Abstract
microRNAs (miRs) serve primary roles in certain human malignancies; however, the detailed regulatory mechanism of miR-200a in glioma progression is yet to be fully elucidated. The current study aimed to assess the expression of miR-200a in glioma as well as the regulatory mechanism of miR-200a in glioma cell proliferation, survival and invasion. RT-qPCR and western blotting were performed to examine mRNA and protein expression. An MTT assay, an EdU incorporation cell proliferation assay and a transwell assay were utilized to assess cell survival, proliferation and invasion. The results indicated that the miR-200a levels were significantly reduced in glioma tissues compared with normal brain tissues. Levels were also downregulated in glioma cell lines when compared with those in normal human astrocyte cells. Furthermore, low miR-200a expression was associated with advanced progression of glioma. The overexpression of miR-200a inhibited glioma cell proliferation, survival and invasion. Results also identified that FOXA1 was a target gene of miR-200a in glioma cells and that the increased expression of FOXA1 was negatively correlated to the decreased expression of miR-200a in glioma tissues. Furthermore, FOXA1 expression was negatively mediated by miR-200a in glioma cells and the overexpression of FOXA1 eliminated the inhibitory effects of miR-200a on the survival, proliferation and invasion of glioma cells. In conclusion, the current study demonstrated that miR-200a functions acts as a tumor suppressor in glioma by directly targeting FOXA1 and may thus be a potential candidate for the treatment of glioma.
Keywords: glioma, microRNA-200a, forkhead box A1, tumor suppressor
Introduction
Glioma is the most common primary brain tumor and the prognosis of patients with advanced glioma is very poor (1). Although a number of studies have focused on exploring the pathogenesis of glioma (2–4), the molecular mechanisms underlying its development and malignant progression remain largely unknown. Therefore, it is necessary to identify novel biomarkers in glioma, as well as reveal the molecular mechanism that underlies glioma progression, which may be beneficial for the early diagnosis and effective treatment of the disease.
microRNAs (miRNAs or miRs), a class of endogenous small RNAs, are primary regulators of gene expression, which function by directly interacting with the 3′untranslational region (3′UTR) of their target mRNAs, leading to translation, repression or mRNA degradation (5–7). Through mediating the expression of their target genes, miRNAs have been demonstrated to participate in various cellular biological processes, including cell proliferation, differentiation, apoptosis, survival, and invasion (8–10). Furthermore, a large number of miRNAs are dysregulated in certain types of human cancer, including glioma and glioblastoma, and many have been reported to serve oncogenic or tumor suppressive roles in the development and malignant progression of glioma, including miR-365, miR-599 and miR-423 (11–13).
miR-200a is a member of the miR-200 family, that has been demonstrated to serve a tumor suppressive or oncogenic role in certain common types of human cancer, including gastric cancer, esophageal cancer, endometrioid endometrial carcinoma, hepatocellular carcinoma, breast cancer and cholangiocarcinoma (14–19). For instance, miR-200a enhanced the tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in gastric cancer cells (17). However, miR-200a promotes esophageal cancer cell proliferation (18). Additionally, certain target genes of miR-200a have been identified in different types of human cancer, including A20 (17), collapsin response mediator protein 1 (18), zinc finger E-box binding homeobox 2 (ZEB2) (15), phosphatase and tens in homolog (14) and yes associated protein 1 (YAP1) (16). For instance, miR-200a suppresses the metastasis of a distinct subpopulation of cells (obtained from flow cytometry) in hepatocellular carcinoma by decreasing ZEB2 (15), but promotes anoikis resistance and metastasis in human breast cancer by targeting YAP1 (16). Su et al (20) previously revealed that miR-200a impairs glioma cell growth, survival and invasion by targeting single-minded family basic helix-loop-helix transcription factor 2 (SIM2). Although a single miRNA generally has many target genes, other targets of miR-200a may also serve primary roles in glioma.
Forkhead box A1 (FOXA1), a member of the fork head transcription factor family, has been demonstrated to participate in the regulation of cell proliferation, differentiation and motility (21,22). The aberrant expression of FOXA1 has been observed in many types of human cancer and its oncogenic role has also been elucidated (23,24). For instance, the knockdown of FOXA1 inhibits the proliferation and migration of pancreatic cancer cells (23). Furthermore, several miRs have been revealed to serve a tumor suppressive role by targeting FOXA1 (25). For instance, miR-212 suppresses the proliferation and migration of osteosarcoma cells via the inhibition of FOXA1 expression (25). However, to the best of our knowledge, no previous studies have assessed whether FOXA1 is associated with miR-200a-mediated glioma cells.
