Abstract
The long non-coding RNA X inactive-specific transcript (XIST) has been implicated in certain human cancers, including osteosarcoma (OS), but the molecular mechanism of XIST underlying OS progression remains to be fully uncovered. In the present study, reverse transcription-quantitative polymerase chain reaction data demonstrated that XIST was significantly upregulated in OS tissues and cell lines (Saos-2, U2OS, HOS and MG63) compared with adjacent non-tumour tissues and normal human osteoblast cell line HFOB1.19. Bioinformatics analysis and luciferase reporter gene assay data demonstrated that XIST could directly target microRNA (miR)-137 and negatively regulate the expression of miR-137 in Saos-2 and U2OS cells. Furthermore, miR-137 was markedly downregulated in OS tissues and cell lines. An inverse association between XIST and miR-137 expression was observed in OS tissues. Knockdown of XIST caused a significant reduction in cell proliferation and invasion and suppressed matrix metalloproteinase (MMP2) and MMP9 protein levels in Saos-2 and U2OS cells. Furthermore, inhibition of miR-137 expression abolished the effects of XIST downregulation on the proliferation and invasion of OS cells. In summary, the present study suggests that XIST promotes OS cell proliferation and invasion by inhibition of miR-137 expression. Thus, XIST may be a potential therapeutic target for the treatment of OS.
Keywords: X inactive-specific transcript, microRNA-137, osteosarcoma, proliferation, invasion
Introduction
Osteosarcoma (OS) is the most prevalent malignant bone tumour in children and adolescents, and it generally occurs in the metaphysis of long bones (1,2). Furthermore, OS is characterized by high relapse, and ~80% of OS patients eventually develop metastatic disease (1,3). Despite the combination of surgery, radiotherapy and chemotherapy, the 5-year survival of advanced OS patients is still poor (1,2). Therefore, it is urgently required to explore the potential molecular mechanism underlying OS progression and to identify effective molecular targets for the treatment of OS.
Long non-coding RNAs (lncRNAs) are a type of regulatory transcript >200 nucleotides in length that mainly function through sponging target mRNAs, microRNAs (miRNAs or miRs) or proteins (4,5). miRNAs are a class of regulatory transcripts containing 22–25 nucleotides and are important gene expression regulators through their interaction with the 3′-untranslated region of their target mRNAs, resulting in mRNA degradation or translation repression (6,7). By regulating the expression of their targets, both lncRNAs and miRNAs have been demonstrated to serve key roles in the development and progression of various human cancers including OS (7–9). For instance, miR-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor 1-α (7). The lncRNA metastasis associated lung adenocarcinoma transcript 1 promotes the proliferation and metastasis of osteosarcoma cells via downregulation of miR-509 and, thus, activation of the Rac1/c-Jun N-terminal kinasepathway (8).
The lncRNA X inactive-specific transcript (XIST) is expressed exclusively from the X inactivation centre of the inactive X chromosome and is essential for the initiation and spread of X-inactivation (10). Previous studies have reported that XIST is frequently upregulated in human cancers, and it functions as an oncogene (11,12). For instance, XIST promotes cell growth and metastasis through regulating the miR-139-5p mediated Wnt/β-catenin signalling pathway in bladder cancer (13). Li et al previously reported that XIST promotes transforming growth factor (TGF)-β-induced epithelial-mesenchymal transition by regulating the miR-367/141-zinc finger E-box binding homeobox 2 axis in non-small-cell lung cancer (14). Recently, XIST has been demonstrated to be upregulated in OS tissues and cell lines, and its upregulation is significantly associated with advanced tumour size, advanced clinical stage, distant metastasis and shorter survival time of OS patients (15). In addition, XIST expression has been demonstrated to be an independent prognostic factor for the survival of patients with OS (15). Furthermore, several miRNAs have been identified to be sponged by XIST in OS cells, including miR-320b (16), miR-193a-3p (17), miR-195-5p (18) and miR-21-5p (19). For example, Lv et al demonstrated that XIST promoted OS progression by targeting RAP2B via miR-320b (16). Yang et al reported that XIST enhanced OS growth by targeting YAP via miR-195-5p (18). However, whether XIST also interacts with other miRNAs in OS cells remains unclear.
The aim of the present study was to explore the molecular mechanism by which XIST promotes OS growth and metastasis in vitro.
