Abstract
In E. coli, under high pH/high salt conditions, a major Na+/H+ antiporter (NhaA) is activated to maintain an internal pH level. Its expression is induced by a specific regulator NhaR, which is also responsible for osmC and pgaA regulation. Here I report that the NhaR regulator affects the carbon storage regulatory Csr system. I found that the expression of all major components of the Csr system–CsrA regulator, CsrB and CsrC small RNAs, and the CsrB and CsrC stability were indirectly affected by nhaR mutation under stress conditions. Using a combination of experimental and in silico analyses, I concluded that the mechanism of regulation included direct and indirect activation of a two-component system (TCS) response regulator–UvrY. NhaR regulation involved interactions with the regulators H-NS and SdiA and was affected by a naturally occurring spontaneous IS5 insertion in the promoter region. A regulatory circuit was proposed and discussed.
Introduction
Living organisms have evolved many genetic mechanisms to deal with different stressful environmental factors. Bacterial pH homeostasis is important for physiology, ecology, and pathogenesis. The effect of low pH on bacterial physiology has been subjected to many studies mostly because an acidic environment is more prevalent in nature [1–5]. Lower pH also plays an important role in human defense systems, starting from the lower pH of the skin and vagina, to the highly acidic stomach environment, and ending with the low pH inside lysosomes in macrophages [6, 7]. High pH environments include natural habitats such as alkaline soda lakes and highly alkaline segments of the hindgut of certain insects, as well as industrial settings such as indigo dye plants, sewage plants, and geochemically unusual groundwater with pH values of >12 [8]. Resistance to base may also enhance survival of bacteria as they pass through the pylorus and enter the upper intestine, where they encounter alkaline pancreatic secretion [9].
Under alkaline and osmotic stress, living cells must maintain an externally directed sodium gradient and a relatively constant intracellular pH [10]. Membrane proteins that exchange Na+ (or Li+) for H+, called Na+/H+ antiporters, play important roles in these processes. NhaA is the key antiporter that protects E. coli against sodium stress, and it is essential for growth in the presence of high sodium concentrations. NhaB, a second antiporter, is necessary only in NhaA mutants [11, 12]. The nhaA gene is located in a two-gene operon, nhaAR, where the nhaR gene encodes a LysR-OxyR family transcriptional regulator [13]. The nhaAR operon is induced by the presence of monovalent cations, high pH, low temperature, and stationary phase [14, 15]. Its expression is driven by two promoters. P1 is an NhaR-dependent, Na+-induced, and H-NS-affected promoter in both the exponential and stationary phases. P2 shows minimal activity during the exponential phase but is somehow induced in the stationary phase and becomes the major promoter. P2 is activated by sigma(S) and is shown to be nonresponsive to Na+, as well as NhaR and H-NS activities [16, 17]. Recent results showed that nhaR is postranscriptionally regulated by the CsrA protein [18]. The expression of nhaAR genes is repressed by the H-NS in the lack of stress; however, in the presence of Na+, NhaR functions as a positive regulator of nhaA and overcomes the repressive effects of H-NS [17]. NhaR also activates the transcription of osmC, which is required for resistance to organic peroxides and long-term survival in the stationary phase [19, 20], as well as the pgaABCD operon required for biofilm formation in E. coli [21, 22]. H-NS is a global regulator [23] that is induced at low temperature and shown to be a common regulator of multiple iron and other nutrient acquisition systems preferentially expressed at 37°C, as well as general stress response, biofilm formation, and cold shock genes highly expressed at 23°C [24].
