Abstract
The abnormal upregulation of transforming growth factor-β (TGF-β) signaling has been demonstrated to initiate keloid formation and progression. Keloid is a type of benign skin tumor that may occur following sustaining skin injury. microRNA-96 (miR-96) serves an important role in the progression of various malignant diseases. Using reverse transcription quantitative polymerase chain reaction (RT-qPCR), the present study demonstrated that miR-96 was overexpressed in keloid-derived fibroblasts (KFs). Luciferase reporter assay revealed mothers against decapentaplegic homolog (Smad)7, which is one of the important inhibitory factors in the TGF-β pathway, as a direct target of miR-96. miR-96 was initially observed to be correlated with the deposition of type I collagen in KFs in vitro. The miR-96 antagomir, was directly added into the keloid organ culture (OC) to find its significant antifibrotic potential, such as keloid OC shrinkage, exhibited by its dry weight loss and improved dermis architecture, exhibited by Masson's staining. Following miR-96 antagomir treatment, a reduction in the mRNA and protein expression levels of collagen type I α 1 chain and collagen type 3 α 1 chain within keloid OC tissues was observed. The present study revealed that miR-96 serves an important role in pathogenic keloid formation, suggesting that miR-96 antagomir has the potential to prevent keloid progression.
Keywords: microRNA-96, mothers against decapentaplegic homolog 7, keloid, benign tumor, skin fibroblasts
Introduction
Keloid formation is a fibroproliferative disorder caused by abnormal cutaneous wound healing that is characterized by the aggressive growth of fibroblasts and excessive extracellular matrix (ECM) deposition (1). Although keloid formation is a nonmalignant disease, keloid-derived fibroblasts (KFs) can undergo malignant manifestations such as hyperproliferation, changes in collagen morphology, and excessive collagen deposition (1,2). Keloid formation in the dermis is primarily driven by upregulated transforming growth factor-β (TGF-β) signaling pathways and the excessive deposition of collagen type I α 1 chain (COL1A1) and collagen type 3 α1 chain (COL3A1), which are the main structural components of the ECM (1–4). These cross-linked long-chain fibers form a strong network in the ECM, thereby maintaining the integrity of the keloid scar. However, this is accompanied with lower flexibility and malformation of the ECM. The etiology and underlying mechanism of keloid formation are still poorly understood (2).
microRNAs (miRNAs or miRs) serve critical roles in fibrosis and in malignant biological processes, such as excessive cell proliferation and ECM deposition (5). The synthesis and degradation of the ECM in fibrotic tissues are controlled by numerous factors, such as cell signaling pathways, certain collagenases and miRNAs (6,7). Certain malignant miRNAs, such as miR-21, miR-181c and miR-196a, have previously been reported to participate in fibrosis and ECM metabolism (8–10). Among them, miR-21 and miR-181c have been demonstrated to mediate collagen deposition in hypertrophic scars (8,9). miR-96 has also been demonstrated to contribute to the malignant features in prostate and colorectal cancer and was found to be associated with TGF-mediated signaling pathways (11,12). In addition, overactivation of TGF-β signaling has been reported to be required for the initiation and progression of keloids (12). The TGF-β pathway modulates tumorigenesis and progression through mothers against decapentaplegic homolog (Smad)2/3 phosphorylation-mediated fibroblast proliferation and collagen deposition in keloids (13). However, the role of miR-96 in keloid pathogenesis and its effect on the TGF-β signaling pathway remains poorly understood.
Smad7, an important inhibitory cytokine in the TGF-β signaling pathway, was predicted to be a target of miR-96, according to the bioinformatics algorithm TargetScan Human7.2. As such, the aim of the present study was to determine the correlation between miR-96 and Smad7, and compare their expression in KFs with that in normal skin-derived fibroblasts (NFs). In addition, an antagomir-treated keloid organ culture (OC) model was used to determine the therapeutic effects of miR-96 downregulation.
Patients and methods
Patient recruitment and keloid OC establishment
Patients with typical keloid characteristics who had no prior treatment were recruited for the present study, as previously described (14). The diagnosis of keloid pathogenesis in the patient specimens was confirmed through routine pathological examination by an experienced dermatopathologist. The present study was conducted with the consent of each recruited patient and with the approval of the Ethics Committee of the Lanzhou General Hospital of Chinese People's Liberation Army (Lanzhou, China). A total of 10 patients with KFs, aged 17–42 years, were recruited from February 2016 to June 2017 at Burns and Plastic Surgery Center of PLA in Lanzhou General Hospital of Chinese People's Liberation Army, and the control group consisted of corresponding tissue sections obtained from the NFs adjacent to the lesion in these 10 keloid cases (Table I). As described previously (15), the excised 5-mm3 tissue biopsies (keloid explants or keloid OC) were embedded in a collagen gel matrix and were then preserved in serum-free William's medium E (WE medium; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) (15).
Table I.
Profile of each sample for primary culture.
| Patient | Sex | Age (years) | Biopsy site | Duration of the lesion (months) | Etiology |
|---|---|---|---|---|---|
| 1 | Male | 21 | Shoulder | 6 | Burn |
| 2 | Female | 17 | Chest | 8 | Scald |
| 3 | Female | 27 | Arm | 13 | Scald |
| 4 | Female | 42 | Shoulder | 11 | Burn |
| 5 | Male | 38 | Chest | 17 | Scald |
| 6 | Female | 24 | Shoulder | 9 | Burn |
| 7 | Male | 36 | Buttock | 7 | Burn |
| 8 | Female | 22 | Cheek | 10 | Burn |
| 9 | Female | 21 | Shoulder | 5 | Scald |
| 10 | Male | 18 | Chest | 14 | Scald |
Cell culture and transfection of miRNA mimics and miRNA inhibitors
The primary fibroblasts were cultured as described previously (9). Fibroblasts that had undergone 0–3 passages were used in the present study. All synthetic miRNAs and miRNA inhibitors (including the scrambled RNA as a negative control) were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). The scrambled RNA was used as a control for mimic or inhibitor transfection experiments. All sequences are presented in Table II. Cells were transduced with miR-96 inhibitor (70 nM) or control molecules (70 nM) with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), 48 h prior to subsequent experiments. miR-96 mimics or control molecules (50 nM) were transfected using Lipofectamine® 2000 (16).
