Abstract
Leptin deficiency is principally linked to metabolic disorders. Leptin knockout (LepΔI14/ΔI14) Sprague Dawley rats created by CRISPR/Cas9 is a new model to study metabolic disorders. We used a whole rat genome oligonucleotide microarray to obtain tissue-specific gene expression profiles of the white adipose tissue, liver and hypothalamus inLepΔI14/ΔI14 and wild-type (WT) rats. We found 1,651 differentially expressed (enriched) genes in white adipose tissue, 916 in the liver, and 306 in the hypothalamus in theLepΔI14/ΔI14 rats compared to WT. Gene ontology category and KEGG pathway analysis of the relationships among differentially expressed genes showed that these genes were represented in a variety of functional categories, including fatty acid metabolism, molecular transducers and cellular processes. The reliability of the data obtained from microarray was verified by quantitative real-time PCR on 14 representative genes. These data will contribute to a greater understanding of different metabolic disorders, such as obesity and diabetes.
Keywords: LepΔI14/ΔI14, microarray analysis, white adipose, liver, hypothalamus
Introduction
Obesity and metabolism-related diseases pose increasing risks to public health[1]. Leptin is a 16 kDa protein and principally produced in white adipose tissues (WAT), as well as the placenta, mammary gland, ovary, skeletal muscle, stomach, and pituitary gland[2]. It functions to reduce feeding and promote energy expenditure, circulating leptin levels are proportional to body fat content[2–3]. The anorexic potential of leptin indicates that this cytokine-like hormone represents a highly promising approach to the treatment of obesity-related metabolic diseases.
Leptin deficiency is closely associated with metabolic disorders, with liver and WAT as the main target tissues. Leptin-deficient obeseob/ob mice exhibit hepatic lipid accumulation[4] and reduced hepatic mitochondrial content and function[5]. Furthermore, in this model, upregulated de novo lipogenesis has also been shown to contribute to obesity-associated nonalcoholic fatty liver disease[5], with increased circulating glycerol levels combined with increased subcutan-eous fat expression of the aquaglyceroporinsAQP3 and AQP7 also observed[6]. In a comparative proteomic analysis of adipose tissue of leptin-deficientob/ob mice treated with leptin or control buffer, 12 differentially expressed functional protein groups were reported[7], which suggesting that leptin-deficiency influences lipid metabolism. Moreover, WAT transplantation normalized metabolic abnormalities (glycemia, alanine aminotransferase, and liver weight) in ob/ob mice and prevented further bodyweight gain[8], thus, demonstrating that leptin deficiency indeed interferes with lipid metabolism, especially in the WAT. The hypothalamus has recently been found to be involved in the phenotypes caused by leptin deficiency. Bouyeret al. demonstrated that neonatal exposure of leptin-deficient mice to leptin improved metabolic status with specific cell-type patterns observed in the arcuate nucleus of the hypo-thalamus[3].
Although numerous studies have been conducted focusing on the metabolic disorders caused by leptin deficiency, the mechanisms underlying these metabolic abnormalities remain to be fully elucidated.Leptin knockout animal models have attracted increasing interest as a means to investigate metabolic diseases, such as obesity, type 2 diabetes and fatty liver disease. Homozygousob/ob and db/db mice have been used in most leptin deficiency studies[9–11]. Until recently, the LepΔI14/ΔI14 rat generated by Cas9 gene knockout technology and zinc finger nuclease technology, was the only animal model of global leptin deficiency[12].
In this study, we performed a comprehensive evaluation of gene expression alterations of WAT, liver and hypothalamus ofleptin knockout (LepΔI14/ΔI14) rats created using Cas9 gene knockout technology. We utilized hematoxylin-eosin staining to examine the histology of fresh white and brown adipose tissue and liver inLepΔI14/ΔI14 and WT rats. Microarray analysis was performed to generate profiles of metabolic gene expression in the WAT, liver and hypothalamus. Furthermore, we performed gene ontology category (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the relationships among differentially expressed genes (DEGs) to explore the possible pathways involved in metabolic diseases associated with leptin deficiency. At last, we selected 14 representative genes on the basis of their functional properties and the fold change in their expression levels to perform quantitative real-time PCR, thus verifying the reliability of the data obtained from the microarray studies.
Materials and methods
Animals
Four-month-old LepΔI14/ΔI14 Sprague Dawley (SD) rats (3 males, 3 females) and littermate wild-type (WT) (3 males, 3 females) were from Zhu's laboratory in Tongji University. They were successfully created by one-step zygotic injection ofCas9 mRNA and sgRNAs targeting rat Lep exons.
The LepΔI14/ΔI14 rats were found to carry homozygous deletions of 3 nucleotides (ATC) which encoded isoleucine at position 14 (I14) in the mature LEP protein. They exhibited similar mutant phenotypes toLEP- and LEPR- null rats. Molecular analyses showed that I14 played a key role for the interaction between LEP and LEPR and the down streaming JAK2-STAT3 pathways[13].
In Zhu's Laboratory, body weight and food consumption of rats were recorded. Glucose and insulin tolerance tests, and serum biochemistry were performed[13].
On arriving at Nanjing Medical University, rats were housed with a 10-hour light, 14-hour dark cycle and with food and water availablead libitum for a week. All animal procedures were conducted in accordance with approval of the local Animal Care and Use committee of Nanjing Medical University.
Sample collection and storage
Rats were euthanized and blood samples were collected by cardiac puncture. Samples of liver, white adipose tissues (visceral), and brown fat (interscapular) were collected. Half of the samples were snapped frozen in liquid nitrogen and stored in -80°C freezer for mRNA analysis later. Half were fixed in 4% paraformaldehyde for at least 24 hours. The brains were also quickly removed from the skull and chilled in powdered ice. Then it was placed (with ventral surface facing up) in an ice-cold stainless steel brain matrix with slots at 1 mm intervals (Ted PellaInc, Redding CA, models 15005 for adult rat 125–185 g, coronal). Since the hypothalamus is related to regulation of metabolism[3], we tried to isolate this area. Razor blades were placed at the appropriate slots of the brain matrix to get a 5 mm slice spanning the whole median eminence, which covered the sections levels 18–30 in Swanson's atlas of the rat brain[16]. An isosceles triangle-shaped piece was cut out with the apex of the triangle positioned at the center of the third ventricle with the basic angle 45°. Hypothalamus pieces were snap frozen and stored at -70°C for further analysis[17].
Serum leptin measurement
Blood samples were centrifuged at 3,000 rpm for 10 minutes at 4°C to obtain the serum. Serum leptin levels were tested using rat leptin ELISA kits (Millipore). Duplicate samples were run at a volume of 10 μL each. The minimum detectable level of leptin was 0.2 ng/mL per tube. The inter-assay coefficients of variation were 3.93%.