Biofilms are associated with various kinds of stress protection, including pH resistance. Biofilm formation is a complicated, multistage process, which is affected by many regulatory systems (see[25]). The RNA-binding protein CsrA plays an important role in many biological processes including quorum sensing and biofilm formation [26]. CsrA is a global regulatory protein that interacts with target mRNAs, changing their stability and/or translation. CsrA activates genes that are necessary for bacterial growth or are expressed in the exponential phase of growth and inhibits genes expressed in stationary phase or encoding secondary metabolites. CsrA inhibits biofilm formation by a posttranscriptional repression of pgaABCD, responsible for synthesis of a polysaccharide adhesin poly-beta-1,6-N-acetyl-d-glucosamine [27]. On the other hand, CsrA activates motility by increasing stability of flhDC mRNA [28]. CsrA expression is increased during entrance into stationary phase. Expression of the csrA gene depends on CsrA activity in the cell [29]. That activity is controlled by two small noncoding RNAs, CsrB and CsrC, which bind to and sequester multiple copies of CsrA [30, 31]. Expression of these small RNAs is affected by the SOS system [32] and by a two-component system (TCS) regulator UvrY [33]. The BarA/UvrY system is activated by an increased pH [34] as well as quorum-sensing regulator SdiA [33]. As SdiA activity is also postransciptionally regulated by the CsrA protein [35], the whole system forms a closed regulatory circuit. A recent paper by Camacho et. al. [36] demonstrated that CsrA is indirectly required for proper uvrY expression and also for activation of the BarA kinase activity.
In this paper, I examined regulatory interactions of the specific NhaR regulator and the CsrA circuit. I was able to show that under some environmental conditions NhaR is necessary for the activity of the CsrA system. Conducted experiments indicated that NhaR indirectly affects expression of csrA, csrB, and csrC and the stability of these small RNAs. A nhaR mutation directly and indirectly affects expression of uvrY genes. In the case of uvrY, the mode of regulation seems to be similar to that proposed for the nhaAR operon where NhaR is necessary to overcome H-NS inhibition. I found that naturally occurring transposition of the IS5 into the uvrY promoter region abolishes regulation of the uvrY gene and affects the whole CsrA circuit. I present and discuss a model of complex interactions between NhaR and the CsrA system.
Materials and methods
Bacterial strains and growth conditions
The E. coli strains, plasmids, and bacteriophage used in this study are listed in Table 1. All mutations except ΔH-NS were transferred into the JEK710 strain from KEIO collection strains by P1 transduction. JEK1553ΔH-NS was constructed by the λRed system and pKD3 plasmid [37]. Bacteria were grown at 37°C or 26°C, shaking at 250 rpm, in Luria-Bertani (LB) medium. LB broth pH 8.4 stabilized with 0.1 mM 3-[N-tris(hydroxymethyl)methylamino]propanesulfonic acid (TAPS) was used for the nhaR gene effect tests. Media were supplemented with antibiotics, as needed, at the following concentrations: kanamycin: 50 μg/ml; gentamicin: 10 μg/ml; ampicillin: 100 μg/ml; chloramphenicol: 25 μg/ml; and tetracycline: 10 μg/ml.
Table 1. Bacterial strains, plasmids and bacteriophage used in this study.
