (B) The blood sample being direct lysed as template at concentrations of 1×108, 1×107, 1×106, 1×105, 1×104, 1×103, 1×102, 1×101 and 1×10° per reaction the sample order from 1×10°−1×108, respectively, with positive plasmids as the template in lane 11 as PC, and nuclease-free water as the template in lane 12 as NC. All the experiments were repeated six times. PC, positive control; NC, negative control; M, marker.
Figure 5.
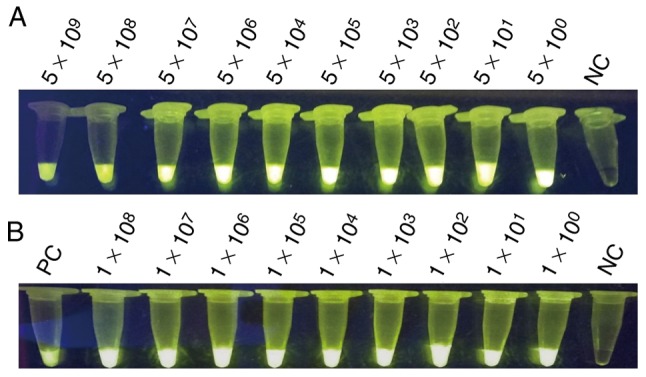
Visualization of LAMP. (A) Sensitivity of the LAMP assay for detecting the target gene of hypothetical protein gene; the templates were the positive plasmids, serially 10-fold diluted as 5×109, 5×108, 5×107, 5×106, 5×105, 5×104, 5×103, 5×102, 5×101 and 5×10° per reaction. (B) Sensitivity of the LAMP assay for detection of the hypothetical protein gene; the templates were mixed with mouse blood, serially 10-fold diluted as 1×108, 1×107, 1×106, 1×105, 1×104, 1×103, 1×102, 1×101 and 1×10° per reaction, respectively, with positive plasmids as the template as PC. LAMP, loop-mediated isothermal amplification; PC, positive control; NC, negative control.
Discussion
P. aeruginosa must be controlled in related infections, and an effective method for the detection must be evaluated and used. Various methods, including culturing and PCR-based amplification method, have been utilized for the detection of this pathogen (7). Conventional methods, such as culturing, are experience-dependent and time-consuming (7). In addition to diagnostic or detection application, PCR is useful for cloning and other molecular biology areas, but has not been broadly established in clinical set-up given the requirement for a high-precision thermal cycler, which cannot be afforded by low-level medical institutions (16).
LAMP is a novel DNA amplification technique (17). This method is widely used in various fields of biological science, including bacterial, viral and parasitic infections, and has the potential to be used as a simple detection assay due to its simplicity, ruggedness and low cost (18). In contrast to PCR technology, wherein a series of alternating temperature steps or cycles is carried out, the LAMP assay requires no thermal cycler and is carried out at a constant temperature of 60–65°C (19). Additionally, compared with the traditional method of isolation and identification and PCR assay (24–48 and 2–3 h, respectively), LAMP is rapid, simple, and convenient (8).
In the present study, a novel specific gene was identified, named hypothetical protein gene (GenBank ID: 882161), which can be used for identification of P. aeruginosa. Hypothetical protein gene was initially used as a target gene for the detection of P. aeruginosa. Hypothetical protein gene was highly conserved in P. aeruginosa, however functional studies have not yet been conducted. In the present study, four oligonucleotide primers were designed for LAMP assay, and the outer primers F3 and B3 were used as the forward and reverse primers, respectively, in PCR assays for the hypothetical protein gene detection; the reaction conditions were optimal (optimizing data not shown). The results demonstrated that the hypothetical protein gene is specific to P. aeruginosa. Therefore, the LAMP system was utilized to detect P. aeruginosa due to its simplicity, and rapid, easy detection, which may appropriately meet the demands in developing countries with insufficient facilities and health resources. Furthermore, it was attempted to use the LAMP assay to detect the antibiotics resistance gene of P. aeruginosa. However, due to the rapid mutation and multiple subtypes of the drug resistance gene, the primers for LAMP reaction cannot correctly match with the target fragment and thus lead to false negative amplification.
P. aeruginosa infection has become one of the most serious nosocomial infections (20). Thus, establishment of a fast and effective detection method is required urgently. The LAMP technology offers the opportunity to resolve such burden for its simplicity, rapidity, and high sensitivity. With the emergence of multidrug-resistant bacteria, the LAMP technology can also be applied for detecting resistant genes (21).