Materials and methods
Clinical tissues
The present study was approved by the Medical Ethics Committee of First Affiliated Hospital of Hunan College of Traditional Chinese Medicine (Zhuzhou, China). OS tissues and matched adjacent non-tumour tissues were collected from 35 patients with primary OS at the Department of Orthopaedics, First Affiliated Hospital of Hunan College of Traditional Chinese Medicine between April 2014 and March 2017. Written informed consent was obtained from all participating patients. These patients included 15 females and 20 males, aged from 11–33 years with a mean age of 18.5 years. The exclusion criterion was that no patient received radiotherapy or chemotherapy prior to surgical resection. These tissues were immediately frozen and stored in liquid nitrogen prior to use.
Cell culture
Human OS cell lines (Saos-2, U2OS, HOS and MG63) and normal human osteoblast cell line HFOB1.19 were purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% foetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). Cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection
For XIST and miR-137 function analysis, Saos-2 and U2OS cells were transiently transfected with 100 µM of negative control (NC) small interfering (si)RNA (4390843; Thermo Fisher Scientific, Inc.), 100 µM of XIST siRNA (4390771; Thermo Fisher Scientific, Inc.), 4 ug pcDNA3.1 vector (V79020; Thermo Fisher Scientific, Inc.), 4 ug of pcDNA-XIST expression plasmid (P00238; Amspring, Changsha, China), 100 µM of miR-137 inhibitor (4464084; Thermo Fisher Scientific, Inc.) or 100 µM of NC inhibitor (4464076; Thermo Fisher Scientific, Inc.) using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following 48 h of transfection, the expression levels of XIST or miR-137 were examined to confirm the transfection efficiency.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Inc.). For XIST expression detection, RT-qPCR was conducted using the OneStep RT-PCR kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. GAPDH was used as an internal reference. For miR-137 expression detection, RT-qPCR was conducted using the Mir-X™ miRNA qRT-PCR SYBR® kit (Clontech Laboratories, Inc., Mountainview, CA, USA) according to the manufacturer's instructions. U6 was used as an internal reference. The reaction conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 30 sec. The relative expression was analysed using the 2−ΔΔCq method (20). Primers used were as follows: XIST, forward 5′-ACGCTGCATGTGTCCTTAG-3′ and reverse 5′-GAGCCTCTTATAGCTGTTTG-3′; GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′; miR-137, forward 5′-GCAGCAAGAGTTCTGGTGGC-3′ and reverse 5′-TGGAACCAGTGCGAAAACAC-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Western blotting
Saos-2 and U2OS cells were lysed in cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.). The protein concentration was examined using a Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific, Inc.). Proteins (50 µg per lane) were separated by 10% SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific, Inc.). The membranes were blocked in 5% non-fat milk in PBS (Thermo Fisher Scientific, Inc.) containing 0.1% Tween-20 (Thermo Fisher Scientific, Inc.) at room temperature for 3 h. Following that, the membranes were incubated with rabbit anti-matrix metalloproteinase (MMP)2 (1:1,000; ab92536; Abcam, Cambridge, MA, USA), rabbit anti-MMP9 (1:500; ab76003; Abcam) or rabbit anti-GAPDH (1:1,000; ab9485; Abcam) primary antibodies at 4°C overnight. Then, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; ab205718; Abcam) at room temperature for 30 min. The protein bands were detected using an Enhanced Chemiluminescence Western Blotting kit (Thermo Fisher Scientific, Inc.) and quantified using Image Lab analysis software version 3.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell counting kit (CCK)-8 assay
Saos-2 and U2OS cells (5,000 cells/well) were seeded into 96-well plates. Following incubation at 37°C for 0, 24, 48 or 72 h, 10 ml CCK-8 reagent (Beyotime Institute of Biotechnology, Haimen, China) was added to each well. The cells were then incubated at 37°C for 2 h. The absorbance was examined at a wavelength of 450 nm using an ELx808 absorbance reader (BioTek Instruments, Inc., Winooski, VT, USA).