| Strain / plasmid/bacteriophage | Genotype | Reference |
|---|---|---|
| E. coli EC100 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG | Epicentre |
| E. coli K12 MG1655 | F- λ- | ATCC |
| GS1114 | CF7787 Δ(λatt-lom)::bla Φ(csrC-lacZ)1(hyb) AmpR | [31] |
| KSB837 | CF7787 Δ(λatt-lom)::blaΦ(csrB-lacZ) 1(hyb) AmpR | [31] |
| KSA712 | CF7787 Δ(λatt-lom)::blaΦ(csrA’-‘lacZ) 1(hyb) AmpR | [39] |
| KSY009 | CF7787 Δ(λatt-lom)::blaΦ(uvrY’-‘lacZ) 1(hyb) AmpR | [33] |
| JEK710 | MG1655 LacZ- GmR | This study |
| JEK736 | JEK710 ΔnhaR GmRKmR | This study |
| JEK821 | JEK710 Δ(λatt-lom)::bla Φ(csrC-lacZ)1(hyb) AmpRGmR | This study |
| JEK825 | JEK710 Δ(λatt-lom)::bla Φ(csrB-lacZ)1(hyb) AmpRGmR | This study |
| JEK829 | JEK736 Δ(λatt-lom)::bla Φ(csrC-lacZ)1(hyb) AmpRGmRKmR | This study |
| JEK833 | JEK736 Δ(λatt-lom)::bla Φ(csrB-lacZ)1(hyb) AmpRGmRKmR | This study |
| JEK879 | JEK710 Δ(λatt-lom)::bla Φ(csrA’-‘lacZ)1(hyb) AmpRGmR | This study |
| JEK881 | JEK736 Δ(λatt-lom)::bla Φ(csrA’-‘lacZ)1(hyb) AmpRGmRKmR | This study |
| JEK1321 | JEK710 Δ(λatt-lom)::bla Φ(uvrY’-‘lacZ)1(hyb) AmpRGmR | This study |
| JEK1315 | JEK736 Δ(λatt-lom)::bla Φ(uvrY’-‘lacZ)1(hyb) AmpRGmRKmR | This study |
| JEK1139 | JEK710 ΔsdiA GmRKmR | This study |
| JEK1553 | JEK710 ΔH-NS GmRCmR | This study |
| JEK1557 | JEK710 ΔH-NSΔnhaR GmRCmRKmR | This study |
| JEK1598 | JEK710 uvrY::uvrY-3xFlag-KmR; GmR | This study |
| JEK1601 | JEK736 uvrY::uvrY-3xFlag-KmR; GmR | This study |
| Plasmids | ||
| pKD3 | Lambda Red mutagenesis plasmid; CmR | [37] |
| pUC19 | Cloning vector; AmpR | |
| pKK223-3 | Cloning vector; AmpR | Amersham Pharmacia Biotech (Uppsala, Sweden) |
| pNhaR | nhaR in pKK223-3 | [22] |
| pNhaA | nhaA cloned under plac promoter in pUC19;AmpR (pJEK550) | This study |
| pG-GFP | Promoter probe vector gfp; AmpR | [40] |
| pJEK1224 | 585 bp of recA promoter fragment (-600,-15) cloned into pG-GFP bp fragment | This study |
| pJEK1263 | 349 bp of uvrY promoter fragment (-358, -9) cloned into pG-GFP | This study |
| pJEK1270 | 625 bp of uvrY promoter fragment (-634, -9) cloned into pG-GFP | This study |
| pJEK1272 | pJEK1270 with a single deletion in H-NS5 motif | This study |
| pJEK1275 | pJEK1270 with a single substitution in H-NS5 motif | This study |
| pJEK1292 | 185 bp of uvrY promoter fragment (-196, -9) cloned into pG-GFP | This study |
| pJEK1295 | 761 bp of uvrY promoter fragment (-770, -9) cloned into pG-GFP | This study |
| pJEK1301 | 251 bp of uvrY promoter fragment (-260, -9) cloned into pG-GFP |
This study |
| Bacteriophage | ||
| P1 vir | Strictly lytic P1 | Carol Gross |
Construction of lacZ and GFP reporter fusions
CsrA, csrB, csrC, and uvrY transcriptional and translational fusions to lacZ were constructed previously [31, 33, 38, 39] and transduced into the JEK710 strain and its derivative mutants. PCR fragments containing uvrY, sdiA, recA, and lexA were amplified with specific primer pairs (Table 2), cloned into EcoRI/KpnI sites of the pG-GFP plasmid [40], and introduced into the JEK710 strain and its derivative mutants. GFP activity (A480-520) was measured using a BioTek Plate Reader (BioTek) and normalized to the optical density of the culture (A600), yielding relative fluorescence units (RFU; A480-520/A600). Student T-test was used to compare results and check statistical significance.
Table 2. Oligonucleotides used in the study.
