Abstract
Peripheral nerve injury often causes neuropathic pain and is associated with changes in the expression of numerous proteins in the dorsal horn of the spinal cord. To date, proteomic analysis method has been used to simultaneously analyze hundreds or thousands of proteins differentially expressed in the dorsal horn of the spinal cord in rats or dorsal root ganglion of rats with certain type of peripheral nerve injury. However, a proteomic study using a mouse model of neuropathic pain could be attempted because of abundant protein database and the availability of transgenic mice. In this study, whole proteins were extracted from the ipsilateral dorsal half of the 4th–6th lumbar spinal cord in a mouse model of spared nerve injury (SNI)-induced neuropathic pain. In-gel digests of the proteins size-separated on a polyacrylamide gel were subjected to reverse-phase liquid-chromatography coupled with electrospray ionization ion trap tandem mass spectrometry (MS/MS). After identifying proteins, the data were analyzed with subtractive proteomics usingProtAn, an in-house analytic program. Consequently, 15 downregulated and 35 upregulated proteins were identified in SNI mice. The identified proteins may contribute to the maintenance of neuropathic pain, and may provide new or valuable information in the discovery of new therapeutic targets for neuropathic pain.
Keywords: proteomics, spinal dorsal horn, neuropathic pain, spared nerve injury, mouse
Introduction
Various peripheral nerve injuries often result in the development of chronic neuropathic pain, which can be characterized by spontaneous pain, allodynia and hyperalgesia[1]. The development and maintenance of neuropathic pain critically involve the immediate and delayed central mechanisms in the spinal cord, associated with peripheral mechanisms[2]. The central mechanisms relate to hypersensitive responses of spinal dorsal horn (DH) neurons to stimuli; therefore, the biochemical molecules that contribute to DH hypersensitivity have been researched intensely. Following peripheral nerve damage, the persistent activity of, or the release of neurotransmitters and proinflammatory cytokines from the central terminals, of dorsal root ganglion (DRG) neurons drives abnormal gene and protein expression in the spinal DH[1]. These processes support the hypersensitive state of the spinal DH, described as "central sensitization"[2]. Therefore, identifying proteins with altered expression in the DH following peripheral nerve damage is essential to elucidate the central mechanisms of neuropathic pain.
Western blotting method and immunohistochemistry are commonly used to identify and quantify changes in protein expression in neuropathic pain; however, they are limited by the inability to assess a large number of proteins simultaneously. Proteomic techniques solve this problem by allowing simultaneous analysis of hundreds or thousands of proteins[3]. Until now, only a few studies have used a proteomic approach to identify spinal cord or DRG proteins; the majority of them use rat models of neuropathic pain[4–8]. Nonetheless, considering abundance of protein database and availability of transgenic mice, a proteomic study using a mouse model could provide more valuable information in neuropathic pain studies.
We sought to elucidate changes in the proteomic profile of the dorsal spinal cord using the mouse spared nerve injury (SNI) model of neuropathic pain[9] by performing tandem mass spectrometry (MS/MS) coupled to liquid chromatography (LC) analysis. Consequently, we identified proteins specific to either sham or SNI mice, which could be further interpreted as decreased or increased expression following surgical nerve injury, respectively.
Materials and methods
SNI-induced neuropathic pain in mice
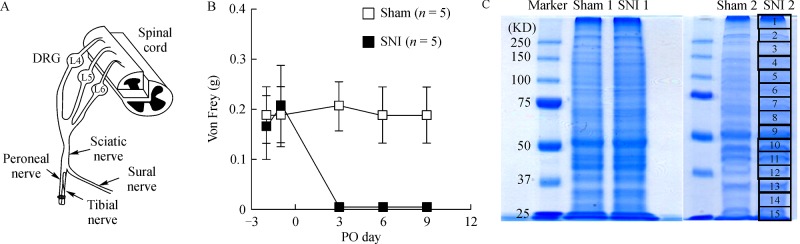
Eight-week-old male C57BL/6 mice (Japan SLC Inc., Shizuoka, Japan) were used. All experiments were approved by the Institutional Animal Care Committee of Kyungpook National University (KNU 2009-38) that follows the guidelines of the International Association for the Study of Pain. The mouse SNI model was created as previously described[9]. Briefly, mice were anesthetized with isoflurane (3% for anesthesia induction; 2% for maintenance), and the three peripheral branches (common peroneal, tibial, and sural nerves) of the sciatic nerve were carefully exposed in the left hind limb without disturbing the sural nerve. Both common peroneal and tibial nerves were ligated tightly and transected distally, and a 2-mm section of the two nerves was removed. Exposure of the three branches of the sciatic nerve occurred during sham control surgery, without ligation or transection. To identify the development of mechanical allodynia in sham and SNI mice, the lateral region of the left hind paw plantar surface was stimulated with calibrated von Frey monofilaments (0.008–4.0 g; Stoelting Co., Wood Dale, IL, USA). A score of two paw withdrawal responses out of ten stimuli was classified as a ‘positive’ response to a certain von Frey monofilament.