Cell invasion assay
The Saos-2 and U2OS cell suspensions (1×105 cells per well) were added to the upper chamber of a Transwell plate that was pre-coated with Matrigel (Chemicon; EMD Millipore, Billerica, MA, USA), and 300 µl DMEM containing 10% FBS was added to the lower chamber. Cells were then incubated at 37°C for 24 h. Following that, the cells on the interior of the inserts were removed using a cotton-tipped swab, while the cells on the lower surface of the membrane were stained with gentian violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at room temperature for 30 min. The invading cells were counted under a light microscope (magnification, ×200; Olympus Corp., Tokyo, Japan).
Bioinformatics analysis and Luciferase reporter gene assay
The target miRNAs of XIST were predicted using RNAhybrid 2.12 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). The fragment from XIST containing the wild-type (WT) or mutant type (MT) miR-137 binding sites was cloned into pmirGLO Dual-luciferase Target Vector (Promega Corporation, Madison, WI, USA), generating WT or MT XIST plasmids. Saos-2 and U2OS cells were co-transfected with miR-137 mimic (4464066; Thermo Fisher Scientific, Inc.) or miRNA-NC (4464058; Thermo Fisher Scientific, Inc.), and WT (or MT) XIST plasmids using Lipofectamine 2000. Following 48 h of transfection, luciferase reporter gene assay was performed using the Dual-Luciferase Reporter Assay System (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation of at least three repeated experiments. SPSS version 20.0 software (IBM Corp., Armonk, NY, USA) was used to conduct statistical analyses. Differences were examined using Student's t-test for two-group comparison. Differences were examined using analysis of variance for comparison of more than two groups followed by Turkey's post hoc test. Pearson correlation analysis was performed to examine the correlation between the XIST and miR-137 expression in OS tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
XIST is upregulated in OS
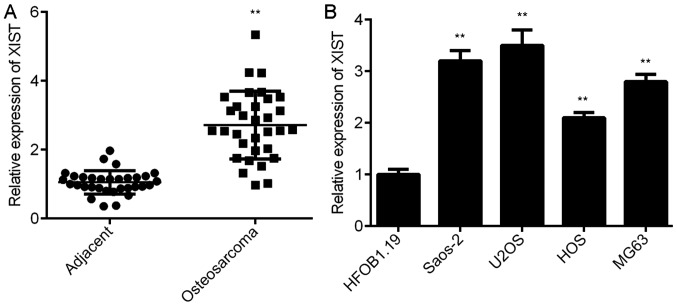
To study the role of XIST in OS, the expression levels of XIST were initially examined in OS tissues and cell lines. RT-qPCR data demonstrated that XIST was significantly upregulated in OS tissues compared with that of matched adjacent non-tumour tissues (Fig. 1A). Similar findings demonstrated that the expression of XIST was increased in OS cell lines (MG63, Saos-2, U2OS and HOS) compared with that of normal human osteoblast cell line HFOB1.19 (Fig. 1B). Therefore, XIST is upregulated in OS.
Figure 1.
XIST is upregulated in OS. (A) RT-qPCR was conducted to examine the expression of XIST in OS tissues compared with adjacent non-tumour tissues. **P<0.01 vs. Adjacent. (B) RT-qPCR was conducted to examine the expression of XIST in OS cell lines compared with the normal human osteoblast cell line HFOB1.19. **P<0.01 vs. HFOB1.19. XIST, X inactive-specific transcript; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