| Primer name | Sequence (5’-3’) | Purpose |
|---|---|---|
| prJEK1 | /5Biosg/AAA GGC GTA AAG TAG CAC CCA TAG | csrC for EMSA. |
| prJEK40 | CAA AGC GGT CGT CTC CGT CAG TC | csrC for EMSA. |
| prJEK6 | /5Biosg/TCG ACG AAG ATA GAA TCG TCT T | csrB for EMSA. |
| prJEK7 | TAA TCC AAA TAC CCC ATC TGG | csrB for EMSA. |
| prJEK66 | AAA AAG CTT GAA ACA TCT GCA TCG ATT CT | nhaA for cloning |
| prJEK67 | AAA GGA TCC ACA TGC TCA TTT CTC TCC CTG | nhaA for cloning |
| prJEK123 | /5Biosg/GTG GTC ATC AAC AAG TAG AAC G | Reverse for uvrY EMSA |
| prJEK122 | GTG GTC ATC AAC AAG TAG AAC G | uvrY primer extension 1 |
| prJEK125 | CAG TTA TGG TCA CGC CCG TC | uvrY primer extension 2 |
| prJEK132 | AAA GAA TTC CAG AAA TAG GGA TAA CG | uvrY (-9) reverse for cloning into pG-GFP. |
| prJEK133 | AAA GGT ACC GAG CGT GAT ATC GGC AGT GC | yvrY (-358) for cloning (pJEK1263) and EMSA. |
| prJEK135 | AAA GGT ACC GCA GCC TGG GTT TCG TCT TC | uvrY (-634) for cloning (pJEK1270). |
| prJEK155 | AAA GAA TTC CCG CTA CTG GCT TAA TTT GAT CTC | uvrY (-770) for cloning (pJEK1270). |
| prJEK156 | AAA GAA TTC GTT ACA TAT TCA GCG GGC TG | uvrY (-260) for cloning (pJEK1301). |
| prJEK172 | CCT CAA CAA ACC ACC CCA ATA TAA GTT TGA GAT TAC TAC AGT GTA GGC TGG AGC TGC TTC | H-NS deletion with pKD3 |
| prJEK173 | GCC GCT GGC GGG ATT TTA AGC AAG TGC AAT CTA CAA AAG AAT GGG AAT TAG CCA TGG TCC | H-NS deletion with pKD3 |
Beta-galactosidase assay
Assay for the activity of all lacZ reporter fusions was conducted as described earlier [41]ith the following modifications. Cells were permeabilized with 100 μl of chloroform and 50 μl of 0.01% (w/v) SDS and reactions were terminated with 0.5 ml of 1 M Na2CO3. LacZ activities were presented as Miller units per OD600 or protein amounts. Protein estimation was done following the bicinchoninic acid method after precipitation with 10% trichloroacetic acid (TCA). Student T-test was used to compare results and check statistical significance.
Electrophoretic mobility shift assays (EMSA)
DNA fragments located upstream of the csrB (220 bp; pos. -215+5), csrC (208 bp; pos. -204+4), and uvrY (388 bp pos. -358+30 and 228 bp pos. -198+30) genes were amplified by PCR using specific primer sets (Table 2). Each set contained a single 3’-end commercially biotinylated primer. These PCR products (1fM) and different amounts of purified recombinant NhaR (NhaR-His6) protein [22] were analyzed using the LightShift Chemiluminescent EMSA Kit as described by the manufacturer (Pierce Biotech., Rockford, IL). A previously described 138-bp promoter region of the pgaABCD genes [22] was used as a specific control and competitor. Binding reactions were separated on 5% PAGE gels, electrotransferred onto a positively charged nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany), and detected using the Chemiluminescent Nucleic Acid Detection Module (Pierce Biotech., Rockford, IL), according to the protocol. Fluorescent bands were visualized by a ChemiDoc XRS+ Imaging System and analyzed using Quantity One Software (BioRad Lab., Hercules, CA).
Northern and Western blotting
Total RNA was isolated using the RNAprotect Bacteria Reagent with the RNeasy kit (Qiagen, Valencia, CA) at different time points from bacterial cultures grown with shaking (250 rpm) at 26°C. For blotting, 1.5 μg of total RNA was separated in denaturing conditions on 5% polyacrylamide gels with urea (7 M) and blotted onto positively charged nylon membranes (Roche Diagnostics GmbH, Mannheim, Germany) by electrotransfer for 40 min. The RNA on the membrane was UV crosslinked, stained with methylene blue solution to control transfer efficiency, and developed following the DIG Northern Starter Kit manual (Roche Diagnostics GmbH, Mannheim, Germany) with DIG-labeled RNA probes specific for the csrB and csrC genes prepared as described previously [42].
The Western blotting protocol was described previously [18]. Bacterial cultures were grown with shaking at 26°C in specified media and cells were harvested at different time points. Cells were mixed with Laemmli sample buffer and lysed by sonication and boiling. Each sample (10 μg protein) was subjected to SDS-PAGE. Proteins were transferred to 0.2-μm PMSF membranes, and NhaR-Flag was detected using anti-Flag monoclonal antibodies as recommended by the manufacturer (Sigma-Aldrich). Quantification analyses were performed using a Syngene GeneTools software.