Sample preparation and proteomic analysis
After the heart was perfused with cold phosphate-buffered saline (PBS), the lumbar segment (L4–L6) of the spinal cord was dissected out by laminectomy. The ipsilateral dorsal half of the lumbar segment, including the spinal DH, was used for protein extraction. The protein concentration in the supernatant was assessed with a protein assay kit, using bovine serum albumin as the standard. Total proteins were separated by size using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining, and each protein lane was cut into 15 gel pieces. Following this, the gel pieces were de-stained by incubation 75 mmol/L ammonium bicarbonate/40% ethanol (1:1), reduced with 5 mmol/L 1,4-dithiothreitol (DTT) (60°C for 30 minutes), alkylated with 55 mmol/L iodoacetamide (room temperature for 30 minutes), and proteolyzed with 20 µg/mL modified sequencing grade trypsin (37°C overnight; Roche Applied Science). The tryptic peptide mixture was eluted from the gel with 0.1% formic acid and subjected to reverse-phase liquid-chromatography coupled with electrospray ionization (ESI) ion trap mass spectrometry (LC-MS/MS). Mass spectrometry was performed with ion-trap mass spectrometers (LTQ; Thermo Fisher Scientific, Waltham, MA, USA). The peptides were eluted onto reversed-phase (RP) Pico Frit columns packed in-house with C18 and then separated on an RP column by gradient elution. The solutions used as the mobile phases were H2O (A) and acetonitrile (B), both containing 0.1% v/v formic acid. The gradient was started at 2% B, reaching 60% B in 50 minutes, 80% B in the next 5 minutes, and 100% A in the final 15 minutes.
All MS/MS data for the first sample set (sham #1 and SNI #1) were searched against the mouse International Protein Index (IPI) database (Version 3.15 consisting of 58,099 protein entries) using the SEQUEST algorithm (Thermo Fisher Scientific, Waltham, MA, USA) in BioWorks (version 3.2). Protein identification was done based on the corresponding peptide identification. After identifying proteins, the data were analyzed with subtractive proteomics usingProtAn, an in-house analytic program. Tandem mass spectra for the second sample set (sham #2 and SNI #2) were extracted by Sorcerer (version 3.4β2). Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using SEQUEST (Thermo Fisher Scientific, Waltham, MA, USA; version 27, rev. 11). Finally, results were imported into Scaffold (version Scaffold-01_07_00, Proteome Software Inc., Portland, OR, USA) for protein identification. The identification of peptides was accepted if they could be established at a probability greater than 95.0%, as specified by the Peptide Prophet algorithm[10]. Protein probabilities were assigned by the Protein Prophet algorithm[11] and contained at least 2 identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Results
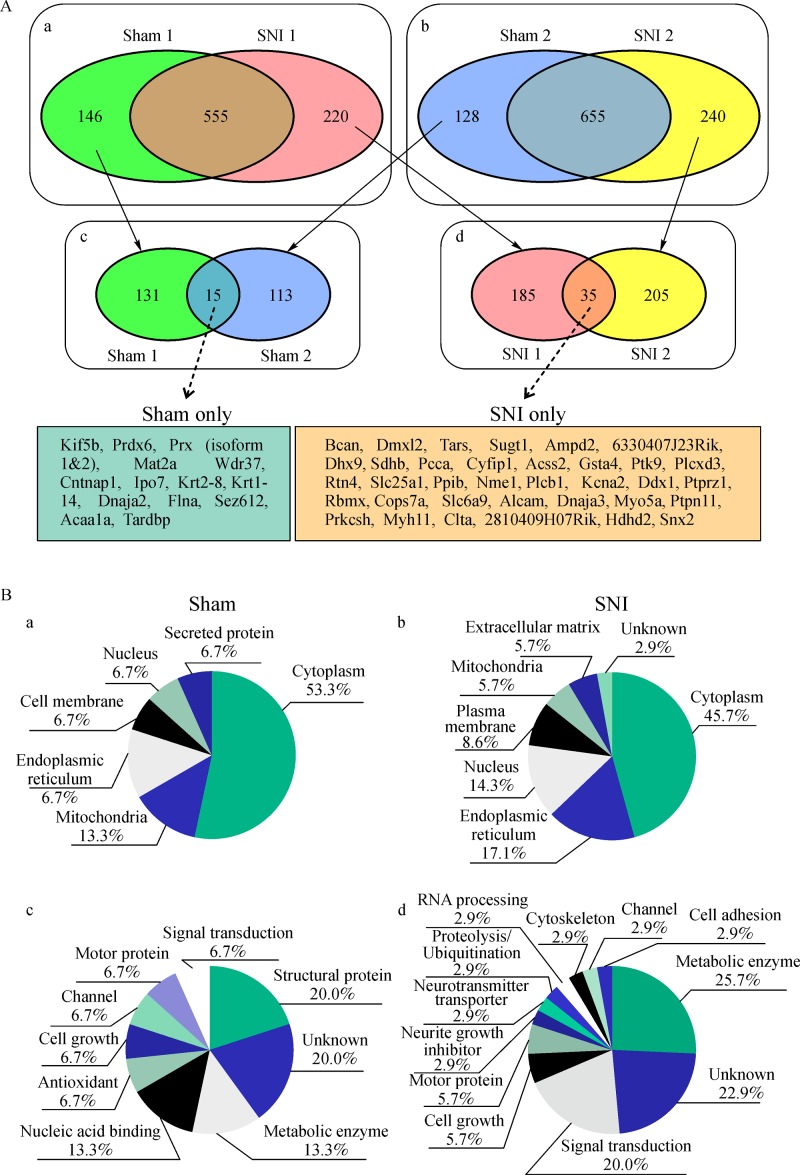