XIST negatively regulates the expression of miR-137 in OS cells by sponging
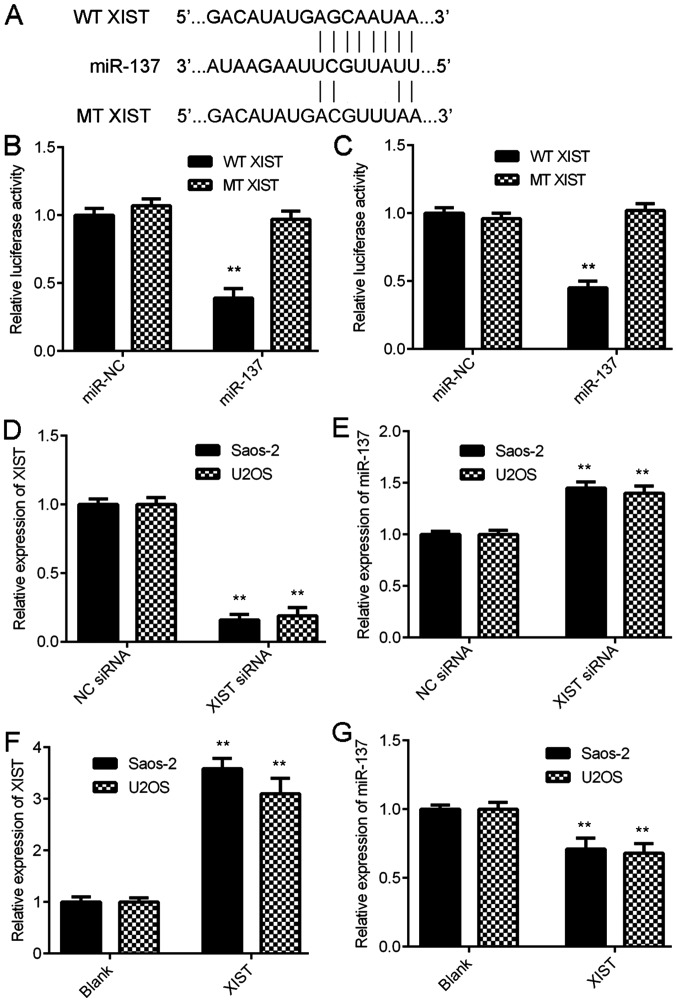
LncRNAs have been demonstrated to function by interacting directly with miRNAs (4,5). Thus, bioinformatics analysis was performed to identify the target miRNAs of XIST. Putative binding sites were identified between XIST and miR-137 (Fig. 2A). To validate this prediction, a luciferase reporter plasmid was constructed containing the WT or MT miR-137 binding sites in XIST (Fig. 2A) and luciferase reporter gene assay was performed using Saos-2 and U2OS cells. The data demonstrated that transfection with the miR-137 mimic caused a significant reduction in the luciferase activity of WT XIST but did not significantly affect the luciferase activity of MT XIST in Saos-2 and U2OS cells (Fig. 2B and C). These data indicate that XIST can directly sponge miR-137 in OS cells.
Figure 2.
XIST negatively regulates the expression of miR-137 in OS cells by sponging. (A) The luciferase reporter plasmid containing the WT or MT miR-137 binding sites in XIST were constructed. (B) Saos-2 and (C) U2OS cells were used for the luciferase reporter gene assay, and transfection with miR-137 mimic significantly inhibited the luciferase activity of WT XIST in OS cells but did not significantly affect the luciferase activity of MT XIST. **P<0.01 vs. miR-NC. (D) Saos-2 and (E) U2OS cells were transfected with XIST siRNA or NC siRNA, and RT-qPCR was conducted to examine the expression of XIST and miR-137. **P<0.01 vs. NC siRNA. (F) Saos-2 and (G) U2OS were transfected with a XIST plasmid or blank vector, and RT-qPCR was conducted to examine the expression of XIST and miR-137. **P<0.01 vs. Blank. XIST, X inactive-specific transcript; miR, microRNA; OS, osteosarcoma; WT, wild-type; MT, mutant type; NC, negative control; siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
The effect of XIST on the miR-137 expression was further studied in OS cells. XIST siRNA or XIST plasmid was used to transfect Saos-2 and U2OS cells. As presented in Fig. 2D, transfection with XIST siRNA significantly inhibited its expression compared with the NC siRNA group. Further experiments demonstrated that inhibition of XIST expression caused a significant increase in miR-137 expression in Saos-2 and U2OS cells (Fig. 2E). In contrast, transfection with XIST plasmid significantly upregulated its expression compared with the blank group (Fig. 2F). Furthermore, overexpression of XIST significantly reduced the expression of miR-137 in Saos-2 and U2OS cells (Fig. 2G). These data suggest that XIST can negatively regulate the miR-137 expression in OS cells by directly sponging it.