Therefore, the present study aimed to assess the clinical significance of miR-200a expression in glioma and to elucidate the regulatory mechanism of miR-200a underlying the development and progression of glioma.
Materials and methods
Clinical tissue samples
The present study was approved by the Ethics Committee of the Brain Hospital of Hunan Province (Changsha, China). Glioma tissues (n=66) were collected from patients with glioma (39 male, 27 female; age, 24–64 years; median age, 49.3 years) by surgical resection at the Brain Hospital of Hunan Province between March 2013 and October 2016. Patients were excluded from the current study if they received chemotherapy or radiotherapy prior to surgical resection. A total of 8 patients with glioma were diagnosed at World Health Organization stage II, 24 at stage III and 34 at stage IV. Furthermore, 15 normal brain tissues were also collected from healthy patients (9 male, 6 female; age, 33–57 years; median age, 46.7 years) via surgical resection at the same hospital between June 2013 and September 2016. Written informed consent was obtained from all patients. The clinicopathologic characteristics of the patients are summarized in Table I. Fresh tissues were stored at −80°C prior to further use.
Table I.
Association between miR-200a expression and clinicopathological characteristics in patients with glioma.
| Variables | Cases (n=66) | Low miR-200a expression (n=36) | High miR-200a expression (n=30) | P-value |
|---|---|---|---|---|
| Age (years) | 0.623 | |||
| <55 | 29 | 17 | 12 | |
| ≥55 | 37 | 19 | 18 | |
| Sex | 0.618 | |||
| Male | 39 | 20 | 19 | |
| Female | 27 | 16 | 11 | |
| WHO stage | 0.003 | |||
| II–III | 32 | 11 | 21 | |
| IV | 34 | 25 | 9 |
WHO, world health organization; miR, microRNA.
Cell culture
Several common human glioma cell lines (U251-MG, T98G, U-87MG Uppsala and U-373MG Uppsala) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Normal human astrocyte (NHA) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C, 5% CO2 and 95% O2.
Cell transfection
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.) was used to perform cell transfection according to the manufacturer's protocol. T98G and U251 cells were transfected with 100 nM miR-200a mimic (cat. no. HmiR0002-MR04; Guangzhou Fulengen Co., Ltd., Guangzhou, China), 100 nM miR-200a inhibitor (cat. no. HmiR-AN0298-SN-10; Guangzhou Fulengen Co., Ltd.) and 100 nM miR negative control (miR-NC; cat. no. CmiR0001-MR04; Guangzhou Fulengen Co., Ltd.). Cells were also co-transfected with 100 nM miR-200a mimic and 100 nM blank pcDNA3.1 vector (cat. no. ZL00008; Yearthbio, Changsha, China) or with 100 nM miR-200a mimic and 100 nM FOXA1 plasmid (cat. no. ZL00371; Yearthbio). Non-transfected cells were used as control. Further experimentation was performed 48 h following transfection.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or U251, T98G, U-87MG Uppsala, U-373MG Uppsala and NHA cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. Total RNA (1 µg) was then reverse transcribed into cDNA using the PrimeScript Reverse Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. qPCR reactions were performed using the SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) with a 7300 plus ABI system (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For the assessment of mRNA expression, GAPDH was used as internal reference. For the expression of miR-200a, U6 was used as internal reference. The thermocycling conditions were as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 15 sec and 60°C for 30 sec. Relative expression was analyzed using the 2−ΔΔCq method (26). The primers for FOXA1 (cat. no. HQP055418), GAPDH (cat. no. HQP006940), miR-200a (cat. no. HmiRQP0298) and U6 (cat. no. HmiRQP9001) were purchased from Guangzhou Fulengen Co., Ltd.
Dual luciferase reporter gene assay
TargetScan 7.1 software (www.targetscan.org) was used to assess the putative target genes of miR-200a. The mutant type (MT) or wild type (WT) 3′UTR of FOXA1 was amplified and cloned into the psiCHECK-2 vector (Promega Corporation, Madison, WI, USA) to construct luciferase reporter plasmids, which were obtained from Yearthbio. U251 and T98G cells were co-transfected with the WT FOXA1 plasmid or the MT FOXA1plasmid and miR-200a mimic or miR-NC, respectively, using Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). Following transfection for 48 h, a dual-luciferase reporter assay system (Promega Corporation) was used to examine the luciferase activity, according to the manufacturer's protocol. Renilla luciferase activity was normalized to Firefly luciferase activity.