Table II.
Sequences of miR-96 mimic, miR-96 inhibitor, scrambled RNA, si-Smad7 and Si-NC.
| Sequence (5′→3′) | |
|---|---|
| miR-96 mimic | UUUGGCACUAGCACAUUUUUGCU |
| miR-96 inhibitor | AGCAAAAAUGUGCUAGUGCCAAA |
| scrambled RNA | CAGUACUUUUGUGUAGUACAA |
| si-Smad7 forward | AAGATAATTCGTTCCCCCTGTCCTGTCTC |
| si-Smad7 reverse | AAACAGGGGGAACGAATTATCCCTGTCTC |
| si-NC forward | GCAAACAUCCCAGAGGUAU′ |
| si-NC reverse | AUACCUCUGGGAUGUUUGC |
si, small interfering RNA; miR, microRNA; NC, negative control.
Masson's staining
The degree of fibrosis was examined using Masson's trichrome staining, according to standard protocols (1,17), and the stained tissue sections were then examined with a light microscope at magnification, ×400 (Olympus Corporation, Tokyo, Japan).
Luciferase reporter assay
TargetScan Human 7.2 (www.targetscan.org/vert_72) was used to predict Smad7 as a target of miR-96. The wild-type (wt) or mutant (mut) Smad7 3′-untranslated region (UTR) was cloned into the pGL3 vector (Promega Corporation, Madison, WI, USA). Subsequently, the pGL3-Smad7-3′UTR-wt or pGL3-Smad7-3′UTR-mut vector, along with the miR-96 mimic or the mimic-control (as described above), were transfected with Lipofectamine® 2000 into 293T cells (ATCC, Rockville, MD, USA). Following 48 h, the luciferase activity was measured with a Luciferase Reporter Gene Assay kit (Promega Corporation), using a GloMax-Multi Jr Single Tube Multimode Reader (Promega Corporation). Renilla luciferase activity was used for normalization of the firefly luciferase activity (18).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the mRNA of cells and tissues, according to the manufacturer's protocol. Universal primer and the miScript reverse transcription kit (both Qiagen GmbH, Hilden, Germany) were used for reverse transcription of miRNA. RETROscript™ reverse transcription kit (cat. no. AM1710; Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used for mRNA reverse transcription. Triplicate RT-qPCR reactions and analyses were performed using a Bio-Rad C1000 Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The qPCR reactions were performed with miScript SYBR Green PCR kit (cat. no. 218200; Qiagen GmbH) using the following thermocycling conditions: Initial denaturation for 15 sec at 95°C; 45 cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and extension at 70°C for 30 sec. The internal loading controls used for the mRNAs and miRNAs were GAPDH and RNU6B, respectively. The PCR primers used for mRNA and miR quantification are listed in Table III. Expression levels were determined using Applied Biosystems 7500 software version 2.0.1 (Applied Biosystems; Thermo Fisher Scientific, Inc.) and analyzed using the 2−ΔΔCq method (19).
Table III.
Primers used in reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence (5′→3′) |
|---|---|
| COL1A1 forward | GTGGAAACCCGAGCCCTGCC |
| COL1A1 reverse | TCCCTTGGGTCCCTCGACGC |
| COL3A1 forward | TCCCACTATTATTTTGGCACAACA |
| COL3A1 reverse | TCATCGCAGAGAACGACGGATCC |
| Smad7 forward | ATGDTGTGCCTTCCTCCGCT |
| Smad7 reverse | CGTCCACGGCTGCTGCATAA |
| GAPDH forward | GTCGCCAGCCGAGCCACATC |
| GAPDH reverse | CCAGGCGCCCAATACGACCA |
| miR-96 forward | TCGTTTTTACACGATCACGGTTT |
| RNU6B RNA | ACGCAAATTCGTGAAGCGTT |
COL1A1, collagen subunit 1α1; COL3A1, collagen subunit 3α1; Smad, mothers against decapentaplegic homolog; miR, microRNA.
Western blotting
The proteins were extracted from KF and NF tissues using a mammalian protein extraction reagent (M-PER™; Thermo Fisher Scientific, Inc.) and concentrations were determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.). The lysates were mixed with laemmli buffer (2X; Bio-Rad Laboratories, Inc.) supplemented with 5% β-mercaptoethanol (1:1 ratio, Thermo Fisher Scientific, Inc.). Samples were boiled for 5 min at 95°C. Proteins (15 µg/lane) were separated using SDS-PAGE on 10% gel and subsequently the proteins were transferred to polyvinylidene fluoride membranes by cold transfer (25V at 4°C overnight). The membranes were blocked by a solution of 5% non-fat dried milk in Tris buffered saline with Tween (TBST; 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20) for 1 h at room temperature (20). The membrane was then incubated at 4°C overnight with primary antibodies for COL1A1 (cat. no. sc-293184, 1:1,000), COL3A1 (cat. no. sc-271249, 1:500), Smad7 (cat. no. sc-365846, 1:300; all Santa Cruz Biotechnology, Inc., Dallas, TX, USA), and β-actin (cat. no. sc-70319, 1:5,000; Sigma-Aldrich; Merck KGaA). All primary antibodies were detected using anti-mouse- or anti-rabbit-horseradish peroxidase conjugated secondary antibodies (1:3,000; cat. nos. sc-390944 and sc-5162, respectively; Santa Cruz Biotechnology, Inc.) at 37°C for 2 h. Membranes were washed three times with TBST and the chemiluminescent signals were detected using Clarity ECL Western Blotting substrate (cat. no. 1705061; Bio-Rad Laboratories, Inc.) and detected using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). The relative protein expression of COL1A1 and COL3A1 was normalized to that of β-actin with Image-Pro Plus software (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA).