miR-137 is downregulated in OS
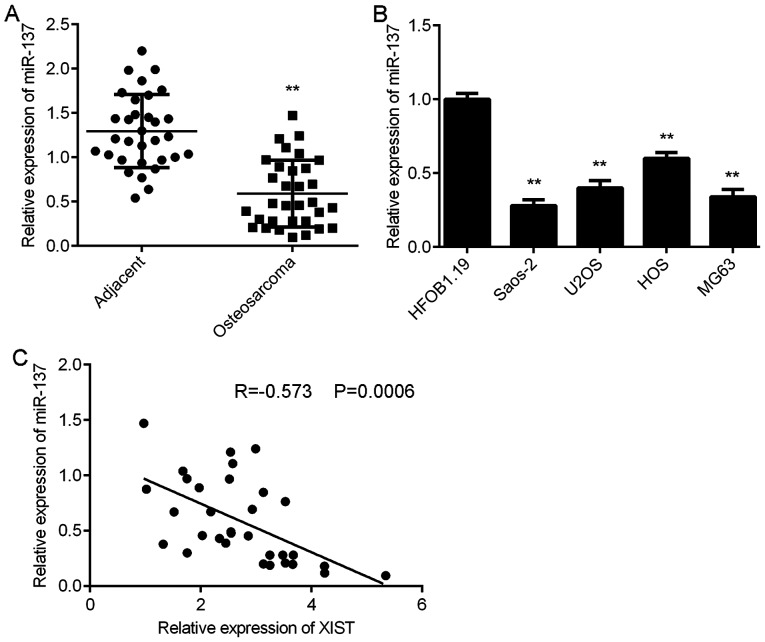
The expression of miR-137 in OS was further examined. RT-qPCR data demonstrated that the expression levels of miR-137 were significantly reduced in OS tissues compared with those of adjacent non-tumour tissues (Fig. 3A). Consistently, miR-137 was also downregulated in OS cell lines compared with that of normal human osteoblasts cell line HFOB1.19 (Fig. 3B). Notably, an inverse correlation was identified between the XIST and miR-137 expression in OS tissues (Fig. 3C). These findings suggest that the reduced expression of miR-137 may be due to the increased expression of XIST in OS tissues.
Figure 3.
miR-137 is downregulated in OS. (A) RT-qPCR was conducted to examine the expression of miR-137 in OS tissues compared with adjacent non-tumour tissues. **P<0.01 vs. adjacent. (B) RT-qPCR was conducted to examine the expression of miR-137 in OS cell lines compared with the normal human osteoblast cell line HFOB1.19. **P<0.01 vs. HFOB1.19. (C) An inverse correlation between the XIST and miR-137 expression was observed in OS tissues. miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; XIST, X inactive-specific transcript.
Inhibition of XIST expression suppresses the proliferation and invasion of OS cells
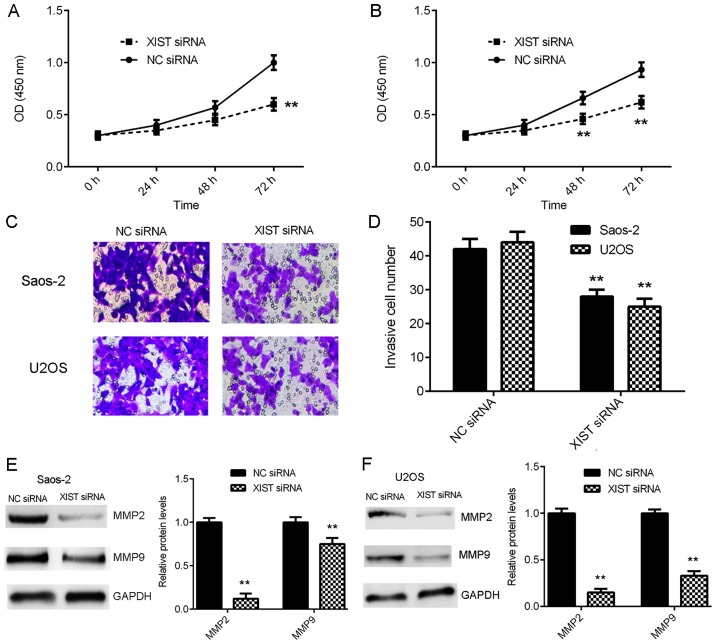
The effects of XIST downregulation on OS cell proliferation and invasion were evaluated. CCK-8 assay data indicated that the proliferation of Saos-2 and U2OS cells was significantly downregulated following knockdown of XIST compared with that of the NC siRNA group (Fig. 4A and B). Transwell assay data demonstrated that the invasion of Saos-2 and U2OS cells was also significantly reduced in the XIST siRNA group compared with the NC siRNA group (Fig. 4C and D). Furthermore, the protein levels of MMP2 and MMP9, which are not direct target genes of XIST and miR-137, but are key factors associated with tumour cell invasion, were also significantly decreased following inhibition of XIST expression (Fig. 4E-F). These findings suggest that XIST may serve a promoting role in OS growth and metastasis.