Histological assessment
Half samples of white and brown adipose and liver were fixed with 4% paraformaldehyde and embedded in paraffin. Four µm-thick paraffin-embedded sections were cut and processed for hematoxylin-eosin (H&E) staining. Samples were examined under 200 × magnification with a light microscope (NIKON ECLIPSE CI-S, Japan). To analyze the adipose tissue further, five sections of white or brown adipose in each rat were chosen and three images of each section were captured with a CCD camera (Carl Zeiss) using AxioVersion 4.5 software (Carl Zeiss) at onetime with same light exposure time. Images were processed and analyzed for adipocyte cell size using Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA). White or brown adipocyte cell size were calculated as mean of five sections.
Microarray analysis
WAT, liver and hypothalamus samples were used to investigate the molecular mechanisms of leptin-associated metabolic diseases as below.
Fabrication of DNA microarrays
The rat mRNA array designed by Agilent (Palo Alto, California, USA) was prepared with eight identical arrays per slide, each array containing probes interrogating 30,003 Entrez Gene RNAs. The arrays also contained 139 Agilent control probes (Supplementary Table 1, available online).
RNA extraction
WAT, liver and hypothalamus samples were sent to CapitalBio (Beijing, China) for microarray hybridization. Total RNA of WAT, liver and hypothalamus samples was extracted using Trizol reagent (Invitrogen) and purified with the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to manufacturers' instructions. The purity and concentration of RNA were determined from OD260/280 readings using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA integrity was determined by capillary electrophoresis using the RNA 6000 Nano Laboratory-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Only RNA extracts with RNA integrity number values greater than 6 underwent further analysis.
RNA amplification, labeling and hybridization
Eberwine's linear RNA amplification method followed by subsequent enzymatic reaction was used to produce cDNA labeled with a fluorescent dye (Cy5 and Cy3-dCTP) as described previously[18–19] and the procedure has been improved by using CapitalBio cRNA Amplification and Labeling Kit (CapitalBio) to yield more labeled cDNA.
In detail, double-stranded cDNAs, which contain the T7 RNA polymerase promoter sequence, were synthesized from 100 ng total RNA using the CbcScript reverse transcriptase with cDNA synthesis system according to the manufacturer's protocol (Capitalbio) with the T7 Oligo (dT). After double-stranded cDNA (dsDNA) synthesis using DNA polymerase and RNase H was completed, the dsDNA products were purified using a PCR NucleoSpin Extract II Kit (MN) and eluted with 30 μL elution buffer. The eluted double-stranded cDNA products were vacum evaporated to 16 μL and subjected to 40 μLin vitro transcription reactions at 37°C for 14 hours using T7 Enzyme Mix. The RNA Clean-up Kit (MN) was used to purifying the amplified cRNA.
Klenow enzyme labeling strategy was adopted after reverse transcription using CbcScript II reverse transcriptase. Briefly, 2 μg amplified RNA was mixed with 4 μg random nanomer, denatured at 65°C for 5 minutes and cooled on ice. Then, 5 μL of 4 × first-strand buffer, 2 μL of 0.1 μmol/L DTT and 1.5 μL of CbcScript II reverse transcriptase were added. The mixtures were incubated at 25°C for 10 minutes, then at 37°C for 90 minutes. The cDNA products were purified using a PCR NucleoSpin Extract II Kit (MN) and vacuum evaporated to 14 μL. The cDNA was mixed with 4 μg random nanomer, heated to 95°C for 3 minutes, and snap cooled on ice for 5 minutes. Then, 5 μL Klenow buffer, dNTP, and Cy5-dCTP or Cy3-dCTP (GE Healthcare) were added, bringing the final concentrations to 240 μmol/L dATP, 240 μmol/L dGTP, 240 μmol/L dTTP, 120 μmol/L dCTP, and 40 μmol/L Cy-dCTP. Then, 1.2 μL Klenow enzyme was added, and the reaction was going on at 37°C for 90 minutes. Labeled cDNA was purified with a PCR NucleoSpin Extract II Kit (MN) and re-suspended in elution buffer. Labeled controls and test samples labeled with Cy5-dCTP and Cy3-dCTP were dissolved in 80 μL hybridization solution containing 3 × SSC, 0.2% SDS, 5 × Denhardt's solution and 25% formamide. DNA in hybridization solution was denatured at 95°C for 3 minutes before loading onto a microarray. Arrays were hybridized in a Agilent Hybridization Oven at a rotation speed of 20 rpm at 42°C overnight and washed with two consecutive solutions (0.2% SDS, 2° SSC at 42°C for 5 minutes, and 0.2° SSC for 5 minutes at room temperature).
Microarray imaging and data analysis
The array data were summarized, normalized and assessed for quality control using the GeneSpring analysis software V12 (Agilent) . To select the DEGs, threshold values of ≥2 and ≤−2 fold change and a Benjamini-Hochberg were used to correctP-value of 0.05. The data was Log2 transformed and median centered by genes using the Adjust Data function of the CLUSTER 3.0 software and then further analyzed for hierarchical clustering with average linkage. Finally, we performed phylogenetic tree visualization using Java Treeview (Stanford University School of Medicine, Stanford, CA, USA).
GO category, pathway analysis and STRING analysis
GO and pathway analysis were conducted using Http://bioinfo.capitalbio.com/mas3/system. The GO project provides a controlled vocabulary which describes genes and gene products attributed to some organisms. The "elim Fisher" algorithm described by Adrian Alexa was used for the GO enrichment test to iteratively remove any genes mapped to significant GO terms from more general (higher level) GO terms, which prevents the general GO terms from masking the significant terms[20]. Gene ontology categories with aP-value<0.05 were shown and organized into three domains: biological process, cellular component and molecular function.
For the pathway analysis, microarray gene pathway annotations (downloaded from KEGG (http://www.genome.jp/kegg/) were used to determine significant pathways for DEGs. To find significant enrichment for pathways, a Fisher exact test was used and the resulting P values were adjusted using the BH FDR algorithm[21]; pathway categories with a FDR smaller than 0.05 were shown.
To analyze DEGs in WAT, liver and hypothalamus from LepΔI14/ΔI14 and WT rats, a database of known and predicted protein interactions called STRING (http://string-db.org/) was used.
Quantitative real-time PCR
Quantitative real-time PCR analysis was performed to validate 14 representative genes (Lep, Adipor2, Ffar4, Fabp3, Fitm1, Cyp2c13, Lcn2, Gpam, Ppara, Wnt9b, Wnt3, Kiss1, Slc6a13, Pomc) from WAT, liver and hypothalamus in LepΔI14/ΔI14 and WT rats, which were selected according to microarray data. Total RNA was reverse transcribed using the PrimeScriptTM reagent Kit (Takara Bio, Tokyo, Japan) according to the manufacturer's instructions, with β-actin serving as an endogenous control. The cDNA samples were used for quantitative real-time PCR analysis, which was performed using an ABI Prism 7900HT System (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex TaqTM (Takara Bio, Tokyo, Japan) according to the manufacturer's instructions. Amplification conditions were 95°C for 10 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 31 seconds. Each sample was assayed in triplicate in parallel with the endogenous control for normalization of the data. After completion of the PCR amplification, the relative fold change in expression after stimulation was calculated based on the 2−ΔΔCT method.