Western blot analysis
T98G and U251 Cells were lysed using radio immunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.). Protein concentration was determined using a BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Protein (50 µg per lane) was separated using 12% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Thermo Fisher Scientific, Inc.). Membranes were then blocked with 5% evaporated milk at room temperature for 1 h and subsequently incubated with rabbit anti-human FOXA1 (1:200; cat. no. ab170933; Abcam, Cambridge, UK)or rabbit anti-human GAPDH (1:200; cat. no. ab9485; Abcam) antibodies overnight at 4°C. Membranes were further incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:5,000; cat. no. ab6721; Abcam) for 40 min at room temperature. Chemiluminescence was detected using the SuperSignal West Femto Maximum Sensitivity substrate (Thermo Fisher Scientific, Inc.). The quantity of protein was analyzed using Image J 1.45 software (National Institutes of Health, Bethesda, MD, USA).
MTT cell proliferation assay
U251 and T98G cells (5×103 cells/well) were plated into 96-well plates and cultured for different durations (0, 24, 48 or 72 h) at 37°C in a humidified atmosphere containing 5% CO2. A total of 10 µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added at different time points (0, 24, 48 or 72 h). Cells were then incubated at 37°C for 4 h. The supernatant was then removed and 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added. The absorbance value (optical density) was measured at 570 nm using a microplate reader.
EdU incorporation cell viability assay
U251 and T98G cells (5×103 cells/well) were seeded into 96-well plates and incubated at 37°C for 24 h. A total of 50 µM EdU (Guangzhou RiboBio Co., Ltd., Guangzhou, China) was then added. Following this, cells were incubated at 37°C for 2 h. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and then treated with 0.5% Triton-X-100 (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. Subsequently, 100 µl Apollo® Reaction Cocktail (Guangzhou RiboBio Co., Ltd.) was added and cells were incubated at room temperature for 30 min. Following staining with DAPI (5 µg/ml; Sigma-Aldrich; Merck KGaA) at room temperature for 30 min, cells were visualized under a fluorescent microscope (IX71; Olympus Corporation, Tokyo, Japan).
Transwell invasion assay
Matrigel pre-coated Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA) were utilized to examine cell invasion. T98G and U251 cell suspensions (3×105 cells/ml) were prepared using DMEM. Subsequently, 300 µl cell suspension was added into the upper chamber and 300 µl DMEM supplemented with 10% FBS was added into the lower chamber. Cells were incubated at 37°C for 24 h and the cells that did not invade through the membrane were carefully removed. The transmigrated cells were then fixed using 4% polyformaldehyde at room temperature for 30 min, stained with crystal violet (Sigma-Aldrich; Merck KGaA) at room temperature for 10 min and counted in five randomly selected microscopic fields using a light microscope (magnification, ×400). All experiments were performed in triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation. GraphPad Prism version 5.01 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform statistical analysis. Differences between two groups were analyzed using a Student's t-test and differences among more than two groups were analyzed using one-way analysis of variance followed by a Tukey's post-hoc test. Glioma patients were divided into high and low miR-200a expression groups, based on its mean expression value, and a χ2 test was used to analyze the results presented in Table I. A Pearson correlation test was used to determine the relationship between miR-200a and FOXA1expression. P<0.05 was considered to indicate a statistically significant result.