Primer extension analysis
Total RNA was prepared from cells grown for 18 h in LB pH 8.4 TAPS at 26°C. Primers prJEK122 and prJEK125 were annealed at positions +30 and -136 relative to the transcript initiation site of uvrY, respectively. Primer extension analyses were performed according to the protocol of Wang et al. [27]. Results are shown in Supplementary materials (S1 Fig).
Results and discussion
NhaR regulates expression of CsrA system at low temperature in high pH/high salt medium
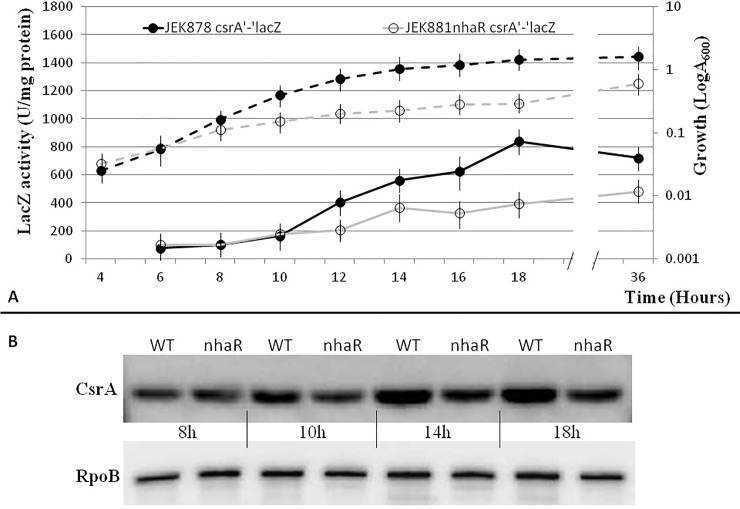
Previous reports have mentioned that viability of E. coli nhaR mutant strains is affected in high salt (<0.1 M) and high pH (>8.4) media; however, detailed information about growth conditions was not supplied [13]. In order to study the effect of NhaR on the CsrA system, I defined growth conditions where nhaR plays its regulatory role affecting the CsrA system. I used previously constructed lacZ gene fusions with csrA, csrB, and csrC genes. These fusions were transduced from the original strains into the E. coli MG1655lacZ- (JEK710) strain and its isogenic nhaR mutant (JEK736). As expected, at 37°C in LB pH 7.4 TAPS, the differences between the wild-type (WT) strain and nhaR mutant were negligible (data not shown). In LB pH 8.4 TAPS, although the specific lacZ activities were slightly lower than in LB pH7.4 medium, no differences between the nhaR mutant and the isogenic WT strain were observed (data not shown). A completely different situation was observed when cells were grown in LB pH 8.4 TAPS at low temperature (26°C). The activity of csrA’-‘lacZ fusion was up to 2.2-fold lower in the nhaR mutant strain than in the WT strain (Fig 1A). To confirm these data, I conducted Western blotting analysis of total proteins isolated from the nhaR mutant and the isogenic WT strain using CsrA-specific antibodies. Hybridization results confirmed that the amount of CsrA protein in the nhaR mutant was lower and the difference level was similar to those for the lacZ fusions (Fig 1B). As one could expect, the biggest difference was observed in early stationary phase (in these conditions 14–18 h) when csrA gene expression is usually induced. The amount of CsrA in the mutant strain was 1.33 to 1.38 lower than in the WT. The EMS experiment with csrA promoter and NhaR protein showed that the interaction is not specific and can be abolished with a specific (pgaA) and nonspecific (dI-dC) competitors (S2 Fig).
Fig 1. Effect of nhaR mutation on csrA gene expression.
Growth curves (dashed lines) and activity of csrA’-’lacA fusion (solid lines) in WT and nhaR mutant of E. coli MG1655 grown at 26C in LB pH8.4 TAPS (A). Western blot analysis of CsrA protein in WT and nhaR mutant (B). RpoB was used as loading control. Data shows representative results from 4 experiments.