Statistical analysis
Results were expressed as mean±SEM. Statistical analysis was conducted using SPSS (version 19.0, Sun Microsystems, USA). Differences in variables between the 2 groups were analyzed using Student'st-test. The criterion for statistical significance was P<0.05.
Results
Leptin knockout induced obesity
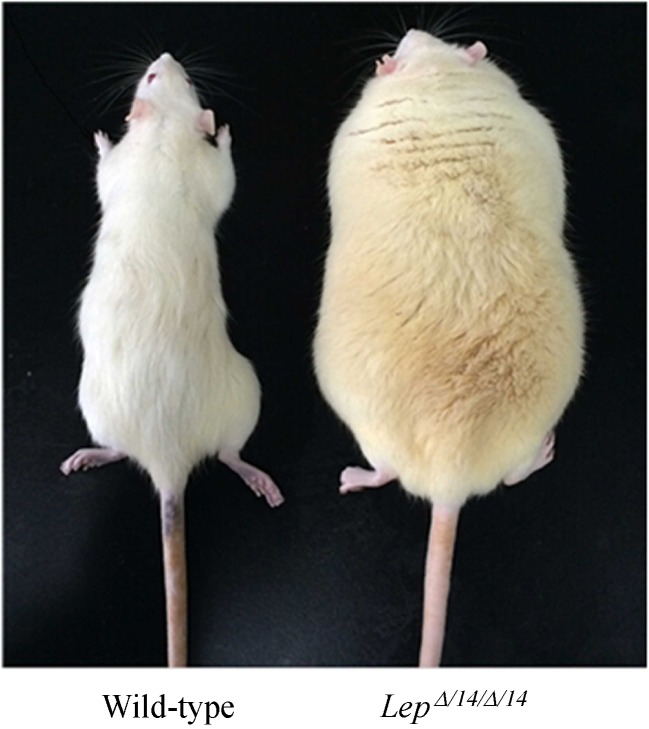
LepΔI14/ΔI14 rats presented significant obesity compared with their WT littermates at the age of four months (Fig. 1). The previous paper showed that LEP-deficiency induced obesity, glucose intolerance, and hyperinsulinemia and significantly increased circulating triglycerides, total cholesterol, high density lipoprotein and low density lipoprotein inLepΔI14/ΔI14 rats[13].
Fig.1.
Representative Wild-type and LepΔI14/ΔI14 rats.
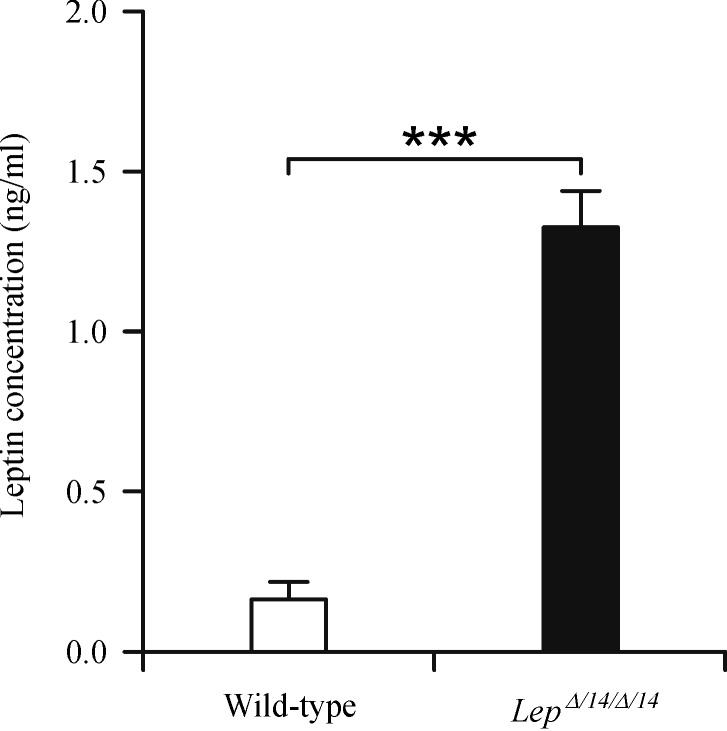
Serum leptin concentration
LepΔI14/ΔI14 rats had significantly higher serum leptin level compared with their WT littermates (P<0.001) (Fig. 2).
Fig.2.
Serum leptin levels in Wild-type and LepΔI14/ΔI14 rats.
Histological analysis of the white adipose, brown adipose and liver
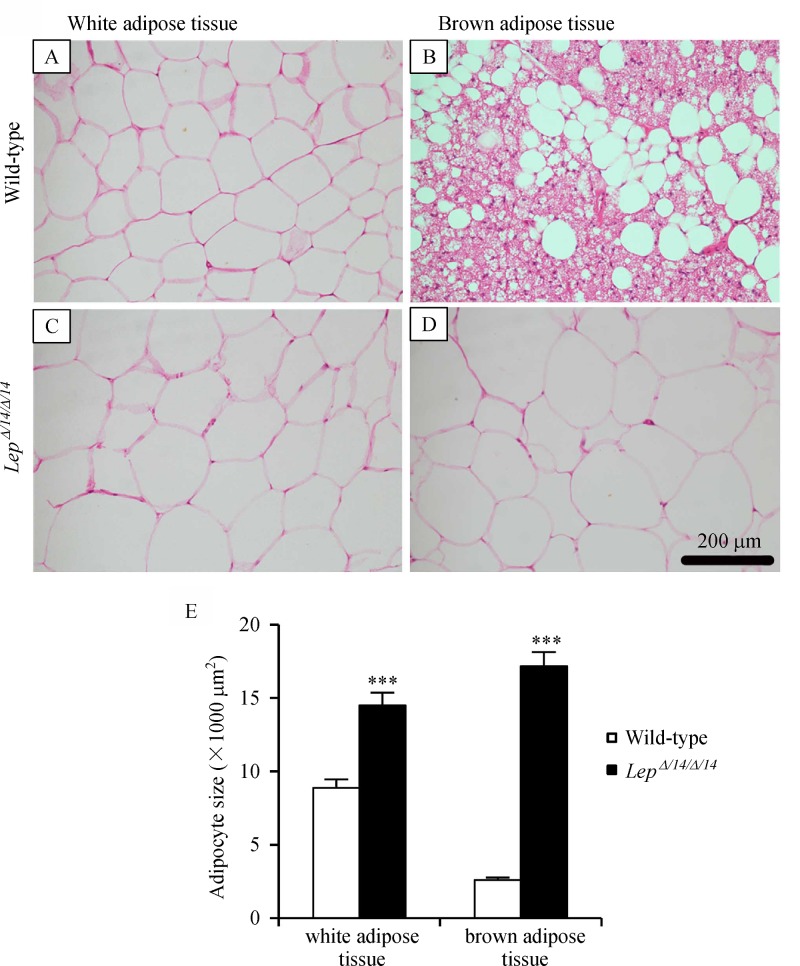
Visual inspection showed that the size of white adipose tissue in LepΔI14/ΔI14 rats was obviously larger than in WT (Fig. 3A&C). Quantitive analysis also found that the average size of white adipose tissue in LepΔI14/ΔI14 rats was significantly larger than in WT (P<0.001) (Fig. 3E).