ELISA
The level of Col1 supernatant protein was assessed using COL1A1 and COL3A1 ELISA kits (cat. nos. ab210966 and ab7778, respectively; Abcam, Cambridge, MA, USA) according to the manufacturer's protocol. Briefly, the conditioned medium from cell culture was added to an ELISA kit plate precoated with an anti-COL1A1 and COL3A1 antibody. Subsequently a biotinylated secondary antibody was added and the plate was incubated at room temperature for 2 h. The optical density was measured at a wavelength of 450 nm (21).
Treatment of keloid OC with the miR-96 inhibitor, antagomir
The chemically synthesized miR-96 inhibitor, antagomir, functions to decrease the miR-96 expression level in the keloid OC (22–24). miR-96 antagomir (100 µg; Guangzhou RiboBio Co., Ltd., Guangzhou, China) was added to 1 ml keloid OC in serum-free WE medium every 72 h for a period of five weeks at 37°C, while the control group received an equal amount of scrambled RNA in the same antagomir medium, and the medium was changed every three days. Keloid shrinkage in vitro was assessed through measuring the dry weight of the 5-µm Masson's trichrome-stained sections on an analytical balance.
siRNA transfection
si-Smad7 sequences were chemically synthesized and purified by high-performance liquid chromatography (Shanghai GenePharma Co., Ltd., Shanghai, China). All the oligonucleotides were 2′-OMe modified. Briefly, cells were transfected with siRNA-Smad7 at a final concentration of 50 nM using Lipofectamine® 2000. siRNA-NC was transfected as a negative control. At 24 h post-transfection, the culture medium was changed according to the protocols of manufacturer. After 48 h, cells were harvested for analysis. All transfections were performed in triplicate (25).
Statistical analysis
Statistical analysis was performed using a two-tailed unpaired Student's t-test, paired Student's t-test, Pearson's linear correlation test and one-way analysis of variance with Tukey's post hoc test. All data were obtained from triplicate experiments and were presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Altered expression of miRNAs and Smad7 in KFs
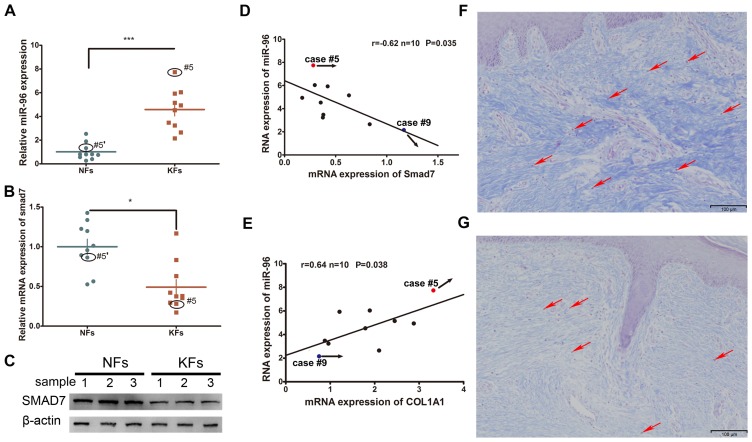
The expression levels of Smad7 and miR-96 were compared between the KFs and NFs obtained from 10 keloid patients. It was demonstrated that the mRNA expression of miR-96 was increased whereas that of Smad7 was decreased in the KFs, as compared with the NFs (Fig. 1A-C). Furthermore, their expression levels were inversely correlated (Fig. 1D). Given that type I collagen is the major component of the ECM that participates in keloid formation, the correlation between miR-96 and COL1A1 was also investigated. Endogenous miR-96 was demonstrated to be positively correlated with the COL1A1 expression levels (Fig. 1E). Masson's staining of KF tissues revealed relatively high COL1A1 and miR-96 expression levels in case #5, yet a low Smad7 expression level (Fig. 1F); whereas case #9 exhibited relatively low expression levels of miR-96 and COL1A1, yet a high Smad7 expression level (Fig. 1G). Examination of the structural characteristics of the keloid scars of cases #5 and #9 revealed that case #5, which had a higher expression level of miR-96 and a lower level of Smad7, exhibited a thicker collagen fiber deposition and a relatively non-uniform collagen density distribution (Fig. 1F). Conversely, case #9, which had a lower level of miR-96 and a higher level of Smad7, exhibited a thinner collagen fiber deposition and a relatively uniform density distribution (Fig. 1G).
Figure 1.
The expression of miR-96, Smad7 and COL1A1 in KFs and NFs. (A) The miR-96 expression level was upregulated in KFs compared with that in NFs. The expression of Smad7 was decreased in KFs compared with that in NFs at both the (B) mRNA and (C) protein levels. Black circles indicate the Smad7 expression level in the keloid of case #5 and in the autologous normal skin control (#5′) of case #5. (D) An inverse correlation of Smad7 and the miR-96 mRNA level was observed in the keloid samples. (E) A positive correlation between Smad7 and miR-96 was revealed in the keloid samples. Masson's staining of two keloid tissue samples: (F) Case #5 and (G) case #9. Case #5 exhibited a relatively thicker dermis and stronger staining of the collagen fibers, a higher miR-96 expression, and a relatively lower Smad7 expression, compared with that of case #9. Red arrows indicate the KFs in the tissue samples of cases #5 and #9. *P<0.05 and ***P<0.001, as indicated. miR, microRNA; Smad, mothers against decapentaplegic homolog; COL1A1, collagen subunit 1α1; KF, keloid-derived fibroblast; NF, normal skin-derived fibroblast.