Results
miR-200a is downregulated in glioma
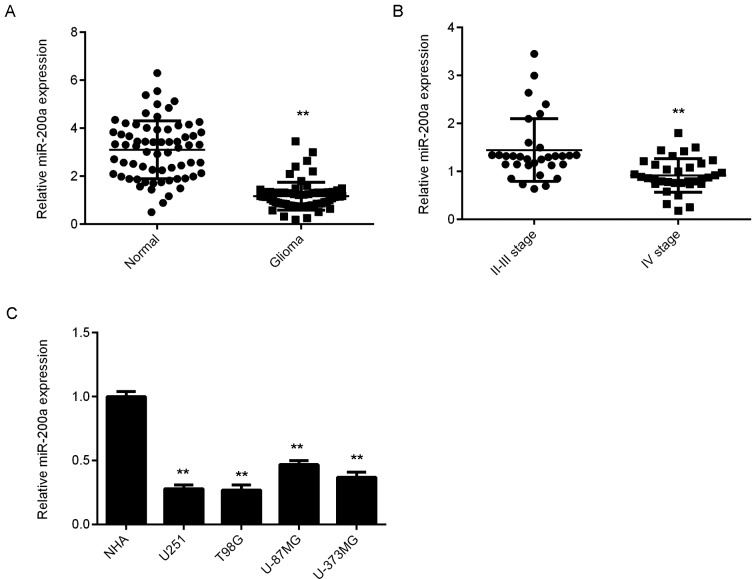
RT-qPCR was used to assess the expression of miR-200a in glioma and normal brain tissues. As presented in Fig. 1A, the levels of miR-200a were significantly lower in glioma tissues compared with normal brain tissues. Furthermore, grade IV glioma tissues demonstrated significantly lower miR-200a levels when compared with grade II–III glioma tissues (Fig. 1B). Subsequently, glioma patients were divided into high and low miR-200a expression groups, based on its mean expression value. As presented in Table I, the low expression of miR-200a was significantly associated with advanced clinical stage in glioma. In addition, miR-200a levels were significantly reduced in several common glioma cell lines compared with those in NHA cells (Fig. 1C). As T98G and U251 exhibited the lowest expression of miR-200a, they were used for the following experiments.
Figure 1.
(A) RT-qPCR data indicated that the expression of miR-200a was lower in glioma tissues compared with those in normal brain tissues. **P<0.01 vs. normal tissues. (B) RT-qPCR data indicated that the levels of miR-200a were lower in glioma tissues at stage IV compared with those at stages II–III. **P<0.01 vs. II–III stage. (C) RT-qPCR demonstrated that miR-200a was significantly downregulated in glioma cell lines compared with NHA cells. **P<0.01 vs. NHA. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; NHA, normal human astrocyte.
miR-200a inhibits glioma cell proliferation, survival, and invasion
T98G and U251 cells were transfected with miR-200a mimics to upregulate the expression of miR200a. The results indicated that miR-200a levels were significantly increased in the miR-200a group compared with the miR-NC group. However, transfection with miR-NC did not significantly affect the expression of miR-200a (Fig. 2A). The effects of miR-200a upregulation on glioma cells proliferation, survival and invasion were then assessed. As presented in Fig. 2B-D, the overexpression of miR-200a markedly reduced T98G and U251 cell proliferation (at 48 and 72 h), viability and invasion. These findings suggest that miR-200a demonstrated suppressive effects on the proliferation, survival and invasion of glioma cells.
Figure 2.
T98G and U251 cells were transfected with an miR-NC or miR-200a mimic (A) A reverse transcription-quantitative polymerase chain reaction assay was performed to determine the expression of miR-200a. Non-transfected cells were used as control. **P<0.01 vs. control. (B) MTT, (C) EdU and (D) Transwell assays were used to determine cell proliferation, viability and invasion, respectively. The magnification used in the EdU and transwell assays were ×40 and ×200, respectively. **P<0.01 vs. miR-NC. miR, microRNA; NC, negative control.
FOXA1 is a direct target gene of miR-200a in glioma cells
The potential downstream target genes of miR-200a in glioma were then assessed. TargetScan software data indicated that the 3′UTR of FOXA1 mRNA contained the binding sequences of miR-200a (Fig. 3A). To confirm this prediction, the present study constructed the WT and MT FOXA1 3′UTR luciferase reporter plasmids (Fig. 3B). Dual luciferase reporter gene assay data indicated that the luciferase activity was significantly reduced in glioma cells co-transfected with miR-200a mimics and WT reporter plasmids, when compared with cells transfected with WT reporter plasmids. However, this result was reversed following transfection with the MT reporter plasmid (Fig. 3C and D). These data indicate that FOXA1 is a target gene of miR-200a in glioma cells.
Figure 3.
(A) TargetScan software data indicated that the 3′UTR of FOXA1 mRNA contained the binding sequences of miR-200a. (B) The WT FOXA1-3′UTR and the MT FOXA1 3′UTR luciferase reporter plasmids were generated. (C) Dual luciferase reporter gene assay was then conducted in T98G cells. **P<0.01 vs. control. UTR, untranslated region; FOXA1, forkhead box A1; miR, microRNA; WT, wild type; MT, mutant type; PCT, the probability of conserved targeting.