While wide distribution of numerous small lipid droplets were observed in brown adipose tissue in WT rats, brown adipose tissue inLepΔI14/ΔI14 rats looked like large white adipose cells (Fig. 3B&D). Quantitive analysis showed that the average size of brown adipose tissue in LepΔI14/ΔI14 rats was significantly larger than in WT (P<0.001) (Fig. 3E).
Fig.3.
Morphological difference of adipose tissue between LepΔI14/ΔI14 and WT rats.
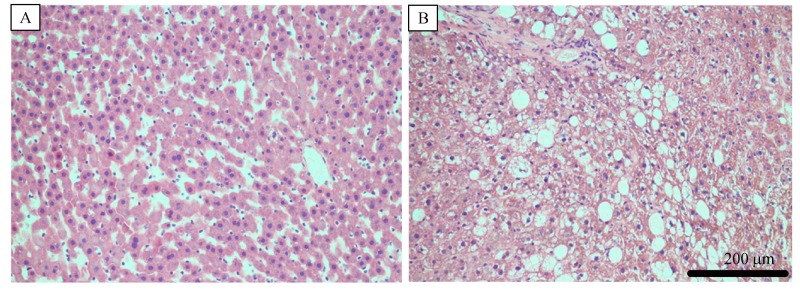
The liver from WT rats exhibited no signs of steatosis with a normal, clear and regular lobular architecture with central veins and radiating hepatic cords (Fig. 4A). In contrast, LepΔI14/ΔI14 rats liver exhibited massive steatosis, with abundant accumulation of fat droplets in hepatocytes, severely damaging central veins and radiating hepatic cords, and numerous infiltrating lymphocytes (Fig. 4B), which provided biochemical evidence of steatohepatitis.
Fig.4.
Representative H&E stained sections of liver of WT (A) and LepΔI14/ΔI14 (B) rats.
Microarray analysis
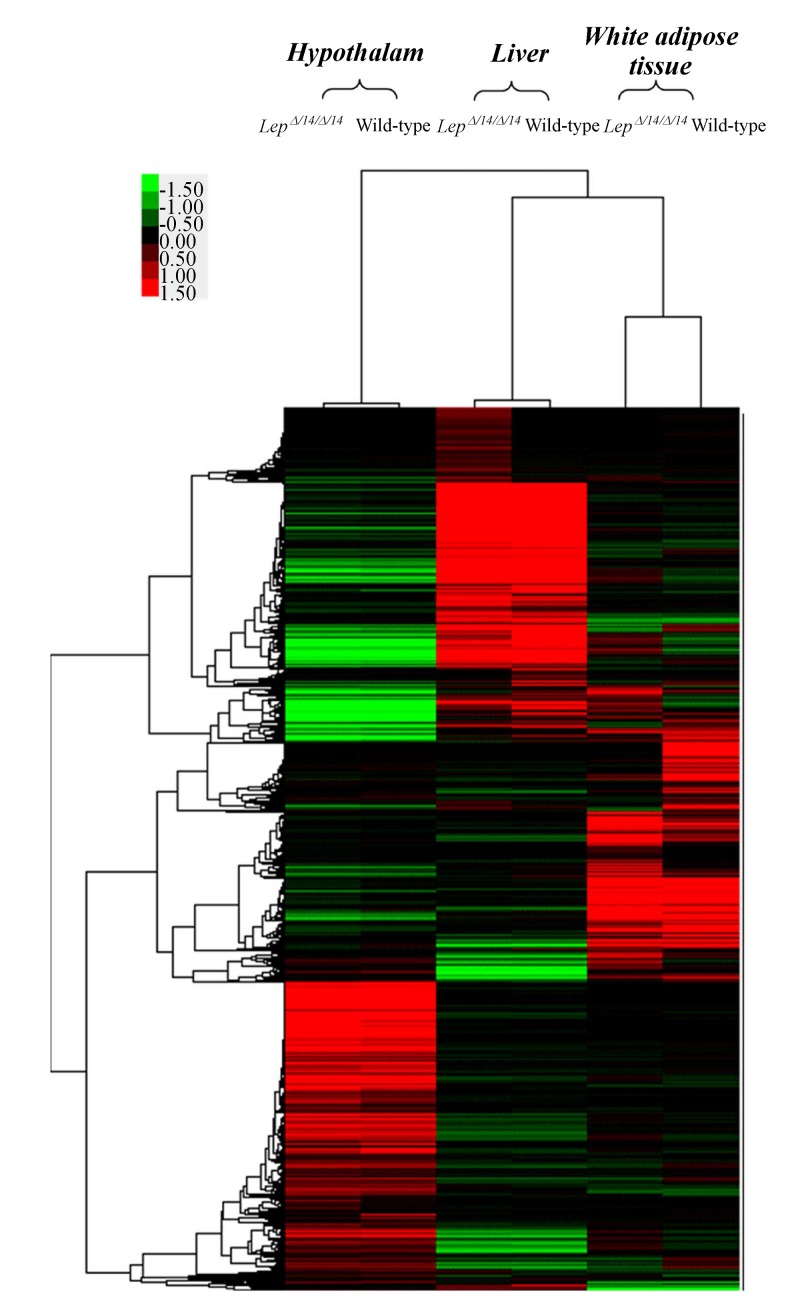
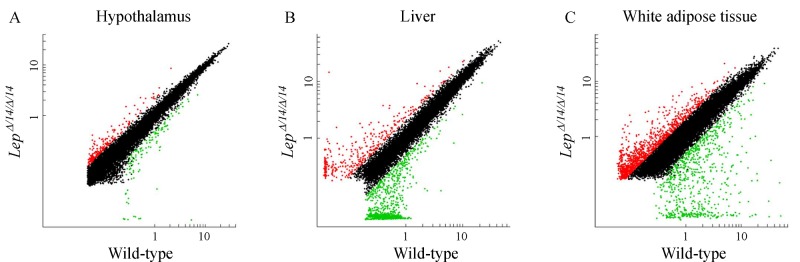
The hierarchical clustering of gene expression profiles revealed a discrepancy between LepΔI14/ΔI14 and WT rats (Fig. 5 and Supplementary File 2) and scatter plots displayed further variations in gene expression in different tissues (Fig. 6). These results confirmed that leptin gene deficiency influences the expressions of other genes. Gene upregulation was defined as a fold change in relative transcription levels log FC ≥1 and FDR ≤ 0.05, while downregulation was defined as a log FC ≤ –1 and FDR ≤ 0.05. Genes with relative transcription levels of –1 ≤ log FC ≤ 1 were considered to show no notable change. Our analysis revealed 1,651 transcripts inLepΔI14/ΔI14 rats differed significantly from that of the WT rats in WAT, 916 in liver and 306 in hypothalamus (Supplementary Table 2-4, available online).