To elucidate the proteomic profile of the spinal DH in a mouse SNI model of neuropathic pain[9], we extracted whole proteins from the ipsilateral dorsal half of the L4–6 spinal cord after the full development of neuropathic pain behavior at 9 days post-SNI surgery (Fig. 1A and B). The extracted protein sample from each mouse was resolved by SDS-PAGE, and each gel was cut into 15 pieces (Fig. 1C). From all MS/MS data of the first sample set, we identified 701 and 775 proteins from sham (control) #1 and SNI #1 samples, respectively. There were 146 and 220 proteins identified in sham #1 or SNI #1 alone, respectively (Fig. 2A). From all MS/MS data from the second sample set, 783 and 895 proteins were identified from sham #2 and SNI #2, respectively, with 128 and 240 proteins identified in sham #2 and SNI #2 alone, respectively (Fig. 2A). Finally, we compared the proteins found only in sham #1 and #2, and found 15 proteins as their intersection. Similarly, the intersection of SNI #1 and #2 samples was 35 proteins. Proteins found in sham and SNI mice alone are based on a detection limit calculated by changes in expression due to SNI surgery; therefore, these results suggested that the 15 proteins found in the sham samples were downregulated (Table 1). Similarly, the 35 proteins detected in the SNI samples were likely to be upregulated by SNI (Table 2).
Fig.1.
Preparation of protein samples for proteomic analysis from the dorsal spinal cord of sham or mice with spared nerve injury (SNI).
Fig.2.
Identified proteins down- or upregulated in a mouse SNI model of neuropathic pain.
Tab.1.
List of fifteen down-regulated proteins after SNI.
|
IPI accession
number |
Protein Name | Symbol | Coverage (%) | Total peptide | M.W. | Function summary | |
|---|---|---|---|---|---|---|---|
| Sorcerer |
Bioworks
(score) |
||||||
| IPI00469952 | Isoform 1 of periaxin* | Prx | 6.40 | 12 | 7 (40.22) | 147668.8 | Structural protein |
| IPI00223331 | Isoform 2 of periaxin* | Prx | 18.20 | 4 | 5 (20.18) | 16106.4 | Structural protein |
| IPI00128454 | Seizure related 6 homolog like 2 | Sez6l2 | 3.47 | 3 | 3 (20.14) | 99061 | Structural protein |
| IPI00338983 | Contactin-associated protein 1 precursor* | Cntnap1 | 2.96 | 5 | 3 (20.21) | 156467.5 | Signal transduction |
| IPI00454016 | S-Adenosylmethionine synthetase isoform type-2* | Mat2a | 6.08 | 2 | 2 (10.21) | 43671.1 | Metabolic enzyme |
| IPI00121833 | 3-Ketoacyl- Co A thiolase A, peroxisomal precusor | Acaa1a | 6.13 | 3 | 3 (20.19) | 43935.2 | Metabolic enzyme |
| IPI00555059 | Peroxiredoxin-6* | Prdx6 | 10.79 | 2 | 2 (10.28) | 24853.7 | Antioxidant |
| IPI00331444 | Importin-7* | Ipo7 | 2.79 | 3 | 4 (28.21) | 119865.8 | Nucleic acid binding |
| IPI00121758 | Tar DNA-binding protein 43 | Tardbp | 9.42 | 5 | 2 (10.24) | 44529.5 | Nucleic acid binding |
| IPI00555125 | Kinesin family member 5b | Kif5b | 3.01 | 3 | 2 (10.17) | 109534.6 | Motor protein |
| IPI00136251 | DNAJ homolog subfamily a member 2 | Dnaja2 | 12.90 | 7 | 3 (20.16) | 45727.6 | Cell growth |
| IPI00131138 | Isoform 1of filamin-A | Flna | 1.17 | 2 | 2 (10.16) | 281166.5 | Channel |
| IPI00322209 | Keratin, type II cytoskeletal 8 | Krt2-8 | 7.96 | 2 | 2 (16.18) | 54549 | Unknown |
| IPI00227140 | Keratin, type I cytoskeletal 14 | Krt1-14 | 4.96 | 3 | 2 (20.15) | 52850.1 | Unknown |
| IPI00387421 | Wd repeat protein 37 | Wdr37 | 5.44 | 2 | 2 (10.21) | 55027.5 | Unknown |
*Bolds are discussed in the text.