FOXA1 is upregulated in glioma and inversely correlates to miR-200a expression
The expression of FOXA1 in glioma was further assessed. The results demonstrated that FOXA1 was significantly upregulated in glioma tissues compared with normal brain tissues (Fig. 4A). A negative correlation between FOXA1 and miR-200a expression in glioma tissues was also identified (Fig. 4B). Therefore, FOXA1was demonstrated to be upregulated in glioma, which may be due to the downregulation of miR-200a.
Figure 4.
(A) Reverse transcription-quantitative polymerase chain reaction data demonstrated that FOXA1 was significantly upregulated in glioma tissues compared with normal brain tissues. (B) A negative correlation was observed between FOXA1 and miR-200a expression in glioma tissues. **P<0.01 vs. normal. FOXA1, forkhead box A1; miR, microRNA.
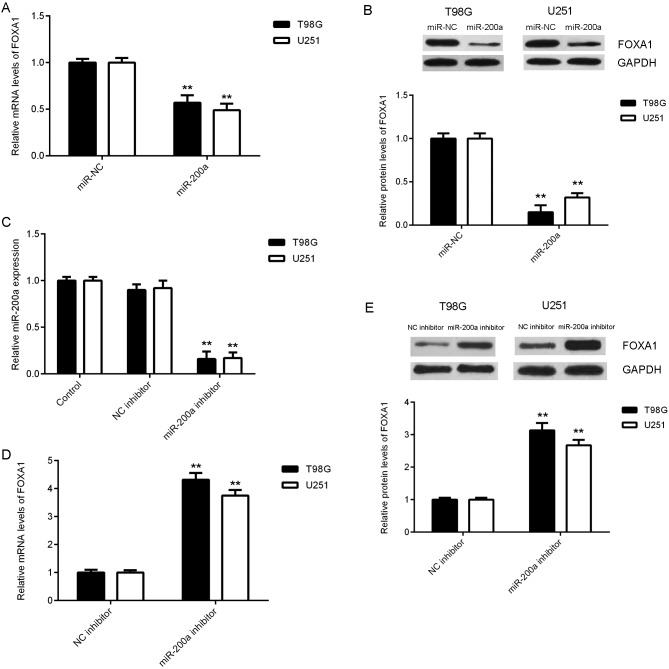
miR-200a negatively regulates FOXA1 expression in glioma cells
The effect of miR-200a on the expression of FOXA1in glioma cells was assessed. The results demonstrated that FOXA1 mRNA and protein expression was significantly reduced following miR-200a overexpression (Fig. 5A and B). miRNAs not only affect mRNA expression, but also function to inhibit protein translation (5–7). Due to this double effect, transfection with miR-200a mimics demonstrated a greater reduction in FOXA1 protein expression when compared with mRNA expression. T98G and U251 cells were transfected with amiR-200a inhibitor to knockdown its expression. RT-qPCR data revealed that miR-200a expression was significantly reduced in the miR-200a inhibitor group compared with the control group. Furthermore, no marked difference in miR-200a expression between the NC inhibitor and control group was identified (Fig. 5C). As presented in Fig. 5D and E, the expression of FOXA1 mRNA and protein was significantly increased in the miR-200a inhibitor group compared with the NC inhibitor group. These findings suggest that the expression of FOXA1 was negatively regulated by miR-200a in T98G and U251 cells.
Figure 5.
T98G and U251 cells were transfected with miR-NCs or miR-200a mimics, respectively. (A) RT-qPCR and (B) western blotting were performed to determine FOXA1 mRNA and protein levels. Non-transfected cells were used as the control group. **P<0.01 vs. miR-NC. (C) RT-qPCR was performed to determine the expression of miR-200a. **P<0.01 vs. control. (D) RT-qPCR and (E) western blotting were used to determine the expression of FOXA1 mRNA and protein, respectively. **P<0.01 vs. NC inhibitor. miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; FOXA1, forkhead box A1.