Fig.5.
Dendrogram of 10,000 tissue-specific differentially expressed genes in the hypothalamus, liver, white adipose tissue ofLepΔI14/ΔI14 and WT rats.
Fig.6.
Scatter plots of gene expression in hypothalamus (A), liver (B) and white adipose tissue (C) of LepΔI14/ΔI14 and WT rats.
The reliability of the data obtained from the microarray analysis was verified by performing quantitative real-time PCR on 14 representative genes selected on the basis of their functional properties and the fold change in their expressions between the two different groups in the microarray analysis (Table 1). The results showed that there were significant difference in the mRNA expressions of the 14 selected genes betweenLepΔI14/ΔI14 rats and WT groups and their upregulations or downregulations were consistent with Microarray analysis.
Profile of metabolism-related genes in the white adipose tissue
Of the 1,651 genes differently expressed in WAT in LepΔI14/ΔI14 rats, 985 genes were upregulated and 666 were downregulated, including a number of metabolism-related genes. Tissue factor pathway inhibitor (Tfpi) (a lipoprotein-associated coagulation inhibitor) and apolipoprotein F (apoF) (a 29 kDa secreted sialoglycoprotein present in the HDL and LDL fractions of human plasma) displayed upregulated expression[22] while fatty acid binding protein 3 (Fabp3), lipopolysaccharide binding protein (Lbp) and fat storage-inducing transmembrane protein 1 (Fitm1) were downregulated (Supplementary Table 2, available online).
Profile of metabolism-related genes in the liver
In the liver, we detected 916 DEGs; 280 were upregulated and 636 were downregulated. These also included some metabolism-related genes:Fabp4 (encoding the fatty acid binding protein found in adipocytes) and glycerol-3-phosphate acyltransferase and mitochondrial (Gpam) (encoding a mitochondrial enzyme which utilizes saturated fatty acids as its preferential substrate for the synthesis of glycerolipids), were both upregulated, while peroxisome proliferator-activated receptor alpha (Ppara) and Fabp12 were downregulated (Supplementary Table 3, available online).
Profile of metabolism-related genes in the hypothalamus
In the hypothalamus, 306 DEGs were identified; 188 were upregulated and 118 were downregulated. The metabolism-related genes, the gamma-aminobutyric acid A (GABAA) receptor, alpha 4 (Gabr~4) was upregulated, while the KiSS-1 metastasis-suppressor and proopiomelanocortin (Pomc) were downregulated (Supplementary Table 4, available online).
Gene ontology category
We further performed GO analysis for the DEGs in the WAT, liver and hypothalamus of LepΔI14/ΔI14 and WT rats to determine gene product enrichment associated with cellular components, molecular functions and biological processes. The DEGs involved in significant GO categories are summarized inSupplementary Table 5–7, available online.
After GO analysis, we identified that these significantly different transcripts were mainly associated with cellular components (cell, cell part, envelope, extracellular region, extracellular region part, macromolecular complex, membrane-enclosed lumen, organelle, organelle part, synapse, synapse part, virion and virion part,) with the highest enriched GO categories in all three tissues being associated with cell, molecular functions (antioxdant, auxiliary transport protein, binding, catalytic, chemoattractant, electron carrier, enzyme regulator, molecular transducer, structural molecule, transcription regulator, translation regular and transporter) with binding taking account of the highest enriched GOs in the three different tissues, biological processes (ranging from anatomical structure formation to viral reproduction) with the cellular process being the most enriched GO category (Supplementary Fig. 1, available online).
KEGG pathway analysis
KEGG pathway analyses of the DEGs in the WAT, liver, and hypothalamus of LepΔI14/ΔI14 and WT rats showed close associations of both upregulated and downregulated genes with various metabolic pathways. DEGs in the WAT were associated with 12 metabolic pathways: the MAPK, wnt, adipocytokine, insulin, PPAR and Jak-STAT signaling pathways, biosynthesis of unsaturated fatty acids, metabolism of galactose, fatty acid, glycerolipid, and ether lipid and type I diabetes mellitus. Among these pathways, the majority of the DEGs were associated with the MAPK signaling pathway, while only five were associated with the fatty acid or ether lipid metabolic pathways (Table 2). DEGs were found to be associated with eight metabolic pathways in the liver: the MAPK, Jak-STAT, adipocytokine, PPAR, wnt and calcium signaling pathways, regulation of actin cytoskeleton, and neuroactive ligand-receptor interaction. Most of the DEGs were associated with the MAPK signaling pathway, while the fewest were associated with the wnt signaling pathway (Table 3). DEGs in the hypothalamus were associated with another seven metabolic pathways: neuroactive ligand-receptor interaction, type I diabetes mellitus, the calcium, wnt and adipocytokine signaling pathways, basal transcription factors, and glycine, serine and threonine metabolism. Most of the DEGs were associated with the neuroactive ligand-receptor interaction, while the fewest were associated with basal transcription factors, glycine, serine and threonine metabolism and the adipocytokine signaling pathway (Table 4).
Tab.1.
Comparison between microarray results and real-time quantitative PCR results on 14 representative genes
| NCBI Reference Sequence | Gene | Microarray LogFC | RT-PCR LogFC |
|---|---|---|---|
| Gene in WAT | |||
| NM_013076.3 | Lep | 2.7937646 | 2.222989333 |
| NM_001037979.1 | Adipor2 | 1.885395 | 1.529276611 |
| NM_001047088.1 | Ffar4 | 1.3467178 | 1.157427444 |
| NM_024162.1 | Fabp3 | -1.5540028 | -1.906396944 |
| NM_001106037.1 | Fitm1 | -4.693125 | -1.497721444 |
| Gene in Liver | |||
| NM_138514.1 | Cyp2c13 | 1.6456175 | 1.218 |
| NM_130741.1 | Lcn2 | 1.2924156 | 3.5169335 |
| NM_017274.1 | Gpam | 1.4904165 | 1.6091255 |
| NM_013196.1 | Ppara | -1.4969883 | -1.043811694 |
| Gene in hypothalamus | |||
| NM_001107055.1 | Wnt9b | 1.6385956 | 2.084756056 |
| NM_001105715.1 | Wnt3 | 1.4795437 | 1.238761833 |
| NM_181692.1 | Kiss1 | -1.2865248 | -1.175188444 |
| NM_133623.1 | Slc6a13 | -1.6948948 | -1.551063111 |
| NM_139326.2 | Pomc | -1.4852152 | -1.528518222 |
Tab.2.