Tab.2.
List of thirty five up-regulated proteins after SNI
|
IPI accession
number |
Protein Name | Symbol | Coverage (%) | Total peptide | M.W. | Function summary | |
|---|---|---|---|---|---|---|---|
| Sorcerer |
Bioworks
(score) |
||||||
| IPI00623371 | Brevican* | Bcan | 4.19 | 6 | 3 (20.24) | 95995.5 | Cell growth |
| IPI00123465 | COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) | Cops7a | 3.28 | 3 | 2 (20.26) | 30206.6 | Cell growth |
| IPI00270767 | Isoform 2 of Reticulon 4* | Rtn4 | 2.49 | 5 | 3 (30.18) | 126669.1 | Neurite growth inhibitor |
| IPI00121378 | Activated leukocyte cell adhesion molecule* | Alcam | 6.69 | 4 | 5 (30.17) | 65075.4 | Cell adhesion |
| IPI00129774 | Potassium voltage-gated channel, shaker-related subfamily, member 2 | Kcna2 | 5.81 | 2 | 4 (10.20) | 56684.5 | Channel |
| IPI00122101 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | Slc6a9 | 3.46 | 4 | 3 (10.13) | 70916.2 | Neurotransmitter transporter |
| IPI00323911 | Glutathione S-transferase, α4* | Gsta4 | 11.30 | 3 | 3 (20.15) | 25547.2 | Metabolic enzyme |
| IPI00468688 | Threonyl-tRNA synthetase | Tars | 3.32 | 3 | 2 (20.18) | 83340.1 | Metabolic enzyme |
| IPI00406639 | Adenosine monophosphate deaminase 2 (isoform L) | Ampd2 | 3.89 | 2 | 2 (20.15) | 92007.7 | Metabolic enzyme |
| IPI00338536 | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | Sdhb | 11.30 | 8 | 3 (20.16) | 31797 | Metabolic enzyme |
| IPI00330523 | Propionyl-Coenzyme A carboxylase, α polypeptide | Pcca | 3.32 | 2 | 4 (40.17) | 79903.9 | Metabolic enzyme |
| IPI00330302 | Acyl-CoA synthetase short-chain family member 2 | Acss2 | 3.98 | 2 | 2 (20.14) | 79002.3 | Metabolic enzyme |
| IPI00230754 | Solute carrier family 25, member 11 | Slc25a1 | 7.01 | 2 | 7 (40.21) | 34138.3 | Metabolic enzyme |
| IPI00131459 | Expressed in non-metastatic cells 1, protein | Nme1 | 12.5 | 3 | 2 (10.21) | 17190 | Metabolic enzyme |
| IPI00111981 | RIKEN cDNA 2810409H07 gene | 2810409H07Rik | 7.07 | 2 | 2 (10.18) | 44713.4 | Metabolic enzyme |
| IPI00330476 | Cytoplasmic FMR1 interacting protein 1 | Cyfip1 | 4.23 | 3 | 2 (20.17) | 145227.6 | Cytoskeleton |
| IPI00118120 | Myosin Va | Myo5a | 0.81 | 3 | 5 (30.19) | 215585 | Motor protein |
| IPI00114894 | Myosin heavy polypeptide 11, smooth muscle | Myh11 | 2.99 | 5 | 2 (20.15) | 227015.2 | Motor protein |
| IPI00114409 | Clathrin light polypeptide (Lca) | Clta | 7.66 | 3 | 4 (30.19) | 25539.1 | Signal transduction |
| IPI00116554 | Protein tyrosine phosphatase, non-receptor type 11 | Ptpn11 | 3.85 | 2 | 5 (30.14) | 68442.1 | Signal transduction |
| IPI00109212 | Sorting nexin 2 | Snx2 | 7.49 | 4 | 4 (20.21) | 58624.5 | Signal transduction |
| IPI00130045 | Phospholipase C, β1* | Plcb1 | 2.06 | 3 | 2 (10.22) | 138311.3 | Signal transduction |
| IPI00314844 | Protein tyrosine kinase 9 | Ptk9 | 8.0 | 2 | 2 (10.28) | 40062.6 | Signal transduction |
| IPI00115680 | Protein kinase C substrate 80K-H | Prkcsh | 4.42 | 2 | 2 (10.16) | 58774.8 | Signal transduction |
| IPI00120414 | DnaJ (Hsp40) homolog, subfamily A, member 3 | Dnaja3 | 7.46 | 2 | 2 (20.19) | 46829.7 | Apoptosis signal transduction |
| IPI00408957 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | Sugt1 | 11.9 | 4 | 2 (10.22) | 38141.9 | Proteolysis/Ubiquitination |
| IPI00127172 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | Ddx1 | 3.24 | 3 | 4 (30.15) | 82483.1 | RNA processing |
| IPI00308851 | Phosphatidylinositol-specific phospholipase C, X domain containing 3 | Plcxd3 | 10.30 | 2 | 2 (10.24) | 36296.3 | Unknown |
| IPI00605677 | Dmx-like 2 | Dmxl2 | 1.18 | 3 | 5 (40.21) | 322567.7 | Unknown |
| IPI00125154 | Protein tyrosine phosphatase, receptor type Z, polypeptide 1 | Ptprz1 | 3.54 | 9 | 6 (40.19) | 175175 | Unknown |