Identification of Smad7 genes as direct targets of miR-96
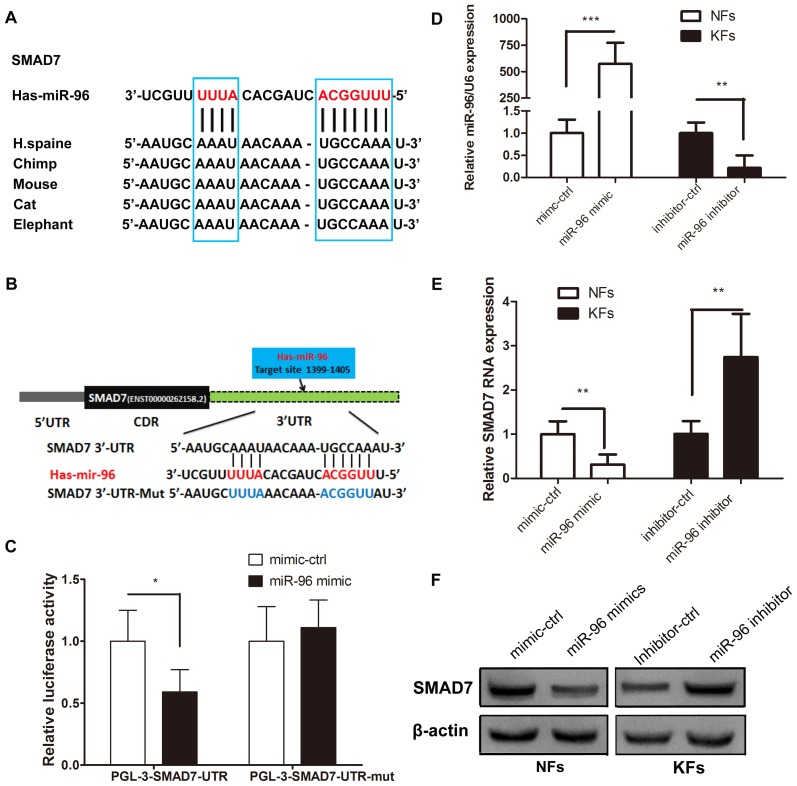
The putative binding sites of miR-96 in the region of the 3′UTR of Smad7 were detected through bioinformatics analysis. Therefore, the predicted region, containing the wt or mut seed sequence of miR-96 in the 3′UTR of Smad7, was cloned into the luciferase reporter plasmid (Fig. 2A and B). The 293T cells transfected with the miR-96 mimic and the Smad7-3′-UTR had a lower luciferase intensity, but the mutant reporter-transfected group did not (Fig. 2C), thus indicating that Smad7 may be a direct target of miR-96. Western blotting and RT-qPCR were used to confirm the role of miR-96 in regulating Smad7 expression in the KFs and NFs. The miR-96 expression level was higher in the KFs than in the NFs (Fig. 1A), thus, the miR-96 mimic was transfected into NFs, whereas the miR-96 inhibitor was transfected into the KFs (Fig. 2D). The results demonstrated that Smad7 was decreased in the miR-96 mimic-transfected NFs at the mRNA and protein levels; however, the Smad7 expression level was increased in the KFs transfected with the miR-96 inhibitor, compared with the corresponding control groups (Fig. 2E and F). These results suggest that miR-96 may regulate Smad7 expression in KFs and NFs at both the mRNA and protein levels.
Figure 2.
miR-96 targets Smad7. (A) The predicted Smad7 binding site on miR-96, which is highly conserved in different species. (B) Using pGL3 vectors, the wild-type luciferase reporter plasmid contains the Smad7 3′UTR binding site, whereas the mutant plasmid does not. (C) Reduced luciferase activity in the wild-type group compared with that of the mutant group at 48 h following transfection into 293T cells. (D) The miR-96 mimic and its control were transfected into NFs, whereas the miR-96 inhibitor and its control were transfected into KFs. (E) Reverse transcription-quantitative polymerase chain reaction and (F) western blotting analyses of the mRNA and protein levels of Smad7 in the NFs comprising the miR-96 mimic and the KFs comprising the miR-96 inhibitor at 48 h following transfection. All values were normalized to their mimic control or inhibitor control, respectively. *P<0.05, **P<0.01 and ***P<0.001, as indicated. miR, microRNA; Smad, mothers against decapentaplegic homolog; UTR, untranslated region; NF, normal skin-derived fibroblast; KF, keloid-derived fibroblast; ctrl, control.