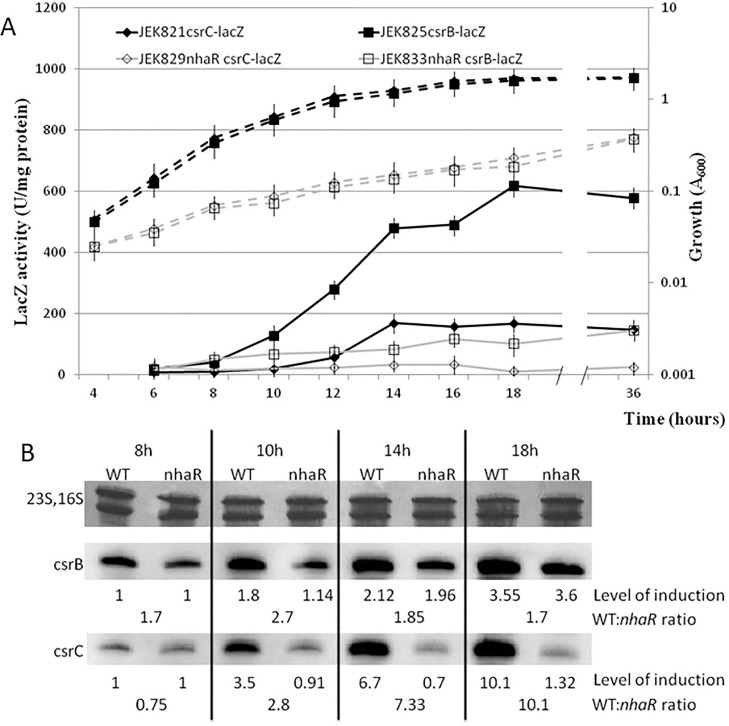
Subsequently, I checked the effect of nhaR mutation on two other components of the Csr system: csrB and csrC. LacZ assay results showed a very strong effect of nhaR mutation on expression of both csrB and csrC genes (Fig 2A). In the WT strain, both the csrB and csrC genes were induced as cells entered the stationary phase (10–12 h). In the nhaR mutant, no induction was observed. The expression of csrB was at a very low level while the csrC promoter showed almost no activity (Fig 2A). The maximal difference in expression activities between the nhaR mutant and isogenic WT strain was 5.2-fold for csrB and up to 12-fold for csrC.
Fig 2. Effect of nhaR on csrB and csrC expression.
Growth curves (dashed lines) and activity of csrB-lacZ and csrC-lacZ fusions (solid lines) in WT and nhaR mutant of E. coli MG1655, grown at 26°C in LB pH 8.4 TAPS (A). Northern blot analysis of csrB and csrC transcripts (B). Total RNA was isolated at different time points from the WT and nhaR mutant strains grown at 26C in LB pH 8.4 TAPS. Level of induction shows amounts of RNA along the growth curve starting with 1 at 8h; WT:nhaR ratio was calculated for each time point. The 16S and 23S rRNA were stained with methylene blue and served as an internal control.
To confirm these differences, a Northern blot hybridization to total RNA isolated from the nhaR mutant and WT strains grown in the same conditions was conducted with csrB and csrC gene probes. Hybridization results confirmed that nhaR is necessary for expression of both small RNAs; however, differences in the csrB and csrC RNA levels between mutant and WT strains were much lower (max. 2.7- and 10.1-fold for csrB and csrC, respectively) than those observed in lacZ fusions (Fig 2B).
NhaR affects stability of csrB and csrC small RNAs
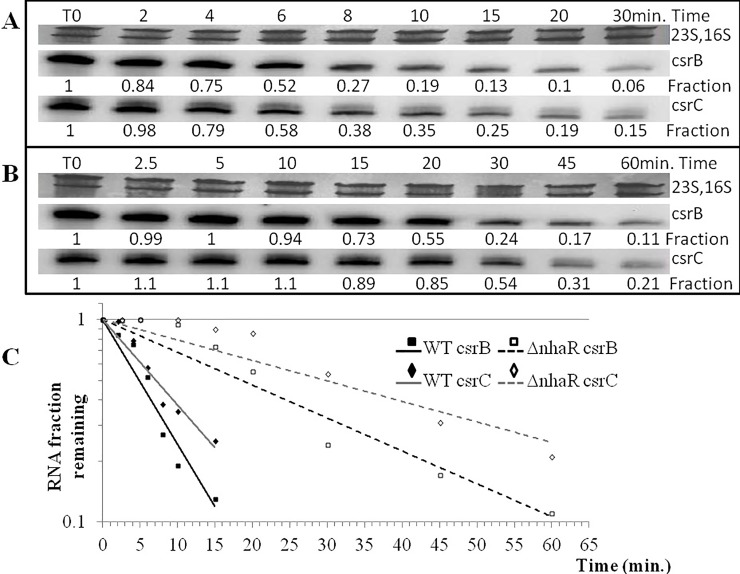
The difference between the expression levels and the actual RNA levels in the cell can be explained by a higher RNA stability in the nhaR mutant strain. To prove this hypothesis, I measured the half-lives of csrB and csrC RNAs in the nhaR and WT strains grown in LB pH 8.4 TAPS at 26°C (Fig 3). The data showed that in the WT strain the turnover for both RNAs was relatively fast (half-lives: 5 and 7 minutes for csrB and csrC, respectively), while in the nhaR mutant strain those RNAs were much more stable with half-lives 2.6 and 3.14 times longer for csrB and csrC, respectively, than in the WT strain. These differences in small RNA stability explained the observed differences between the transcriptional activity measured by lacZ fusions and the actual RNA levels shown by Northern hybridization.