| IPI00124979 | RNA binding motif protein, X chromosome | Rbmx | 6.14 | 4 | 2 (10.24) | 42284.7 | Unknown |
| IPI00339468 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | Dhx9 | 2.60 | 3 | 3 (20.19) | 149604.3 | Unknown |
| IPI00403618 | RIKEN cDNA 6330407J23 gene | 6330407J23Rik | 2.43 | 2 | 8 (50.18) | 103464.3 | Unknown |
| IPI00111166 | Haloacid dehalogenase-like hydrolase domain containing 2 | Hdhd2 | 11.20 | 4 | 4 (20.28) | 28713.1 | Unknown |
| IPI00135686 | Peptidylprolyl isomerase B | Ppib | 12 | 2 | 2 (10.15) | 23695.8 | Unknown |
*Bolds are discussed.
Subcellular localization and function of the proteins are shown inFig. 2B. Most of detected proteins in both sham and SNI groups are cytoplasmic proteins, and the rest of proteins are categorized into endoplasmic reticulum, cell membrane, mitochondria or nucleus groups (a and b inFig. 2B), thus showing a similar distribution in both groups. On the other hand, there was a discrepancy in the distributions for functions of proteins. For example, proteins relating antioxidant and nucleic acid binding were found only in the sham group, whereas those relating cell adhesion and neurite growth inhibition only in the SNI group (c and d inFig. 2B).
Discussion
The present study, using proteomic analysis, identified 35 differentially expressed proteins in the dorsal spinal cord 9 days after SNI surgery in mice, and 15 differentially expressed proteins in that of sham-operated control mice. The lists were obtained by duplicate sampling from two different mice for each group. Because a detection limit determines the presence of proteins in each group, the 15 proteins detected only in the sham samples can be interpreted as downregulated proteins by SNI, and the 35 proteins only in the SNI samples as upregulated proteins by SNI. Although this list needs to be further compared with Western blot and immunohistochemistry analyses in the spinal DH, it provides a novel protein list for exploring new targets to treat neuropathic pain.
Recently, proteomic approaches have been attempted to identify spinal cord or DRG proteins in rat models of neuropathic pain. Chronic constriction-induced sciatic nerve injury (CCI) regulated 5 proteins in the rat spinal cord[7]. In addition, 38 proteins were identified as differentially expressed in the rat dorsal spinal cord following spinal nerve ligation (SNL)-induced peripheral nerve injury[6]. In the DRG, approximately 1,300 protein spots have been detected, and 67 proteins of these were tightly modulated by SNL injury[4]. The proteins reported rarely overlap in these studies or with our study, suggesting that proteomic data could vary depending on the species used and injury type. In addition, experimental schemes, such as sampling area (whole or dorsal spinal cord), time points, and proteomic tools, could affect the proteomic data produced. To ensure consistency of animal-to-animal and batch-to-batch samples, we used two animals in each group and only proteins found in both animals were regarded as positive identification.
Among the 15 proteins identified in the sham group, periaxin is a cytoskeleton-associated protein of myelinating Schwann cells[12] and plays a key role in stabilization of the myelin sheath in the subsequent stage of myelination[13]. Recently, it has been demonstrated that mice lacking the periaxin gene develop late-onset demyelination, slower electrical conduction of sciatic nerves, and neuropathic pain behaviors[14]. Interestingly, proteomic analysis of the DRG showed downregulation of the periaxin protein in a rat SNL model of neuropathic pain[4]. Demyelination of peripheral nerves gives rise to neuropathic pain behaviors[15]; therefore, the downregulation of spinal periaxin may contribute to development of neuropathic pain.