miR-96 inhibition leads to decreased type I and Ш collagen production in KFs, which may be reversed by Smad7 reintroduction
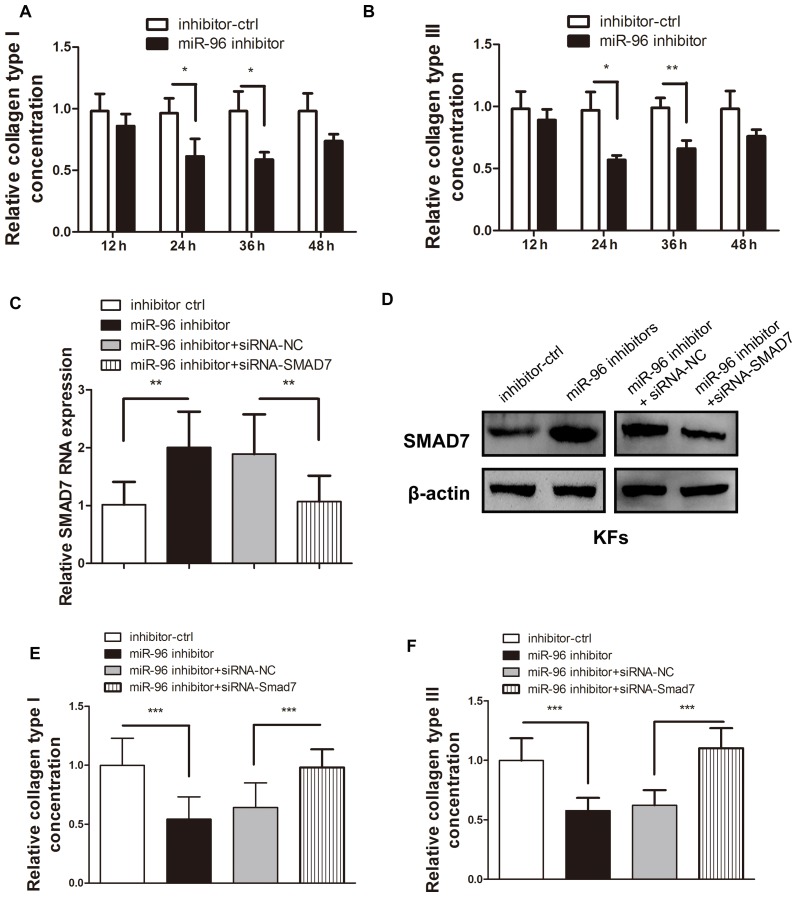
Type I and Ш collagen are the most common types of collagen that are deposited in the abnormal ECM of KFs (1–3,10,26). Therefore, these two collagen types were chosen to evaluate the regulatory effect of miR-96 on ECM deposition. As Smad7 is a well-known inhibitory cytokine in the TGF-β pathway and is under the regulation of miR-96, it was assumed that miR-96 inhibition may reduce the COL1A1 and COL3A1 expression levels in KFs. ELISA was used to confirm that transfection of the miR-96 inhibitor into KFs led to a reduction in COL1A1 and COL3A1 expression (Fig. 3A and B). However, the introduction of siRNA-Smad7 was demonstrated to significantly reverse the miR-96-inhibitor-induced Smad7 upregulation (Fig. 3C and D), thereby leading to increased COL1A1 and COL3A1 production (Fig. 3E and F).
Figure 3.
Smad7 inhibition reversed the upregulation of COL1A1 and COL3A1 expression induced by the miR-96 inhibitor in the KFs. Following transfection of the miR-96 inhibitor, both the (A) type I and (B) type Ш collagen in the supernatant of the cultured KFs were detected by ELISA. The addition of siRNA-Smad7 reversed the effect of the upregulated (C) mRNA and (D) protein expression of Smad7, which was initially upregulated by the miR-96 inhibitor. The miR-96 inhibitor-mediated reduction of (E) COL1A1 and (F) COL3A1 expression levels in the supernatant of the cultured KFs was reversed by transfection of siRNA-Smad7. *P<0.05, **P<0.01 and ***P<0.001, as indicated. Smad, mothers against decapentaplegic homolog; COL1A1, collagen subunit 1α1; COL3A1, collagen subunit 3α1; miR, microRNA; KF, keloid-derived fibroblast; siRNA, small interfering RNA; ctrl, control.
Direct delivery of miR-96 antagomir into the keloid OC decreased the expression level of miR-96
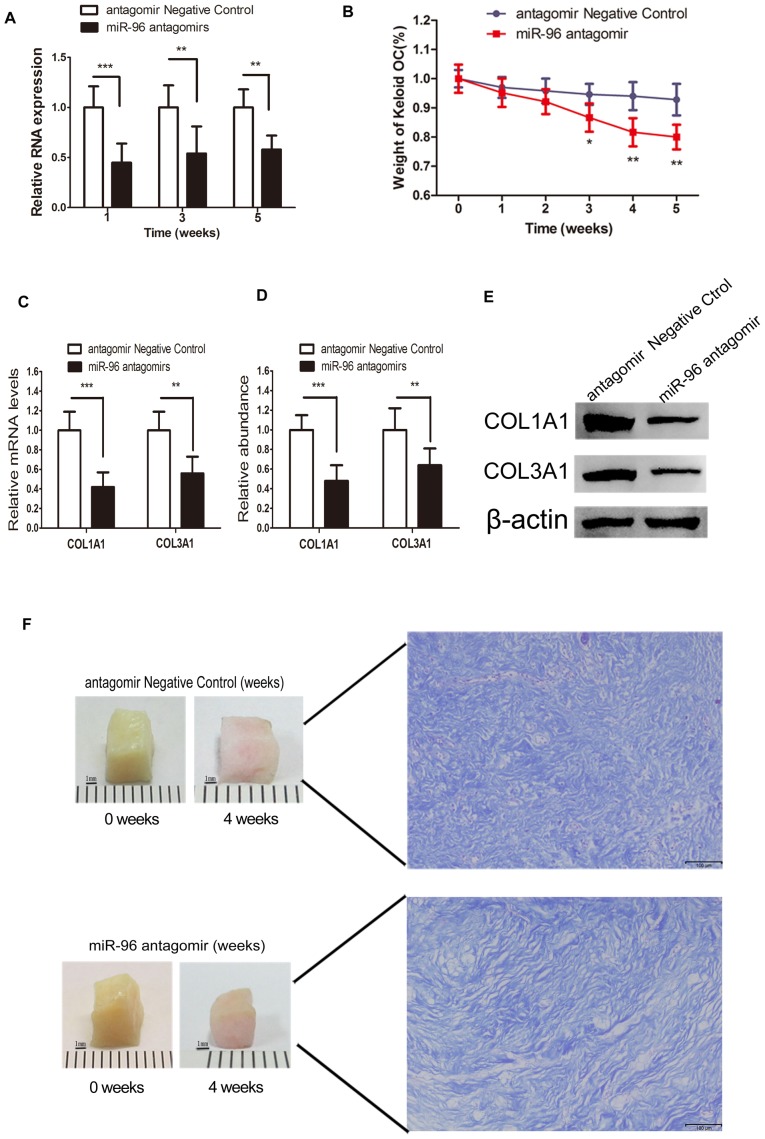
Local gene therapies targeted at reducing fibrosis through the application of liposome-complex small interfering RNA or an miRNA mimic, or through the direct administration of plasmid, have been demonstrated to be effective and successful (22). Given the validation of the degradation ability of antagomirs (23), miRNA antagomir was added to the keloid OC media, as described previously (22–24). The RT-qPCR data demonstrated that the miR-96 expression level was barely changed at each time point in the control group. However, the miR-96 antagomir-treated keloid OC tissues exhibited decreased miR-96 expression levels at each time point, compared with the control groups (Fig. 4A). These findings demonstrated that the direct delivery of miR-96 antagomir into the keloid OC may lead to a reduction in miR-96 expression.