Fig 3.
Stability of csrB and csrC in WT (A) and nhaR mutant (B) of E. coli MG1655, grown for 18 h at 26°C in LB pH 8.4 TAPS. RNA synthesis was inhibited by rifampicin (300μg/ml) and RNA was isolated at different time points after inhibition. Fraction of remaining RNA at each time point is shown. The 16S and 23S rRNA were stained with methylene blue and served as an internal control. The RNA half-lives were determined from the linear portions of the decay curves (C). The CsrB half-life in isogenic MG1655 (wild type) and nhaR mutant strains was 5 and 13 min., respectively. The CsrC half-life in the same strains was 7 and 22 min., respectively.
To check if NhaR directly activates transcription of csrB and csrC genes, I used an electrophoretic mobility shift assay (EMSA; See Methods section). Obtained results suggested that NhaR does not interact directly with the csrB and csrC promoters (data not shown).
NhaR regulates uvrY expression
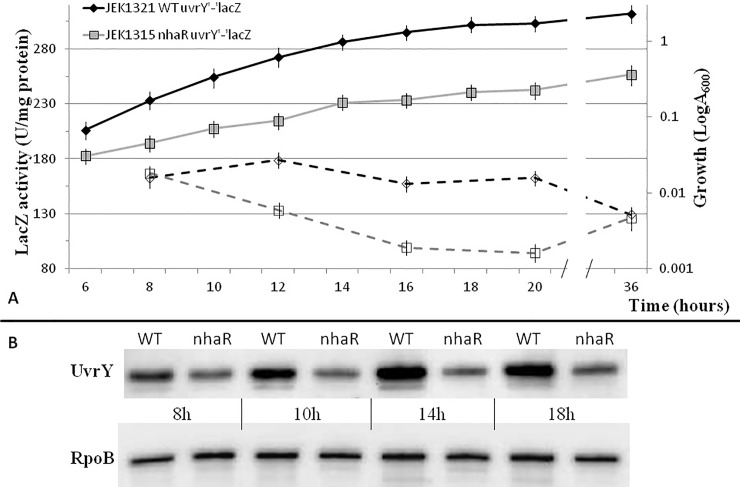
Expression of csrB and csrC RNAs depends on the presence of the TCS response regulator UvrY [33]; therefore, I analyzed if mutation in the nhaR gene somehow affects expression of the uvrY gene. The previously constructed translational uvrY’-‘lacZ fusion (KSY0009) was transferred into E. coli MG1655 JEK710 and its isogenic nhaR mutant strains. A time course experiment showed that the expression of uvrY gene was up to 1.7-fold lower in the mutant strain in early stationary phase (Fig 4A).
Fig 4. Effect of nhaR mutation on uvrY gene expression.
Growth curves (solid lines) and activity of uvrY’-’lacA fusion (dashed lines) in WT (JEK1321) and nhaR mutant (JEK1315) of E. coli MG1655 grown at 26C in LB pH8.4 TAPS (A). Western blot analysis of the UvrY-Flag protein in WT and nhaR mutant (B). RpoB was used as loading control. Data shows representative results from 3 experiments.