KEGG pathway analysis for differentially expressed genes in white adipose of LepΔI14/ΔI14 and WT rats.
| KEGG pathways | P-value | Gene |
|---|---|---|
| MAPK signaling pathway | 3.729E-12 | Fgf9;Hspa1l;Ppm1a;Tgfb1;Nfkb2;Fgf1;Ntrk2;Fgf21;Prkcb;Il1r2;Mras;Cdc25b;Mknk1;Dusp3;Cacng1;Myc;Nr4a1;Hspa1b;LOC682930;Pla2g12a;Stmn1;Tgfb3;Fos;Cacnb3;Mapk11;Cd14;Mapkapk3;Cacna2d1 |
| Wnt signaling pathway | 5.462E-05 | Sfrp4;Camk2b;Tcf3;Rock2;Mmp7;Camk2g;Prkcb;Wnt5a;Fzd4;Myc;Ctbp2;Fzd6 |
| Adipocytokine signaling pathway | 7.689E-05 | Irs3;Lep;Cpt1b;Adipor2;Tnfrsf1b;Pomc;Nfkbie;Slc2a1 |
| Insulin signaling pathway | 6.402E-04 | Irs3;Gys2;Pklr;Shc1;Mknk1;Pde3b;Pik3r1;Pfkm;Pfkl;Pygm |
| Galactose metabolism | 1.883E-07 | Hk3;Glb1;Akr1b1;Lalba;Gale;Pfkm;Pfkl |
| PPAR signaling pathway | 3.446E-07 | Cpt1b;Olr1;Acox1;Scd4;Apoa5;Slc27a1;LOC681458;Fabp3;Scd;Gk;Dbi |
| Biosynthesis of unsaturated fatty acids | 9.699E-07 | Hadha;Acox1;Acot3;Scd4;LOC681458;Scd;Fads1 |
| Fatty acid metabolism | 1.813E-03 | Hadha;Cpt1b;Acox1;Aldh3a2;Aldh1a7 |
| Type I diabetes mellitus | 3.471E-03 | Prf1;RT1-N1;RT1-Bb;Ica1;RT1-Cl;RT1-CE5 |
| Glycerolipid metabolism | 1.142E-07 | Glb1;Akr1b1;Aldh3a2;Mgll;Ppap2a;Gk;Ppap2c;Aldh1a7;Cel |
| Ether lipid metabolism | 2.076E-04 | Pla2g7;Ppap2a;Pafah1b3;Ppap2c;Pla2g12a |
| Jak-STAT signaling pathway | 9.520E-04 | Lep;Csf2rb;Ghr;Il7;Myc;Stat5a;Jak3;Spry2;Pik3r1;Pim1 |
Tab.3.
KEGG pathway analysis for differentially expressed genes in the liver of LepΔI14/ΔI14 and WT rats.
| KEGG pathways | P-value | Gene |
|---|---|---|
| MAPK signaling pathway | 1.6E-08 | Pla2g4a;Fgf21;Mapk11;Cdc25b;Myc;Hspa1b;Jun;Pla2g12a;Dusp1;Cacng4;Rela;Map2k4;Fgfr3;Mapk4;Map3k10 |
| Regulation of actin cytoskeleton | 2.71E-06 | Fgf21;Myh10;Pip5k1b;Pip5k1a;Arhgef12;Gsn;Pip5k1c;Mylk;Fgfr3;Fn1;Pak4 |
| Jak-STAT signaling pathway | 2.78E-04 | Il3ra;Myc;Cish;Socs7;Il13ra2;Spred1;Lifr |
| Adipocytokine signaling pathway | 2.95E-04 | Irs3;Slc2a1;Ppara;Irs2;Rela |
| Neuroactive ligand-receptor interaction | 4.17E-04 | Grin1;Adora1;Gpr77;P2ry6;Prlhr;Nmu;Trpv1;Ntsr1;Taar7a;Gabrg3 |
| PPAR signaling pathway | 4.44E-04 | Me1;Fabp4;Fads2;Ppara;Ppard |
| Wnt signaling pathway | 3.7342E-02 | Myc;Jun;Ppard;Nkd2 |
| Calcium signaling pathway | 5.82E-05 | Grin1;Atp2b2;Tnnc2;Plce1;Plcd3;Mylk;Ntsr1;Atp2b4;Itpr1 |
Tab.4.
KEGG pathway analysis for differentially expressed genes in hypothalamus ofLepΔI14/ΔI14 and WT rats.
| KEGG pathways | P-value | Gene |
|---|---|---|
| Neuroactive ligand-receptor interaction | 4.8E-08 | Nmu;Trhr2;Adra1d;Nmbr;Tshb;Ptger3;Vip;Gabra4;Gabrd;Prl;Pomc |
| Type I diabetes mellitus | 8.78E-06 | H2-Ea;RT1-CE11;RT1-CE4;RT1-Cl;RT1-CE15 |
| Calcium signaling pathway | 1.109E-03 | Trhr2;Adra1d;Ptger3;Atp2a1;Pln |
| Basal transcription factors | 8.934E-03 | Taf5l;Taf6l |
| Glycine,serine and threonine metabolism | 1.2118E-02 | Alas2;Aoc3 |
| Wnt signaling pathway | 1.9626E-02 | Wnt9b;Wnt3;Lef1 |
| Adipocytokine signaling pathway | 2.8857E-02 | Camkk2;Pomc |
STRING analysis of the relationships between DEGs
DEGs in the WAT, liver, and hypothalamus were analyzed using STRING (http://string90.embl.de), a database of known and predicted protein interactions. The results indicated that the following groups of associated genes: 1.Ppard, Ppara, Hnf4a, lep, Stat5a, Ghr, Jak3, Irs2, Aldoc, Pfkm, Shc1, Prkcb, Pik3r1, Prkcd, Akr1b1, Pla2g4a, Pklr and Jun (associated with involvement in many signaling pathways and other responses); 2. Gstm4, Gstt1, Gsta3, Cyp2b2 and Cyp2e1; 3. Wnt5a, Fzd4, Fzd6, Strp4, Wnt9b and Wnt3; 4. RT1-M10-1, RT1-CE7, RT1-CE10 and RT1-N1. The key genes in this protein interaction network included Hnf4a, Stat5a, Jak3, Pik3r1, Prkcd, Pla2g4a, Jun and Pklr, which were linked to Plcd3, Lef1, POMC, Ppara, Adipor2, Fabp3, Aldoc and many downstream genes. All of these genes are inter-related, forming a large network. However, many genes were not involved in this network, indicating unrelated or unknown functions.
Discussion
In this study, we identified the different gene expression profile of WAT, liver and hypothalamus of LepΔI14/ΔI14 rats and WT to screen genes involved in metabolic alterations in this CRISPR/Cas9-created model of leptin-deficiency. According to the results of the microarray analysis, we selected 14 representative genes on the basis of their functional properties and the fold change in their expression to perform quantitative real-time PCR, thus verifying the reliability of the data obtained from the microarray studies.