FOXA1 reduces the inhibitory effects of miR-200a on the malignant phenotypes of glioma cells
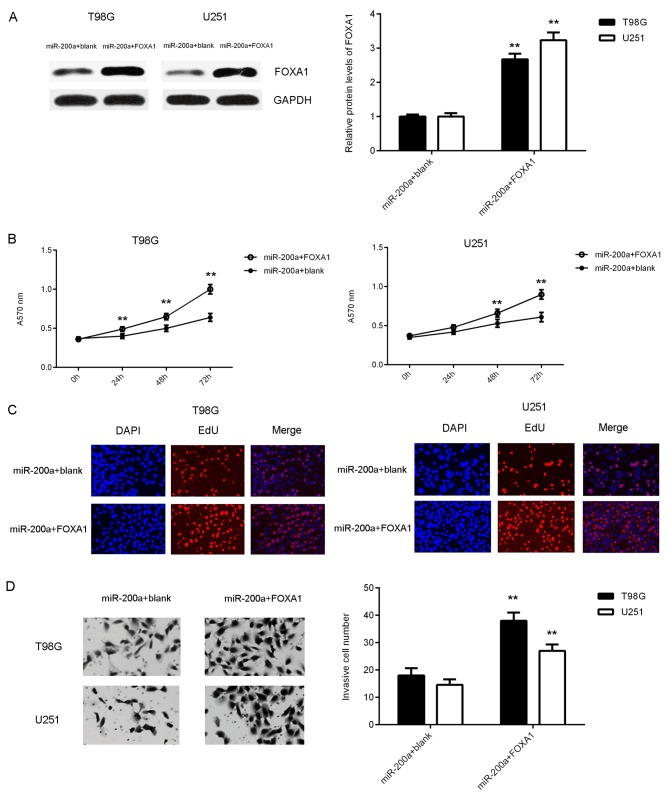
Rescue experiments were performed to assess whether FOXA1 acted as a downstream effecter of miR-200a in glioma cells. miR-200a-overexpressing T98G and U251 cells were transfected with FOXA1 expression plasmid or blank vectors (acting as the negative control group). Following transfection, FOXA1 protein levels were markedly higher in the miR-200a+FOXA1 group compared with those in the miR-200a+blank group. It was also demonstrated that the proliferation (at 48 and 72 h), viability and invasion of glioma cells were significantly increased in the miR-200a+FOXA1 group compared with those in the miR-200a+blank group (Fig. 6B-D). These results indicated that FOXA1 rescued the inhibitory effects of miR-200a by affecting the proliferation, viability and invasion of glioma cells.
Figure 6.
T98G and U251 cells which overexpressed miR-200a were transfected with a FOXA1 expression plasmid or blank vector. Following transfection, (A) western blotting was performed to examine the expression of FOXA1 protein. (B) An MTT, (C) EdU and (D) transwell assay were then performed to assess cell proliferation, viability and invasion, respectively. The magnification used for the EdU and transwell assays were ×40 and ×200, respectively. **P<0.01 vs. miR-200a+blank. miR, microRNA; FOXA1, forkhead box A1.
Discussion
The molecular mechanism of miR-200a underlying glioma progression remains largely unknown. However, the present study demonstrated that the level of miR-200a was significantly reduced in glioma tissues and cell lines. Low miR-200a expression was also negatively correlated with advanced stages of glioma. Furthermore, miR-200a overexpression inhibited glioma cell proliferation, survival and invasion. Results also identified that FOXA1 was a target gene of miR-200a in glioma cells and the increased expression of FOXA1 was negatively correlated with the decreased expression of miR-200a in glioma tissues. FOXA1 expression was also negatively regulated by miR-200a in glioma cells and the overexpression of FOXA1 rescued the inhibitory effects of miR-200a on the malignant phenotypes of glioma cells.
Previous studies have demonstrated that miR-200a serves key roles in multiple types of human cancer; however, studies that focus on miR-200a function in glioma are limited (20,27,28). Berthois et al (28) revealed that miR-200a was downregulated in glioblastoma multiforme (grade IV glioma) when compared with grade II–III glioma. These results were consistent with the data of the present study. Furthermore, Berthois et al (28) demonstrated that miR-200a levels were significantly higher in Timozolamine (TMZ)-responsive glial tumor cells compared to TMZ-unresponsive glial tumor cells in primary culture and that the overexpression of miR-200a in glioblastoma (GBM) cells promoted TMZ-sensitivity. Therefore, miR-200a may serve as a promising therapeutic target for GBM treatment. The reduced expression of miR-200a may also be due to the increased expression of the DNA repair enzyme O(6)-methylguanine methyltransferase (28) and long non-coding RNA activated by TGF-β, which has been demonstrated to serve an oncogenic role in glioma (27).