Figure 4.
Inhibition of XIST expression suppresses the proliferation and invasion of OS cells. Saos-2 and U2OS were transfected with XIST siRNA or NC siRNA. Following transfection, (A and B) CCK-8 and (C and D) Transwell assays were conducted to evaluate cell proliferation and invasion (original magnification, ×200). (E and F) Western blotting was used to determine the protein levels of MMP2 and MMP9. **P<0.01 vs. NC siRNA. XIST, X inactive-specific transcript; OS, osteosarcoma; siRNA, small interfering RNA; NC, negative control; MMP, matrix metalloproteinase; OD, optical density.
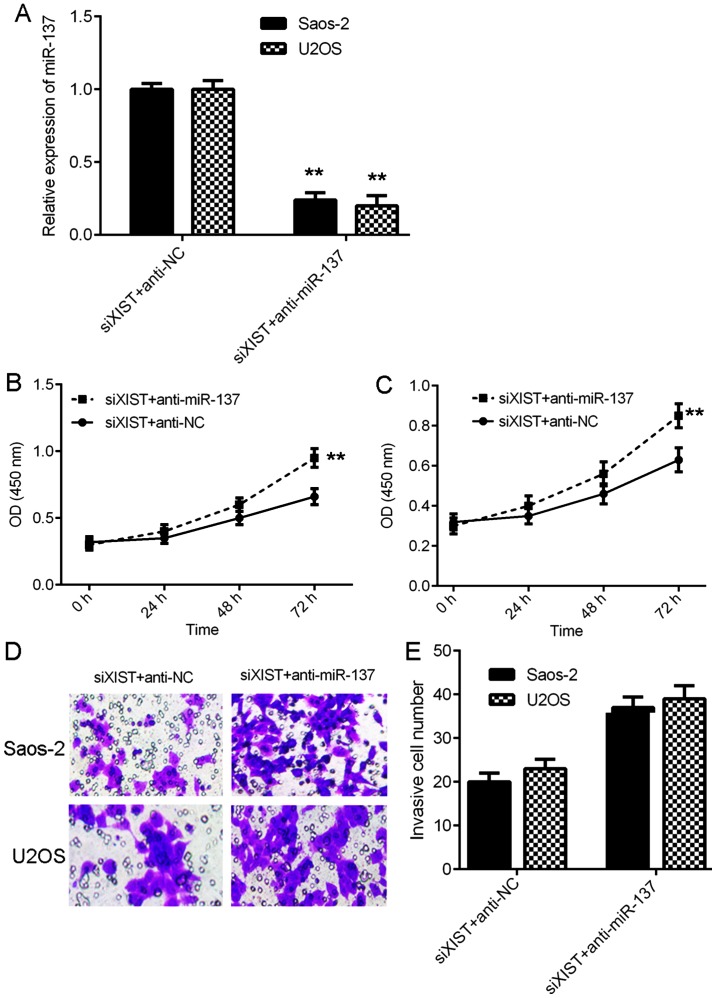
miR-137 is associated with XIST-mediated OS cell proliferation and invasion
The above findings suggest that miR-137 may be associated with XIST-mediated OS cell proliferation and invasion. To test this hypothesis, XIST siRNA transfected cells were then transfected with an miR-137 inhibitor to downregulate the expression of miR-137 in OS cells. RT-qPCR data demonstrated that miR-137 was significantly downregulated in the siXIST+anti-miR-137 group compared with the siXIST+anti-NC group (Fig. 5A). Furthermore, CCK-8 assay data demonstrated that the proliferation of Saos-2 and U2OS cells was significantly increased in the siXIST+anti-miR-137 group compared with the siXIST+anti-NC group (Fig. 5B and C). Similarly, Transwell assay data demonstrated that the invasion of Saos-2 and U2OS cells was also upregulated in the siXIST+anti-miR-137 group compared with the siXIST+anti-NC group (Fig. 5D-E). Therefore, the present findings suggest that XIST serves a promoting role in the regulation of OS cell proliferation and invasion by directly sponging miR-137.