Figure 4.
Therapeutic effects of miR-96 antagomir on the keloid OC. (A) Reduction in the miR-96 expression at 1, 3, and 5 weeks following miR-96 antagomir administration. (B) The mean weight of the miR-96 antagomir-treated keloid OC was decreased at week 3 (8%) and week 5 (14%), as compared with the negative control. (C) The relative mRNA levels of COL1A1 and COL3A1 in the keloid OC at week 3 following miR-96 antagomir treatment were significantly decreased (n=10). (D) Relative quantitative analysis of western blotting indicated reduced COL1A1 and COL3A1 protein levels in the miR-96 antagomir-treated group (n=10). (E) Western blots for COL1A1 and COL3A1. (F) Representative images of the keloid OC volume shrinkage at week 4. The dermal architecture was demonstrated using Masson's staining. Loosened and thinner collagen bundles were observed in the miR-96 antagomir-treated group. *P<0.05, **P<0.01 and ***P<0.001 vs. the antagomir negative control group. miR, microRNA; OC, organ culture; COL1A1, collagen subunit 1α1; COL3A1, collagen subunit 3α1.
miR-96 antagomir promotes type I and Ш collagen degradation and keloid shrinkage in the keloid OC model
Compared with the scrambled RNA-treated control, the dry weight of the keloid OC was significantly reduced by 8% at week 3, and by 14% at week 5 (Fig. 4B). The keloid OC models were treated with miR-96 antagomir to investigate its effect on collagen degradation. In the miR-96 antagomir-treated group, both the COL1A1 and COL3A1 mRNA expression levels in the keloid tissues at the end of the 4th week of OC were significantly reduced (Fig. 4C). Similarly, the protein expression levels of COL1A1 and COL3A1 (Fig. 4D and E) were also significantly inhibited in the miR-96 antagomir-treated group. These findings indicated that the addition of miR-96 antagomir to the keloid OC decreased the quantity of type I and Ш collagen. The weight of the keloid OC was measured weekly for a period of 5 weeks post-treatment to assess the antifibrotic potential of miR-96 antagomir. Visible keloid shrinkage was observed at the end of week 4 (Fig. 4F).
Improved dermal architecture in the miR-96 antagomir-treated keloid OC
At the end of 4 weeks of OC, the harvested keloid tissues were subjected to Masson's staining. Thinner and more loosely arranged collagen fibers were observed in the miR-96 antagomir-treated keloid tissues, compared with the control (Fig. 4F).
Discussion
Keloid pathogenesis is characterized by the aggressive proliferation and excessive deposition of type I and Ш collagen in the ECM (27). Furthermore, ECM proteins, including collagen, are synthesized by dermal fibroblasts; therefore, KFs were used for the present study (28). During wound healing, the expression of type I and Ш collagen increases significantly, thereby accelerating wound repair (17). However, the persistent overexpression of collagen can lead to skin fibrosis, which can eventually result in keloid pathogenesis.
It has been indicated that miRNAs serve an important role in fibrotic diseases (29) by promoting aggressive fibroblast proliferation and collagen deposition. Our previous study revealed that miR-21 overexpression within the hypertrophic scar, which is one of the two types of pathological scars, may promote fibroblast proliferation and inhibit apoptosis (8). Conversely, a number of previous studies have reported that certain miRNAs have an antifibrotic ability, thereby preventing organ fibrosis. For example, a lower miR-29 expression level is observed in the fibrotic tissues of the lungs, myocardium, and skin; hence, fibrotic diseases may be ameliorated by miR-29 overexpression (29–31). miR-10a attenuated collagen type I generation in hypertrophic scars by targeting PAI-1 (9). miR-196a overexpression decreases the expression of type I and Ш collagens in keloid fibroblasts (10).
miR-96 has been the focus of previous studies on diseases associated with aggressive proliferation, particularly cancers. One previous study has demonstrated the correlation between miR-96 upregulation and chemoresistance in non-small cell lung cancer cells through the downregulation of Smad9 (32). The miR-200c-mediated regulatory mechanism of keloid formation, which is a well-known antitumor miRNA correlated with the TGF-β pathway, has been elucidated in our previous study (13). However, the biological relevance and mechanism of action of miR-96 in keloid pathogenesis have not yet been investigated, to the best of our knowledge. Therefore, the present study focused on the investigation of miR-96 and the TGF-β pathway in keloid formation.