LepΔI14/ΔI14 SD rats were produced by knocking out one amino acid of the leptin protein which inactivated leptin's function. Although significantly higher serum leptin level inLepΔI14/ΔI14 rats was detected compared to the one in their WT littermates, it was "inactivated leptin". We also observed nonalcoholic steatohepatitis and increased lipid area in brown adipose inLepΔI14/ΔI14 rats, which were consistent with previous reports[5,23–24]. In our study DEGs in the liver were found to be associated withPPAR , which stimulates fatty acid-oxidation and can modulate hepatic amino acid oxidation during the transition of energy fuels. DEGs were also found to be associated withWnt3. It has been reported that adenoviral vector-mediated overexpression of Wnt3 could suppress apoptosis of hepatic progenitor cells, possibly through the Bcl-2 pathway[25]. Examination of the relationships among the identified DEGs revealed a large network of associations.
The sample size of LepΔI14/ΔI14 or WT rats was only six for each group, which was slightly small for statistical analysis. However, that was the maximum number we could get from Zhu's laboratory since this was a newly created rat model.
Gene expression profiling in mouse models lacking leptin signaling have been reported[9–11,26–29]. The gene expression profile in our rat model was provided for further comparison. Although some gene identified from microassay analysis were originally thought to be metabolism-unrelated, they were involved in other leptin deficiency rat model studies. For example, our results showed that bone morphogenetic protein 3 (Bmp3) gene was upregulated in white adipose tissue, which had been originally identified as inducers of bone formation[30]. This is consistent with the fact that mouse models (including leptin deficient (ob/ob), leptin receptor-deficient, and lipodystrophic mice) which had decreased leptin signaling always had high bone mass phenotype[31].
The metabolism-related genes finally verified in liver in our studies were different from those identified by Aizawa-Abe and her colleagues[32]. These discrepancies may be due to variations in experimental methods. Compared to their model, the technique we explored provided a complete knockout of leptin, thus avoiding unnecessary interference caused by endogenous leptinin vivo.
Our present study demonstrated upregulation of GABAA receptor mRNA expression in LepΔI14/ΔI14 rat hypothalamus, which could be confirmed by Turenius' study[33]. Compared our results of the rat hypothalamic microassay analysis to the work using inob/ob mice, the downregulation of proopiomelanocortin (POMC)-1 mRNA expression was found in both LepΔI14/ΔI14 rat and ob/ob mice, whereas mRNA expressions of urocortin (UCN)-3 and agouti-related protein (AGRP) expression were opposite[34]. This may be due to the difference in sensitivity to leptin signaling in different species.
After gene ontology analysis, we discovered that the DEGs were all primarily targeted to cell (ontology: cellular component), DNA binding (ontology: molecular function), and cellular process (ontology: biological process). Marcelin reported leptin-deficient mice exhibited increased adipose triglyceride lipase (ATGL) content in adipocytes, along with increased lipolysis and fatty acid oxidation[35], indicating the existence of DEGs in adipocytes, which is consistent with our results. In addition, Altintas demonstrated that leptin-deficiency-induced obesity was accompanied by alterations in the density of mast cells in abdominal fat depots[36], indicating that some cellular processes are influenced by genetic disorders. However, some GO analyses have yielded contradictory results. These discrepancies may be due to different species.
Some DEGs (both upregulated and downregulated) were found to be associated with a variety of metabolic pathways, especially wnt and adipocytokine signaling pathways, which were shown to be associated with DEGs in WAT, liver and hypothalamus. These two signaling pathways were reported to be related to metabolism. Koziński reported that the wnt signaling pathway was involved in the regulation of gluconeogenesis, glycolysis, maintains carbohydrate and lipid homeostasis, the disruption of which has been found in many pathophysiological states such as different types of cancer, neurodegenerative diseases and metabolic disorders[37–38]. Research related to eating disorders showed thatLEP (leptin), LEPR (leptin receptor), ADIPO (adiponectin), POMC (proopio-melanocortin), AGRP (agouti-related protein homolog) and NPY (neuropeptide Y), were annotated to the adipocytokine signaling pathway[29]. In our study although wnt and adipocytokine signaling pathways were detected to be DEG-related in WAT, liver and hypothalamus, the DEGs identified in the two pathways in three different tissues were not completely consistent (data are not given). Further investigations are needed to clarify these differences.
We also found some tissue-specific metabolic pathways. The MAPK signaling pathway appeared in both WAT and liver. MAPK has been known to be involved in matrix metalloproteinase production and the regulation of cartilage cell proliferation, apoptosis, differentiation, metabolism and homeostasis[39–40]. Similarly, the JAK-STAT signaling pathway was found to participate metabolism in WAT and liver in our study. It had been reported that JAK-STAT dysregulation led to multiple metabolism disorders[41]. Therefore, it might be postulated that some metabolic pathways were tissue-specific, highlighting the importance of identifying tissue-targeted pathways in the development of novel therapeutic strategies for metabolic disorders.
In conclusion, we used microarray analysis to investigate gene expression in the new leptin knockout LepΔI14/ΔI14 rats and used RT-PCR to confirm the data obtained from the microarray. The results indicated that the metabolic disorders associated with leptin-deficiency might be induced by gene dysregulation in some key metabolic signaling pathways. In this study, a new rat model created by updated technique had been utilized. It provided data to study gene function and discern molecular mechanisms in leptin-related diseases and first supplied microarray analysis baseline data of Cas9 leptin-knockoutLepΔI14/ΔI14 rat model for biomedical and pharmacological research later. Further investigations are required to elucidate the influence of DEGs on different tissues and provide important information for the development of targeted strategies for the prevention and treatment of leptin-associated metabolic diseases.
Acknowledgments
We gratefully acknowledge the technical assistance of Ling Song. This work was supported by grants from the National Natural Science Foundation of China (No. 81470150).