Contactin-associated protein (Caspr) 1 precursor, which exists as a complex with contactin and is enriched in the paranodal region of mature myelinated axons in the central (CNS) and peripheral nervous system[16–17], was also downregulated by SNI. Caspr is important during myelination and in the maintenance of myelination[16–17]. Recently, it has been shown that the expression of Caspr is reduced in the axons of demyelinating lesions in patients with multiple sclerosis[18] and in lesions in infraorbital nerve injury[19]. Therefore, the downregulation of Caspr precursors in the present study may result from demyelination following peripheral nerve injury and contribute to the induction or maintenance of neuropathic pain.
Among the metabolic enzymes downregulated by SNI, S-adenosylmethionine synthetase (isoform type-2), also called methionine adenosyltransferase IIα, is known to catalyze the formation ofS-adenosylmethionine from methionine and ATP. It has been reported that S-adenosylmethionine synthetase is sensitive to active oxygen and nitrogen species and regulates the consumption of ATP in the liver, thus preserving cellular viability under different stress conditions[20]. Reactive oxygen species (ROS) in the spinal DH are thought to contribute to neuropathic pain[21–22]. Interestingly, peroxiredoxin-6 was also downregulated by SNI in the present study. Peroxiredoxin-6 reduces hydrogen peroxide or other hydroperoxides to water or their corresponding alcohols during the oxidation of glutathione[23]. Therefore, the downregulation of two proteins above suggests a less protective environment in the spinal DH against the overproduction of oxygen free radicals in neuropathic pain[20].
The nucleic acid binding proteins identified in the sham group included importin-7. Importin-7 binds to the transcription factor c-Jun which is phosphorylated and activated by c-Jun N-terminal kinase (JNK)[24], promoting neuropathic pain via upregulation of monocyte chemoattractant protein (MCP)-1[25]. Therefore, it would be interesting to assess whether the downregulation of importin-7 plays a role in JNK-MCP-1 signaling in neuropathic pain.
Of the 35 proteins identified in the SNI samples, brevican, a chondroitin sulfate proteoglycan, is an extracellular matrix molecule that is widely expressed throughout the developing and adult CNS. After spinal cord injury, neurocan, brevican and versican immuno-labelings are increased within days in the injured spinal cord parenchyma surrounding the lesion site with expression peaking at 2 weeks post-injury[26–27]. However, the total expression level of brevican protein is decreased in the broad epicenter area of the injured spinal cord, or is not significantly changed in the distant above- or below-levels of the injured spinal cord[27], indicating an area-specific change in the expression level of brevican. In addition, the proteoglycan, DSD-1-PG, is upregulated within the denervated outer molecular layer after lesioning of the entorhinal cortex and contributes to the re-patterning process[28]. Therefore, the upregulation of brevican by SNI may be related to a molecular process that induces structural reorganization in the spinal cord after peripheral nerve injury[29].
Reticulon-4 (known as Nogo) is expressed as one of three isoforms, Nogo-A (isoform 1), Nogo-B (isoform 2) and Nogo-C (isoform 3), by transcriptional processes and alternative splicing[30]. Nogo-A mediates the inhibition of axonal outgrowth in the CNS, including the spinal cord[31]. Recently, it has been shown that the expression of Nogo-A is increased in the injured spinal cord[32], indicating its role in structural plasticity[33]. Therefore, it would be interesting to assess whether the upregulation of reticulon-4 isoform 2 (Nogo-B) that was shown in the present study, also plays a role in axonal sprouting and targeting of primary afferents following peripheral nerve injury[29], in addition to neuropathic pain.
Activated leukocyte cell adhesion molecule (ALCAM) is a member of the immunoglobulin superfamily. It is a CD6 ligand expressed on activated leukocytes, lymphocytes and monocytes[34]. Currently, there is no literature regarding the role of ALCAM in neuropathic pain; however, its expression is increased in active multiple sclerosis and experimental autoimmune encephalomyelitis lesions, and its upregulation facilitates leukocyte invasion into inflamed CNS tissues[35]. Leukocyte invasion occurs in the spinal cord DH following peripheral nerve injury[36]. Therefore, the upregulated ALCAM that we have found in this proteomic study may contribute to neuropathic pain at a certain stage of its development.
Among metabolic enzymes identified in the SNI samples, glutathione S-transferase A4 (GSTA4) is important for scavenging electrophilic substrates, such as hydroperoxides and ROS, by catalyzing glutathione conjugation[37]. Interestingly, GST was increased in the spinal cord after CCI[38], which is in agreement with our proteomic results. Therefore, the upregulation of GSTA4 could be related to the outcome of neuropathic pain development.
Recently, it has been shown that SNI induces expression of phospholipase (PLC) β3 in mouse and human DRG neurons with mainly small and non-peptidergic phenotypes; systemic treatment with the PLC inhibitor U73122 increases the pain threshold in mice in a rapid and long-lasting (48 hours) manner[39]. Therefore, the upregulation of PLCβ1 that was shown in this study may contribute to neuropathic pain following SNI.