In the present study, miR-96 upregulation was observed in KFs and compared with that of NFs. Furthermore, the positive correlation between miR-96 and COL1A1 as well as the negative correlation between miR-96 and Smad7 were revealed for the first time, to the best of our knowledge. Furthermore, a luciferase reporter assay demonstrated the direct targeting of Smad7 by miR-96. The overexpression or inhibition of miR-96 in the primary fibroblast cells and the keloid OC models validated the results of the reporter assay. These findings supported the hypothesis that Smad7 is a direct target of miR-96 and that it is regulated by miR-96 in the keloid. Also, it was demonstrated that miR-96 inhibited the production of COL1A1 and COL3A1 proteins, which could be reversed with the transfection of siRNA-Smad7, indicating that miR-96 in KFs can regulate collagen deposition through the targeting of Smad7 expression. However, it was noted that the expression level of Smad7 was slightly decreased following transfecting the miR-96 mimic into the KFs, but the difference was not statistically significant (data not shown).
The lack of an in vivo animal model that can mimic human keloid scars has limited the study of potential keloid therapeutic agents (33,34). Therefore, the ex vivo OC of skin tissue and keloids according were evaluated, according to reference protocols (15,24,35). Treatment with the miRNA inhibitor, miR-96 antagomir, revealed shrinkage of the keloid OC. In addition, a reduction in COL1A1 and COL3A1 expression was observed following treatment.
In summary, the present study revealed the upregulation of miR-96 expression in the keloid, and that Smad7 is a direct target of miR-96. Furthermore, the addition of miR-96 antagomir into the keloid OC model revealed an effective reduction of type I and Ш collagen expression, which further led to keloid shrinkage. These findings suggest that miR-96 upregulation is correlated with keloid formation and that miR-96 may be a therapeutic target in keloid pathogenesis.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ECM
extracellular matrix
- COL1A1
collagen subunit 1α1
- COL3A1
collagen subunit 3α1
- TGF-β
transforming growth factor β
- Smad7
mothers against decapentaplegic homolog 7
Funding
The present study was conducted with the support of the National Natural Science Foundation of China (grant nos. 81501684 and 1506RJZA303).
Availability of data and materials
The datasets used and/or analyzed in this study can be obtained from the corresponding author upon request. Genbank (www.ncbi.nlm.nih.gov/genbank) is recommended for performing the DNA and RNA sequence searches.
Authors' contributions
LC made substantial contributions to the conception and design of experiments and was a major contributor in the writing of the manuscript. H-YZ was involved in the writing of the manuscript and revisions to ensure important intellectual content. W-DB performed data acquisition and prepared the figures. BX performed histological examination of the keloid and the skin. MS performed the cell culture and transfection experiments. D-HH conceived the study, analyzed data, and wrote the manuscript. YL analyzed and interpreted the data. All the authors read and approved the final manuscript. Each author participated actively in the study and takes responsibility for the respective work conducted.
Ethics approval and consent to participate
The present study was conducted with the approval of the Medical Ethics Committee of Lanzhou General Hospital of Chinese People's Liberation Army (approval no. LB1706472). All patients provided written informed consent.
Patient consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Verhaegen PD, van Zuijlen PP, Pennings NM, van Marle J, Niessen FB, van der Horst CM, Middelkoop E. Differences in collagen architecture between keloid, normotrophic scar, and normal skin: An objective histopathological analysis. Wound Repair Regen. 2009;17:649–656. doi: 10.1111/j.1524-475X.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- 2.Niessen FB, Spauwen PH, Schalkwijk J, Kon M. On the nature of hypertrophic scars and keloids: A review. Plast Reconstr Surg. 1999;104:1435–1458. doi: 10.1097/00006534-199910000-00031. [DOI] [PubMed] [Google Scholar]
- 3.Friedman DW, Boyd CD, Mackenzie JW, Norton P, Olson RM, Deak SB. Regulation of collagen gene expression in keloids and hypertrophic scars. J Surg Res. 1993;55:214–222. doi: 10.1006/jsre.1993.1132. [DOI] [PubMed] [Google Scholar]
- 4.Cheng J, Wang Y, Wang D, Wu Y. Identification of collagen 1 as a post-transcriptional target of miR-29b in skin fibroblasts: Therapeutic implication for scar reduction. Am J Med Sci. 2013;346:98–103. doi: 10.1097/MAJ.0b013e318267680d. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015–2021. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chau BN, Brenner DA. What goes up must come down: The emerging role of microRNA in fibrosis. Hepatology. 2011;53:4–6. doi: 10.1002/hep.24071. [DOI] [PubMed] [Google Scholar]
- 8.Zhu HY, Li C, Bai WD, Su LL, Liu JQ, Li Y, Shi JH, Cai WX, Bai XZ, Jia YH, et al. MicroRNA-21 regulates hTERT via PTEN in keloid fibroblasts. PLoS One. 2014;9:e97114. doi: 10.1371/journal.pone.0097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Zhu HY, Bai WD, Su LL, Liu JQ, Cai WX, Zhao B, Gao JX, Han SC, Li J, Hu DH. MiR-10a and miR-181c regulate collagen type I generation in hypertrophic scars by targeting PAI-1 and uPA. FEBS Lett. 2015;589:380–389. doi: 10.1016/j.febslet.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Kashiyama K, Mitsutake N, Matsuse M, Ogi T, Saenko VA, Ujifuku K, Utani A, Hirano A, Yamashita S. miR-196a downregulation increases the expression of type I and Ш collagens in keloid fibroblasts. J Invest Dermatol. 2012;132:1597–1604. doi: 10.1038/jid.2012.22. [DOI] [PubMed] [Google Scholar]