Su et al (20) demonstrated that SIM2 was a target gene of miR-200a in glioma cells, as the inhibition of miR-200a resulted in the upregulation of SIM2 in glioma cells and promoted glioma cell motility. It was also revealed that miR-200a knockdown promoted glioma growth in a mouse model (20). As one miRNA has many target genes (5), other target genes of miR-200 a may also exist and serve important roles in glioma. The current study identified FOXA1 as a novel target gene of miR-200a and revealed that the expression of FOXA1 was negatively regulated by miR-200a in glioma cells. FOXA1 has previously been reported to function as an oncogene in certain types of common human cancer, including prostate (23), endometrial (21) and breast cancer (29). For instance, FOXA1 enhanced the proliferation and migration of prostate cancer cells by modulating ELL-associated factor 2 regulation of androgen receptor transcriptional activity (23). Furthermore, Wang et al (30) previously revealed that the expression of FOXA1 in glioma tissues was significantly higher than those in corresponding non-neoplastic brain tissues, which was consistent with the results of the current study. In addition, the present study demonstrated that the expression of FOXA1 was upregulated in high-grade glioma tissue compared with low-grade tissue and that the overall survival of patients with glioma that exhibited high FOXA1 expression was shorter than those with low FOXA1 (30). This indicates that the upregulation of FOXA1 may serve a key role in glioma progression (30). The present study identified a negative correlation between FOXA1 and miR-200a expression in glioma tissues. These findings indicate that the reduced expression of miR-200a may contribute to the increased expression of FOXA1 in glioma tissues. Further investigation revealed that FOXA1 overexpression rescued the inhibitory effects of miR-200a on the proliferation, viability and invasion of glioma cells. Thus, FOXA1 may act as a key downstream effecter of miR-200a in glioma cells.
To the best of our knowledge, this is the first study which demonstrates that miR-200a inhibits glioma cell survival, proliferation and invasion partly through the inhibition of FOXA1 expression. These results indicate that miR-200a may be a potential therapeutic candidate for glioma.
Acknowledgements
Not applicable.
Funding
Not funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
KL and XC designed the present study and wrote the manuscript. BL, YZ and BZ collected clinical tissues. XC, LY, PY, WK, HH, and ET performed clinical and cell experimentation.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Brain Hospital of Hunan Province (Changsha, China). Written informed consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained for all patients.
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Song H, Zhang Y, Liu N, Wan C, Zhang D, Zhao S, Kong Y, Yuan L. miR-92b regulates glioma cells proliferation, migration, invasion, and apoptosis via PTEN/Akt signaling pathway. J Physiol Biochem. 2016;72:201–211. doi: 10.1007/s13105-016-0470-z. [DOI] [PubMed] [Google Scholar]
- 3.Gu J, Xu R, Li Y, Zhang J, Wang S. MicroRNA-218 modulates activities of glioma cells by targeting HMGB1. Am J Transl Res. 2016;8:3780–3790. [PMC free article] [PubMed] [Google Scholar]
- 4.Marumoto T, Saya H. Molecular biology of glioma. Adv Exp Med Biol. 2012;746:2–11. doi: 10.1007/978-1-4614-3146-6_1. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol Res. 2017;25:397–405. doi: 10.3727/096504016X14743337535446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang M, Zhai X, Ge T, Yang C, Lou G. MiR-181a-5p promotes proliferation and invasion, and inhibits apoptosis of cervical cancer cells via regulating inositol polyphosphate-5-phosphatase A (INPP5A) Oncol Res. 2018;26:703–712. doi: 10.3727/096504017X14982569377511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wang Y, Li J, Xu C, Zhang X. MicroRNA-139-5p inhibit cell proliferation and invasion by targeting RHO-associated Coiled-coil containing protein kinase 2 in ovarian cancer. Oncol Res. 2017 Jun 14; doi: 10.3727/096504017X14974343584989. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, Zhou B, Liu M, Liu Y, Gao R. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25:463–470. doi: 10.3727/096504016X14685034103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Zhao H, Rao M, Xu S. MicroRNA-365 inhibits proliferation, migration and invasion of glioma by targeting PIK3R3. Oncol Rep. 2017;37:2185–2192. doi: 10.3892/or.2017.5458. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Ma G, Zhang Y, Huo H, Zhao Y. miR-599 inhibits proliferation and invasion of glioma by targeting periostin. Biotechnol Lett. 2017;39:1325–1333. doi: 10.1007/s10529-017-2365-7. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Zeng A, Hu Q, Yan W, Liu Y, You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55–65. doi: 10.1093/neuonc/now129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama K, Ishibashi O, Kawase R, Kurose K, Takeshita T. miR-200a, miR-200b and miR-429 are Onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Anticancer Res. 2015;35:1401–1410. [PubMed] [Google Scholar]
- 15.Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao Y, Ma B, Wang X, Wu N, Li X, et al. MicroRNA-200a suppresses metastatic potential of side population cells in human hepatocellular carcinoma by decreasing ZEB2. Oncotarget. 2015;6:7918–7829. doi: 10.18632/oncotarget.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, Wu J, Shen ZZ, Song HY, Shao ZM. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389–1399. doi: 10.1158/1078-0432.CCR-12-1959. [DOI] [PubMed] [Google Scholar]
- 17.Guo T, Zhang Y, Qu X, Che X, Li C, Fan Y, Wan X, Ma R, Hou K, Zhou H, et al. miR-200a enhances TRAIL-induced apoptosis in gastric cancer cells by targeting A20. Cell Biol Int. 2018;42:506–514. doi: 10.1002/cbin.10924. [DOI] [PubMed] [Google Scholar]
- 18.Zang Y, Tai Y, Wan B, Jia X. miR-200a-3p promotes the proliferation of human esophageal cancer cells by post-transcriptionally regulating cytoplasmic collapsin response mediator protein-1. Int J Mol Med. 2016;38:1558–1564. doi: 10.3892/ijmm.2016.2758. [DOI] [PubMed] [Google Scholar]
- 19.Peng F, Jiang J, Yu Y, Tian R, Guo X, Li X, Shen M, Xu M, Zhu F, Shi C, et al. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br J Cancer. 2013;109:3092–3104. doi: 10.1038/bjc.2013.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, He Q, Deng L, Wang J, Liu Q, Wang D, Huang Q, Li G. MiR-200a impairs glioma cell growth, migration, and invasion by targeting SIM2-s. Neuroreport. 2014;25:12–17. doi: 10.1097/WNR.0000000000000032. [DOI] [PubMed] [Google Scholar]
- 21.Qiu M, Bao W, Wang J, Yang T, He X, Liao Y, Wan X. FOXA1 promotes tumor cell proliferation through AR involving the Notch pathway in endometrial cancer. BMC Cancer. 2014;14:78. doi: 10.1186/1471-2407-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson JL, Carroll JS. FoxA1 is a key mediator of hormonal response in breast and prostate cancer. Front Endocrinol (Lausanne) 2012;3:68. doi: 10.3389/fendo.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo W, Keener AL, Jing Y, Cai L, Ai J, Zhang J, Fisher AL, Fu G, Wang Z. FOXA1 modulates EAF2 regulation of AR transcriptional activity, cell proliferation, and migration in prostate cancer cells. Prostate. 2015;75:976–987. doi: 10.1002/pros.22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horimoto Y, Arakawa A, Harada-Shoji N, Sonoue H, Yoshida Y, Himuro T, Igari F, Tokuda E, Mamat O, Tanabe M, et al. Low FOXA1 expression predicts good response to neo-adjuvant chemotherapy resulting in good outcomes for luminal HER2-negative breast cancer cases. Br J Cancer. 2015;112:345–351. doi: 10.1038/bjc.2014.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Chen B, Yue B, Yang J. MicroRNA-212 suppresses the proliferation and migration of osteosarcoma cells by targeting forkhead box protein A1. Exp Ther Med. 2016;12:4135–4141. doi: 10.3892/etm.2016.3880. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthois Y, Delfino C, Metellus P, Fina F, Nanni-Metellus I, Al Aswy H, Pirisi V, Ouafik L, Boudouresque F. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: Evidence that miR200a-3p is regulated by O6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol Ther. 2014;15:938–950. doi: 10.4161/cbt.28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, Qian B, Tian D, Tang T, Wan S, Wang L, Zhu L, Geng X. FOXA1 positively regulates gene expression by changing gene methylation status in human breast cancer MCF-7 cells. Int J Clin Exp Pathol. 2015;8:96–106. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Qin H, Li L, Feng F, Ji P, Zhang J, Li G, Zhao Z, Gao G. Forkhead-box A1 transcription factor is a novel adverse prognosis marker in human glioma. J Clin Neurosci. 2013;20:654–658. doi: 10.1016/j.jocn.2012.03.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.