In conclusion, our proteomic data provide the expression pattern of dorsal spinal cord proteins, which have diverse functions, such as cell/neurite growth, cell adhesion, structure, channels, transporters, metabolism, signal transduction, following SNI. Particularly, as mentioned above, proteins regulated by the SNI are related to functional roles in anti-oxidation (S-adenosylmethionine synthetase, peroxiredoxin-6, GSTA4), axon regeneration/myelination (periaxin, Caspr 1 precursor, brevican, reticulon-4), leukocyte invasion (ALCAM), and signal transduction (importin-7, PLCβ3). Some of the proteins that were identified in sham or SNI mice may be important new therapeutic targets for neuropathic pain. This should be assessed with future systemic studies.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2015R1D1A1A01059432).
References
- 1. Zimmermann M. Pathobiology of neuropathic pain[J].Eur J Pharmacol, 2001,429(1-3):23–37. [DOI] [PubMed] [Google Scholar]
- 2. Scholz J, Woolf CJ. Can we conquer pain[J] Nat Neurosci, 2002,5(Suppl):1062–1067. [DOI] [PubMed] [Google Scholar]
- 3. Mallick P, Kuster B. Proteomics: a pragmatic perspective[J].Nat Biotechnol, 2010,28(7):695–709. [DOI] [PubMed] [Google Scholar]
- 4. Komori N, Takemori N, Kim HK, et al. Proteomics study of neuropathic and nonneuropathic dorsal root ganglia: altered protein regulation following segmental spinal nerve ligation injury[J].Physiol Genomics, 2007,29(2):215–230. [DOI] [PubMed] [Google Scholar]
- 5. Zou W, Zhan X, Li M, et al. Identification of differentially expressed proteins in the spinal cord of neuropathic pain models with PKCgamma silence by proteomic analysis[J].Brain Res, 2012,1440:34–46. [DOI] [PubMed] [Google Scholar]
- 6. Sui P, Watanabe H, Ossipov MH, et al. Proteomics of neuropathic pain: proteins and signaling pathways affected in a rat model[J].J Proteome Res, 2014,13(9):3957–3965. [DOI] [PubMed] [Google Scholar]
- 7. Kunz S, Tegeder I, Coste O, et al. Comparative proteomic analysis of the rat spinal cord in inflammatory and neuropathic pain models[J].Neurosci Lett, 2005,381(3):289–293. [DOI] [PubMed] [Google Scholar]
- 8. Lee SC, Yoon TG, Yoo YI, et al. Analysis of spinal cord proteome in the rats with mechanical allodynia after the spinal nerve injury[J].Biotechnol Lett, 2003,25(24):2071–2078. [DOI] [PubMed] [Google Scholar]
- 9. Bourquin A.F., Suveges M., Pertin M., et al. , Assessment and analysis of mechanical allodynia-like behavior induced by spared nerve injury (SNI) in the mouse[J]. Pain 122 (2006)14 e11–14. [DOI] [PubMed] [Google Scholar]
- 10. Keller A, Nesvizhskii AI, Kolker E, et al. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search[J].Anal Chem, 2002,74(20):5383–5392. [DOI] [PubMed] [Google Scholar]
- 11. Nesvizhskii AI, Keller A, Kolker E, et al. A statistical model for identifying proteins by tandem mass spectrometry[J].Anal Chem, 2003,75(17):4646–4658. [DOI] [PubMed] [Google Scholar]
- 12. Gillespie CS, Sherman DL, Blair GE, et al. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment[J].Neuron, 1994,12(3):497–508. [DOI] [PubMed] [Google Scholar]
- 13. Scherer SS, Xu YT, Bannerman PG, et al. Periaxin expression in myelinating Schwann cells: modulation by axon-glial interactions and polarized localization during development[J].Development, 1995,121(12):4265–4273. [DOI] [PubMed] [Google Scholar]
- 14. Gillespie CS, Sherman DL, Fleetwood-Walker SM, et al. Peripheral demyelination and neuropathic pain behavior in periaxin-deficient mice[J].Neuron, 2000,26(2):523–531. [DOI] [PubMed] [Google Scholar]
- 15. Wallace VC, Cottrell DF, Brophy PJ, et al. Focal lysolecithin-induced demyelination of peripheral afferents results in neuropathic pain behavior that is attenuated by cannabinoids[J].J Neurosci, 2003,23(8):3221–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Einheber S, Zanazzi G, Ching W, et al. The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination[J].J Cell Biol, 1997,139(6):1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peles E, Salzer JL. Molecular domains of myelinated axons[J].Curr Opin Neurobiol, 2000,10(5):558–565. [DOI] [PubMed] [Google Scholar]