- 11.Siu MK, Tsai YC, Chang YS, Yin JJ, Suau F, Chen WY, Liu YN. Transforming growth factor-β promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene. 2015;34:4767–4776. doi: 10.1038/onc.2014.414. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Mao Y, Zhu J, Meng F, Chen Q, Tao L, Li R, Fu F, Liu C, Hu Y, et al. TGF-β1 promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget. 2016;7:67196–67211. doi: 10.18632/oncotarget.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu HY, Bai WD, Li C, Zheng Z, Guan H, Liu JQ, Yang XK, Han SC, Gao JX, Wang HT, Hu DH. Knockdown of lncRNA-ATB suppresses autocrine secretion of TGF-β2 by targeting ZNF217 via miR-200c in keloid fibroblasts. Sci Rep. 2016;6:24728. doi: 10.1038/srep24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang ZF, Zhang YG, Hu DH, Shi JH, Liu JQ, Zhao ZT, Wang HT, Bai XZ, Cai WX, Zhu HY, Tang CW. Smad interacting protein 1 as a regulator of skin fibrosis in pathological scars. Burns. 2011;37:665–672. doi: 10.1016/j.burns.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Bagabir R, Syed F, Paus R, Bayat A. Long-term organ culture of keloid disease tissue. Exp Dermatol. 2012;21:376–381. doi: 10.1111/j.1600-0625.2012.01476.x. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Xu H, Pan X, Wu W, Wang H, Yan L, Zhang M, Liu X, Xia S, Shao Q. miR-34a and miR-125b are upregulated in peripheral blood mononuclear cells from patients with type 2 diabetes mellitus. Exp Ther Med. 2017;14:5589–5596. doi: 10.3892/etm.2017.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Reilly S. MicroRNAs in fibrosis: Opportunities and challenges. Arthritis Res Ther. 2016;18:11. doi: 10.1186/s13075-016-0929-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W, He L, Dai Y, Zhang Y, Wang J, Liu B. MicroRNA-124 inhibits cell proliferation, invasion and migration by targeting CAV1 in bladder cancer. Exp Ther Med. 2018;16:2811–2820. doi: 10.3892/etm.2018.6537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Xing T, Du L, Zhuang X, Zhang L, Hao J, Wang J. Upregulation of microRNA-206 induces apoptosis of vascular smooth muscle cells and decreases risk of atherosclerosis through modulating FOXP1. Exp Ther Med. 2017;14:4097–4103. doi: 10.3892/etm.2017.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie JM, Li HF. Therapeutic effects of Salvia miltiorrhiza injection combined with telmisartan in patients with diabetic nephropathy by influencing collagen IV and fibronectin: A case-control study. Exp Ther Med. 2018;16:3405–3412. doi: 10.3892/etm.2018.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang LL, Liu JQ, Bai XZ, Fan L, Han F, Jia WB, Su LL, Shi JH, Tang CW, Hu DH. Acute downregulation of miR-155 at wound sites leads to a reduced fibrosis through attenuating inflammatory response. Biochem Biophys Res Commun. 2014;453:153–159. doi: 10.1016/j.bbrc.2014.09.077. [DOI] [PubMed] [Google Scholar]
- 23.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 24.Syed F, Bagabir RA, Paus R, Bayat A. Ex vivo evaluation of antifibrotic compounds in skin scarring: EGCG and silencing of PAI-1 independently inhibit growth and induce keloid shrinkage. Lab Invest. 2013;93:946–960. doi: 10.1038/labinvest.2013.82. [DOI] [PubMed] [Google Scholar]
- 25.Zhou D, Wang J, He LN, Li BH, Ding YN, Chen YW, Fan JG. Prolyl oligopeptidase attenuates hepatic stellate cell activation through induction of Smad7 and PPAR-γ. Exp Ther Med. 2017;13:780–786. doi: 10.3892/etm.2017.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidgwick GP, Bayat A. Extracellular matrix molecules implicated in hypertrophic and keloid scarring. J Eur Acad Dermatol Venereol. 2012;26:141–152. doi: 10.1111/j.1468-3083.2011.04200.x. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda M, Naitoh M, Kubota H, Ishiko T, Yoshikawa K, Yamawaki S, Kurokawa M, Utani A, Nakamura T, Nagata K, Suzuki S. Elastic fiber assembly is disrupted by excessive accumulation of chondroitin sulfate in the human dermal fibrotic disease, keloid. Biochem Biophys Res Commun. 2009;390:1221–1228. doi: 10.1016/j.bbrc.2009.10.125. [DOI] [PubMed] [Google Scholar]
- 28.Aoki M, Miyake K, Ogawa R, Dohi T, Akaishi S, Hyakusoku H, Shimada T. siRNA knockdown of tissue inhibitor of metalloproteinase-1 in keloid fibroblasts leads to degradation of collagen type I. J Invest Dermatol. 2014;134:818–826. doi: 10.1038/jid.2013.396. [DOI] [PubMed] [Google Scholar]
- 29.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 30.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer B, Stanczyk J, Jüngel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI, Li K. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams FN, Herndon DN, Branski LK. Where we stand with human hypertrophic and keloid scar models. Exp Dermatol. 2014;23:811–812. doi: 10.1111/exd.12506. [DOI] [PubMed] [Google Scholar]
- 34.van den Broek LJ, Limandjaja GC, Niessen FB, Gibbs S. Human hypertrophic and keloid scar models: Principles, limitations and future challenges from a tissue engineering perspective. Exp Dermatol. 2014;23:382–386. doi: 10.1111/exd.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Z, Hasse S, Bodo E, Rose C, Funk W, Paus R. Towards the development of a simplified long-term organ culture method for human scalp skin and its appendages under serum-free conditions. Exp Dermatol. 2007;16:37–44. doi: 10.1111/j.1600-0625.2006.00510.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in this study can be obtained from the corresponding author upon request. Genbank (www.ncbi.nlm.nih.gov/genbank) is recommended for performing the DNA and RNA sequence searches.