Figure 5.
miR-137 is associated with the XIST-mediated osteosarcoma cell proliferation and invasion. siXIST-transfected Saos-2 and U2OS cells were transfected with NC inhibitor or miR-137 inhibitor, respectively. Following transfection, (A) reverse transcription-quantitative polymerase chain reaction was conducted to examine the expression of miR-137. (B and C) Cell Counting Kit-8 and (D and E) Transwell assays were performed to evaluate cell proliferation and invasion (original magnification, ×200). **P<0.01 vs. siXIST+anti-NC. miR, microRNA; XIST, X inactive-specific transcript; siXIST, XIST small interfering RNA; NC, negative control; OD, optical density.
Discussion
XIST has been implicated in OS, but the underlying molecular mechanism remains largely unclear. In the present study it was reported that XIST was significantly upregulated in OS tissues and cell lines compared with adjacent non-tumour tissues and normal human osteoblast cells. Bioinformatics analysis and luciferase reporter gene assay data demonstrated that XIST could directly target miR-137, and the expression of miR-137 was negatively regulated by XIST in Saos-2 and U2OS cells. Furthermore, miR-137 was markedly downregulated in OS tissues and cell lines, and an inverse association was observed between XIST and miR-137 expression in OS tissues. Knockdown of XIST caused a significant reduction in cell proliferation and invasion and suppressed MMP2 and MMP9 protein levels in Saos-2 and U2OS cells. Furthermore, inhibition of miR-137 expression abolished the effects of XIST downregulation on the proliferation and invasion of OS cells.
Recently, the increased expression of XIST has been observed in different human cancers, and previous studies have demonstrated that XIST serves an oncogenic role (21,22). For instance, Sun et al reported that XIST was significantly upregulated in pancreatic cancer, and it exerts oncogenic functions via miR-34a-5p (21). Jiang et al demonstrated that XIST was significantly upregulated in NSCLC, and inhibition of XIST expression effectively suppressed NSCLC cell viability and invasion by regulating the miR-137/paxillin axis (22). In the present study, it was demonstrated that the expression levels of XIST were significantly upregulated in OS tissues and cells compared with adjacent non-tumour tissues and normal human osteoblast cell line HFOB1.19. These findings were consistent with several previous studies (15,16). It was also demonstrated that inhibition of XIST expression by siRNA significantly reduced the proliferation and invasion of Saos-2 and U2OS cells. Similarly, Yang et al (18) also demonstrated that knockdown of XIST inhibited OS cell proliferation and invasion in vitro and tumour growth in vivo.
Previous studies have demonstrated that lncRNAs including XIST can negatively regulate the expression of miRNAs through sponging (23,24). For instance, Zhang et al demonstrated that XIST promoted gastric cancer progression through TGF-β1 via targeting miR-185 (23). Cheng et al reported that XIST suppressed nasopharyngeal carcinoma progression by activating miR-491-5p (24). In OS, several miRNAs have been identified as the targets of XIST, such as miR-195-5p (18), miR-21-5p (19) and miR-320b (16). In the present study, bioinformatics analysis data demonstrated that miR-137 was a potential target of XIST, which was confirmed by luciferase reporter gene assay. The expression of miR-137 was negatively regulated by XIST in OS cells. Furthermore, it was observed that miR-137 was significantly downregulated in OS tissues and cell lines, and the miR-137 levels were inversely correlated to the XIST levels in OS tissues. These findings suggest that the upregulation of XIST may contribute to the downregulation of miR-137 in OS tissues.
A previous study has demonstrated that miR-137 functions as a tumour suppressor in OS via targeting EZH2 (25). In addition, Li et al also reported that miR-137 is downregulated in OS tissues and cell lines, consistent with the present findings, and can inhibit OS cell proliferation and migration through directly targeting FXYD6 (26). However, the molecular mechanism of miR-137 underlying OS progression remains largely unclear. In the present study, it was identified that inhibition of miR-137 reversed the suppressive effects of XIST downregulation on proliferation and invasion of Saos-2 and U2OS cells. These findings suggest that miR-137 functions as a downstream effector of XIST in OS cells.
To the best of our knowledge, this is the first study to report that XIST has promoting effects on proliferation and invasion of OS cells by sponging miR-137. Therefore, these finding suggest suggest that XIST may become a potential therapeutic target for the treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
HL collected clinical tissues, examined the expression in tissues, and wrote the manuscript. SH and HY designed the study and revised the manuscript. JC, BX and LL performed the in vitro experiments.