- 18. Wolswijk G, Balesar R. Changes in the expression and localization of the paranodal protein Caspr on axons in chronic multiple sclerosis[J].Brain, 2003,126(Pt 7):1638–1649. [DOI] [PubMed] [Google Scholar]
- 19. Henry MA, Freking AR, Johnson LR, et al. Sodium channel Nav1.6 accumulates at the site of infraorbital nerve injury[J].BMC Neurosci, 2007,8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Avila MA, Corrales FJ, Ruiz F, et al. Specific interaction of methionine adenosyltransferase with free radicals[J].Biofactors, 1998,8(1-2):27–32. [DOI] [PubMed] [Google Scholar]
- 21. Guedes RP, Araújo AS, Janner D, et al. Increase in reactive oxygen species and activation of Akt signaling pathway in neuropathic pain[J].Cell Mol Neurobiol, 2008,28(8):1049–1056. [DOI] [PubMed] [Google Scholar]
- 22. Park ES, Gao X, Chung JM, et al. Levels of mitochondrial reactive oxygen species increase in rat neuropathic spinal dorsal horn neurons[J].Neurosci Lett, 2006,391(3):108–111. [DOI] [PubMed] [Google Scholar]
- 23. Phelan SA, Wang X, Wallbrandt P, et al. Overexpression of Prdx6 reduces H2O2 but does not prevent diet-induced atherosclerosis in the aortic root[J].Free Radic Biol Med, 2003,35(9):1110–1120. [DOI] [PubMed] [Google Scholar]
- 24. Gao YJ, Ji RR. Activation of JNK pathway in persistent pain[J].Neurosci Lett, 2008,437(3):180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji RR, Gereau RW 4th, Malcangio M, et al. MAP kinase and pain[J].Brain Res Rev, 2009,60(1): 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones LL, Margolis RU, Tuszynski MH. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury[J].Exp Neurol, 2003,182(2):399–411. [DOI] [PubMed] [Google Scholar]
- 27. Andrews EM, Richards RJ, Yin FQ, et al. Alterations in chondroitin sulfate proteoglycan expression occur both at and far from the site of spinal contusion injury[J].Exp Neurol, 2012,235(1):174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deller T, Haas CA, Frotscher M. Sprouting in the hippocampus after entorhinal cortex lesion is layer- specific but not translaminar: which molecules may be involved[J].Restor Neurol Neurosci, 2001,19(3-4):159–167. [PubMed] [Google Scholar]
- 29. Woolf CJ, Shortland P, Coggeshall RE. Peripheral nerve injury triggers central sprouting of myelinated afferents[J].Nature, 1992,355(6355):75–78. [DOI] [PubMed] [Google Scholar]
- 30. Oertle T, Schwab ME. Nogo and its paRTNers[J].Trends Cell Biol, 2003,13(4):187–194. [DOI] [PubMed] [Google Scholar]
- 31. Bandtlow CE, Schwab ME. NI-35/250/nogo-a: a neurite growth inhibitor restricting structural plasticity and regeneration of nerve fibers in the adult vertebrate CNS[J].Glia, 2000,29(2):175–181. [DOI] [PubMed] [Google Scholar]
- 32. Hunt D, Coffin RS, Prinjha RK, et al. Nogo-A expression in the intact and injured nervous system[J].Mol Cell Neurosci, 2003,24(4):1083–1102. [DOI] [PubMed] [Google Scholar]
- 33. Schwab ME. Increasing plasticity and functional recovery of the lesioned spinal cord[J]. Prog Brain Res, 2002,137:351–359. [DOI] [PubMed] [Google Scholar]
- 34. Bowen MA, Patel DD, Li X, et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand[J].J Exp Med, 1995,181(6):2213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cayrol R, Wosik K, Berard JL, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system[J].Nat Immunol, 2008,9(2):137–145. [DOI] [PubMed] [Google Scholar]
- 36. Hu P, Bembrick AL, Keay KA, et al. Immune cell involvement in dorsal root ganglia and spinal cord after chronic constriction or transection of the rat sciatic nerve[J].Brain Behav Immun, 2007,21(5):599–616. [DOI] [PubMed] [Google Scholar]
- 37. Salinas AE, Wong MG. Glutathione S-transferases—a review[J].Curr Med Chem, 1999,6(4):279–309. [PubMed] [Google Scholar]
- 38. Horst A, Kolberg C, Moraes MS, et al. Effect of N-acetylcysteine on the spinal-cord glutathione system and nitric-oxide metabolites in rats with neuropathic pain[J].Neurosci Lett, 2014,569:163–168. [DOI] [PubMed] [Google Scholar]
- 39. Shi TJ, Liu SX, Hammarberg H, et al. Phospholipase Cβ3 in mouse and human dorsal root ganglia and spinal cord is a possible target for treatment of neuropathic pain[J].Proc Natl Acad Sci U S A, 2008,105(50):20004–20008. [DOI] [PMC free article] [PubMed] [Google Scholar]