References
- 1. Narayan KM, Boyle JP, Thompson TJ, et al. Lifetime risk for diabetes mellitus in the United States[J].JAMA, 2003,290(14):1884–1890. [DOI] [PubMed] [Google Scholar]
- 2. Fernéndez-Formoso G, Párez-Sieira S, González-Touceda D, et al. Leptin, 20 years of searching for glucose homeostasis[J].Life Sci, 2015,140:4–9. [DOI] [PubMed] [Google Scholar]
- 3. Bouyer K, Simerly RB. Neonatal leptin exposure specifies innervation of presympathetic hypothalamic neurons and improves the metabolic status of leptin-deficient mice[J].J Neurosci, 2013,33(2):840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu J, Donepudi AC, More VR, et al. Deficiency in Nrf2 transcription factor decreases adipose tissue mass and hepatic lipid accumulation in leptin-deficient mice[J].Obesity (Silver Spring), 2015,23(2):335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perfield JW 2nd, Ortinau LC, Pickering RT, et al. Altered hepatic lipid metabolism contributes to nonalcoholic fatty liver disease in leptin-deficient Ob/Ob mice[J].J Obes, 2013,2013:296537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodríguez A, Moreno NR, Balaguer I, et al. Leptin administration restores the altered adipose and hepatic expression of aquaglyceroporins improving the non-alcoholic fatty liver of ob/ob mice[J].Sci Rep, 2015,5:12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang W, Ambati S, Della-Fera MA, et al. Leptin modulated changes in adipose tissue protein expression in ob/ob mice[J].Obesity (Silver Spring), 2011,19(2):255–261. [DOI] [PubMed] [Google Scholar]
- 8. Sennello JA, Fayad R, Pini M, et al. Transplantation of wild-type white adipose tissue normalizes metabolic, immune and inflammatory alterations in leptin-deficient ob/ob mice[J].Cytokine, 2006,36(5-6):261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang B, Chandrasekera PC, Pippin JJ. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes[J].Curr Diabetes Rev, 2014,10(2):131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmström MH, Tom RZ, Björnholm M, et al. Effect of leptin treatment on mitochondrial function in obese leptin-deficient ob/ob mice[J].Metabolism, 2013,62(9):1258–1267. [DOI] [PubMed] [Google Scholar]
- 11. Hao Z, Münzberg H, Rezai-Zadeh K, et al. Leptin deficient ob/ob mice and diet-induced obese mice responded differently to Roux-en-Y bypass surgery[J].Int J Obes (Lond), 2015,39(5):798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaira S, Yang C, McCoy A, et al. Creation and preliminary characterization of a leptin knockout rat[J].Endocrinology, 2012,153(11):5622–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu S, Zhu X, Li H, et al. The 14th Ile residue is essential for Leptin function in regulating energy homeostasis in rat[J].Sci Rep, 2016,6:28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J].Science, 2013,339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering[J].Cell, 2013,153(4):910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swanson LW. Brain Maps: Structure of the Rat Brain. 3rd ed. Academic Press. (2003).
- 17. Srivastava VK, Hiney JK, Dees WL. Short-term alcohol administration alters KiSS-1 gene expression in the reproductive hypothalamus of prepubertal female rats[J].Alcohol Clin Exp Res, 2009,33(9):1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo Y, Guo H, Zhang L, et al. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells[J].J Virol, 2005,79(22):14392–14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project[J].Nat Biotechnol, 2006,24(9):1140–1150. [DOI] [PubMed] [Google Scholar]
- 20. Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure[J].Bioinformatics, 2006,22(13):1600–1607. [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate A Practical and Powerful Approach to Multiple Testing[J].Journal of the Royal Statistical Society Series., 1995,57:289–300. [Google Scholar]
- 22. Lagor WR, Fields DW, Khetarpal SA, et al. The effects of apolipoprotein F deficiency on high density lipoprotein cholesterol metabolism in mice[J].PLoS One, 2012,7(2):e31616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singhal NS, Patel RT, Qi Y, et al. Loss of resistin ameliorates hyperlipidemia and hepatic steatosis in leptin-deficient mice[J].Am J Physiol Endocrinol Metab, 2008,295(2):E331–E338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dinh CH, Szabo A, Yu Y, et al. Bardoxolone Methyl Prevents Fat Deposition and Inflammation in Brown Adipose Tissue and Enhances Sympathetic Activity in Mice Fed a High-Fat Diet[J].Nutrients, 2015,7(6):4705–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang X, Hu D, Zhang C, et al. [Overexpression of Wnt3 inhibits apoptosis of hepatic progenitor cells in vitro] [In Chinese]. Nan Fang Yi Ke Da Xue Xue Bao, 2014,34(1):46–50. [PubMed] [Google Scholar]
- 26. Bao D, Ma Y, Zhang X, et al. Preliminary Characterization of a Leptin Receptor Knockout Rat Created by CRISPR/Cas9 System[J].Sci Rep, 2015,5:15942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bray GA. The Zucker-fatty rat: a review[J].Fed Proc, 1977,36(2):148–153. [PubMed] [Google Scholar]
- 28. Duan J, Choi YH, Hartzell D, et al. Effects of subcutaneous leptin injections on hypothalamic gene profiles in lean and ob/ob mice[J].Obesity (Silver Spring), 2007,15(11):2624–2633. [DOI] [PubMed] [Google Scholar]
- 29. Zhao M, Li X, Qu H. EDdb: a web resource for eating disorder and its application to identify an extended adipocytokine signaling pathway related to eating disorder[J].Sci China Life Sci, 2013,56(12):1086–1096. [DOI] [PubMed] [Google Scholar]
- 30. Ducy P, Karsenty G. The family of bone morphogenetic proteins[J].Kidney Int, 2000,57(6):2207–2214. [DOI] [PubMed] [Google Scholar]
- 31. Takeda S, Elefteriou F, Levasseur R, et al. Leptin regulates bone formation via the sympathetic nervous system[J].Cell, 2002,111(3):305–317. [DOI] [PubMed] [Google Scholar]
- 32. Aizawa-Abe M, Ebihara K, Ebihara C, et al. Generation of leptin-deficient Lepmkyo/Lepmkyo rats and identification of leptin-responsive genes in the liver[J].Physiol Genomics, 2013,45(17):786–793. [DOI] [PubMed] [Google Scholar]
- 33. Turenius CI, Htut MM, Prodon DA, et al. GABA(A) receptors in the lateral hypothalamus as mediators of satiety and body weight regulation[J].Brain Res, 2009,1262:16–24. [DOI] [PubMed] [Google Scholar]
- 34. Duan J, Choi YH, Hartzell D, et al. Effects of subcutaneous leptin injections on hypothalamic gene profiles in lean and ob/ob mice[J].Obesity (Silver Spring), 2007,15(11):2624–2633. [DOI] [PubMed] [Google Scholar]
- 35. Marcelin G, Liu SM, Li X, et al. Genetic control of ATGL-mediated lipolysis modulates adipose triglyceride stores in leptin-deficient mice[J].J Lipid Res, 2012,53(5):964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altintas MM, Nayer B, Walford EC, et al. Leptin deficiency-induced obesity affects the density of mast cells in abdominal fat depots and lymph nodes in mice[J].Lipids Health Dis, 2012,11:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koziński K, Dobrzyń A. Wnt signaling pathway--its role in regulation of cell metabolism[J]. Postepy Hig Med Dosw (Online), 2013,67:1098–1108. [DOI] [PubMed] [Google Scholar]
- 38. Sherwood V. WNT signaling: an emerging mediator of cancer cell metabolism[J]? Mol Cell Biol, 2015,35(1):2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao SC, Yin HB, Liu HX, et al. Research progress on MAPK signal pathway in the pathogenesis of osteoarthritis[J]. Zhongguo Gu Shang, 2014,27(5):441–444. [PubMed] [Google Scholar]
- 40. Martínez-Soto D, Ruiz-Herrera J. Regulation of the expression of the whole genome of Ustilago maydis by a MAPK pathway[J].Arch Microbiol, 2015,197(4):575–588. [DOI] [PubMed] [Google Scholar]
- 41. Xu D, Yin C, Wang S, et al. JAK-STAT in lipid metabolism of adipocytes[J].JAKSTAT, 2013,2(4):e27203. [DOI] [PMC free article] [PubMed] [Google Scholar]