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from Yunnan Science and Technology Commission (grant nos. 2015BC001 and 2015DH010).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CL wrote the manuscript and performed experiments. YSh constructed the positive plasmids. GY performed the specificity of PCR assay experiments. X-SX performed the BLAST experiments and proofread the manuscript. XM collected the strains and performed the sensitivity of LAMP experiments. YF performed data analysis. YSo and A-MZ designed the study.
Ethics approval and consent to participate
The current study was approved by the ethics committee of Kunming University of Science and Technology.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3:32. doi: 10.1186/2047-2994-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sana TG, Berni B, Bleves S. The T6SSs of Pseudomonas aeruginosa strain PAO1 and their effectors: Beyond bacterial-cell targeting. Front Cell Infect Microbiol. 2016;6:61. doi: 10.3389/fcimb.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker J, Moore G. Pseudomonas aeruginosa in hospital water systems: Biofilms, guidelines, and practicalities. J Hosp Infect. 2015;89:324–327. doi: 10.1016/j.jhin.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Ding C, Yang Z, Wang J, Liu X, Cao Y, Pan Y, Han L, Zhan S. Prevalence of Pseudomonas aeruginosa and antimicrobial-resistant Pseudomonas aeruginosa in patients with pneumonia in mainland China: A systematic review and meta-analysis. Int J Infect Dis. 2016;49:119–128. doi: 10.1016/j.ijid.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa Acinetobacter baumannii Mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Jami Al-Ahmadi G. Ann Burns Fire Disasters. Vol. 29. Zahmatkesh Roodsari R; 2016. Fast and specific detection of Pseudomonas aeruginosa from other pseudomonas species by PCR; pp. 264–267. [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh R, Nagavardhini A, Sengupta A, Sharma M. Development of Loop-Mediated Isothermal Amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris-wilt pathogen of chickpea. BMC Res Notes. 2015;8:40. doi: 10.1186/s13104-015-0997-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. 1986. Biotechnology. 1992;24:17–27. [PubMed] [Google Scholar]
- 10.Duś I, Dobosz T, Manzin A, Loi G, Serra C, Radwan-Oczko M. Role of PCR in Helicobacter pylori diagnostics and research-new approaches for study of coccoid and spiral forms of the bacteria. Postepy Hig Med Dosw (Online) 2013;67:261–268. doi: 10.5604/17322693.1044005. [DOI] [PubMed] [Google Scholar]
- 11.Lee PL. DNA amplification in the field: Move over PCR, here comes LAMP. Mol Ecol Resour. 2017;17:138–141. doi: 10.1111/1755-0998.12548. [DOI] [PubMed] [Google Scholar]
- 12.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Agarwal RK, Manjunathachar HV, Dhama K. Loop-mediated isothermal amplification (LAMP) test for specific and rapid detection of Brucella abortus in cattle. Vet Q. 2014;34:174–179. doi: 10.1080/01652176.2014.966172. [DOI] [PubMed] [Google Scholar]
- 14.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 15.Brewster JD, Paoli GC. DNA extraction protocol for rapid PCR detection of pathogenic bacteria. Anal Biochem. 2013;442:107–109. doi: 10.1016/j.ab.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Seki M, Yamashita Y, Torigoe H, Tsuda H, Sato S, Maeno M. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J Clin Microbiol. 2005;43:1581–1586. doi: 10.1128/JCM.43.4.1581-1586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray L, Edwards L, Tuppurainen ES, Bachanek-Bankowska K, Oura CA, Mioulet V, King DP. Detection of capripoxvirus DNA using a novel loop-mediated isothermal amplification assay. BMC Vet Res. 2013;9:90. doi: 10.1186/1746-6148-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtsuka K, Yanagawa K, Takatori K, Hara-Kudo Y. Detection of Salmonella enterica in naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl Environ Microbiol. 2005;71:6730–6735. doi: 10.1128/AEM.71.11.6730-6735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhai S, Liu C, Zhang Q, Tao C, Liu B. Detection of two exogenous genes in transgenic cattle by loop-mediated isothermal amplification. Transgenic Res. 2012;21:1367–1373. doi: 10.1007/s11248-012-9614-2. [DOI] [PubMed] [Google Scholar]
- 20.Poole K. Pseudomonas aeruginosa Resistance to the max. Front Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng J, Tang S, Liu L, Kuang X, Wang X, Hu S, You S. Development of a loop-mediated isothermal amplification (LAMP) assay for rapid and specific detection of common genetically modified organisms (GMOs) Int J Food Sci Nutr. 2015;66:186–196. doi: 10.3109/09637486.2014.979318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.