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of First Affiliated Hospital of Hunan College of Traditional Chinese Medicine (Zhuzhou, China) and written informed consents was obtained from all patients.
Patient consent for publication
Written informed consents was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4:25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Maximov VV, Aqeilan RI. Genetic factors conferring metastasis in osteosarcoma. Future Oncol. 2016;12:1623–1644. doi: 10.2217/fon-2016-0014. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Hui Y, Li X, Jia Q. Silencing of lncRNA-CCDC26 restrains the growth and migration of glioma cells in vitro and in vivo via targeting miR-203. Oncol Res. 2018;26:1143–1154. doi: 10.3727/096504017X14965095236521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Li J, Zi Y, Wang W, Li Y. LncRNA MEG3 inhibits cell proliferation and metastasis in chronic myeloid leukemia via targeting MiR-184. Oncol Res. 2018;26:297–305. doi: 10.3727/096504017X14980882803151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu K, He Y, Xia C, Yan J, Hou J, Kong D, Yang Y, Zheng G. MicroRNA-15a inhibits proliferation and induces apoptosis in CNE1 nasopharyngeal carcinoma cells. Oncol Res. 2016;24:145–151. doi: 10.3727/096504016X14611963142290. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1alpha. Oncol Res. 2017;25:397–405. doi: 10.3727/096504016X14743337535446. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhang Y, Dai Q, Zeng F, Liu H. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the Rac1/JNK pathway via targeting miR-509. Oncol Res. 2018 Apr 27; doi: 10.3727/096504017X14957939026111. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem. 2000;275:36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 11.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, Xu RH. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Xu J, Lu H, Lin B, Cai S, Guo J, Zang F, Chen R. LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol Rep. 2017;38:3659–3667. doi: 10.3892/or.2017.6040. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Deng C, Zhang H, Zhang J, Peng B, Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/β-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8:94554–94568. doi: 10.18632/oncotarget.21791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Li C, Wan L, Liu Z, Xu G, Wang S, Su Z, Zhang Y, Zhang C, Liu X, Lei Z, Zhang HT. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 15.Li GL, Wu YX, Li YM, Li J. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur Rev Med Pharmacol Sci. 2017;21:2829–2834. [PubMed] [Google Scholar]
- 16.Lv GY, Miao J, Zhang XL. Long non-coding RNA XIST promotes osteosarcoma progression by targeting ras-related protein RAP2B via miR-320b. Oncol Res. 2018;26:837–846. doi: 10.3727/096504017X14920318811721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, Nie X, Ma C, Liu X, Liang X, An Y, Zhao B, Wu X. RSF1 functions as an oncogene in osteosarcoma and is regulated by XIST/miR-193a-3p axis. Biomed Pharmacother. 2017;95:207–214. doi: 10.1016/j.biopha.2017.08.068. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–5656. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 19.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51:1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Sun Z, Zhang B, Cui T. Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep. 2018;39:1591–1600. doi: 10.3892/or.2018.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. doi: 10.1016/j.ijbiomac.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-beta1 via targeting miR-185. J Cell Biochem. 2018;119:2787–2796. doi: 10.1002/jcb.26447. [DOI] [PubMed] [Google Scholar]
- 24.Cheng Q, Xu X, Jiang H, Xu L, Li Q. Knockdown of long non-coding RNA XIST suppresses nasopharyngeal carcinoma progression by activating miR-491-5p. J Cell Biochem. 2018;119:3936–3944. doi: 10.1002/jcb.26535. [DOI] [PubMed] [Google Scholar]
- 25.Feng Q, Wu Q, Liu X, Xiong Y, Li H. MicroRNA-137 acts as a tumor suppressor in osteosarcoma by targeting enhancer of zeste homolog 2. Exp Ther Med. 2017;13:3167–3174. doi: 10.3892/etm.2017.4435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Li ZM, Zhang HY, Wang YX, Wang WB. MicroRNA-137 is downregulated in human osteosarcoma and regulates cell proliferation and migration through targeting FXYD6. J Drug Target. 2016;24:102–110. doi: 10.3109/1061186X.2015.1057149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.