To confirm these results, I analyzed the actual level of UvrY protein. The chromosomal uvrY gene was modified in situ such that it produced a protein containing a 3xFlag epitope tag at the C terminus. Western immunoblotting studies with an anti-Flag monoclonal antibody revealed that the UvrY-Flag protein level was higher (up to 7-fold) in the WT than in the isogenic nhaR mutant (Fig 4B).
IS5 insertion in the sdiA-uvrY intergenic region abolishes the effect of nhaR mutation
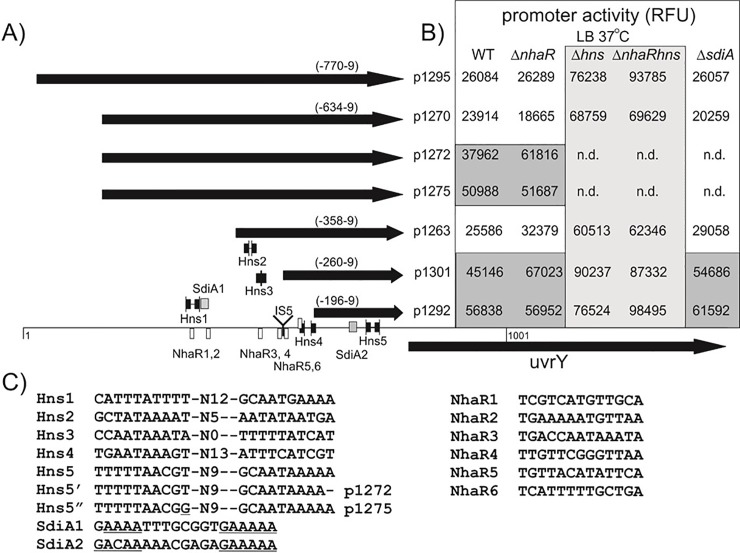
During my experiments with csrC-lacZ gene fusions and Northern hybridizations, I noticed that some of my clones behaved differently–showing no effect of nhaR mutation on both LacZ activity and the csrB and csrC levels (data not shown). Amplification of the sdiA-uvrY intergenic region revealed that the DNA fragment amplified from those atypical strains was ~1.3 kb bigger than in the WT E. coli MG1655 strain (data not shown). The DNA sequence analysis showed an insertion of IS5 element in that region, 260 bp upstream of the uvrY start codon (Fig 5).
Fig 5. Analysis of uvrY promoter region.
Fragments cloned into pG-GFP reporter plasmid are represented by arrows (A). Predicted binding sites for H-NS, SdiA and NhaR are marked as linked black arrows, gray and white boxes, respectively. Expression activity of each clone in a given strain in standard LB (18h cultures at 37°C) is presented in the table (B). DNA sequence for each binding site including two mutations in H-NS5, is shown. Conserved motifs in SdiA sites are underlined (C).
In silico analysis of regulatory motifs in uvrY promoter region
Back in the mid 1980’s when the UvrY gene was originally described, three promoters were identified upstream of the uvrY gene: one promoter in front of the current sdiA gene (P1), and two promoters (P2a and P2b) in front of the uvrY gene [43]. My in silico and primer extension analyses revealed the possible presence of additional promoter sequences in front of the uvrY gene (See S1 Fig). Primer extension analysis showed that the only difference between the WT and nhaR mutant strains was lack of a single extension product (T-542) in the mutant strain (See S1 Fig).
The DNA sequence motif that is recognized by the NhaR protein is not well characterized. According to the PRODORIC Database [44], the consensus position weight matrix constructed for the NhaR binding site contained 11 nucleotides 5’- TcgtaAaAAac-3’. I noticed that this motif was not correct as all LysR-type regulators recognize a consensus sequence (T-N11-A) [45]. Using D-MATRIX software [46] and available sequence data [43, 47], I constructed a new nhaR consensus sequence 5’–TCgaAAAAatCtA-3’ and identified at least 6 hypothetical NhaR binding sites in front of uvrY gene (Fig 5). I noticed that three of the NhaR binding sites were located immediately upstream of the T-542 transcription start, which was not detected in the nhaR mutant strain (see above).
As some reports showed that uvrY gene expression is regulated by the SdiA protein, I also searched for the presence of a DNA motif that might be responsible for binding of that regulator. SdiA belongs to the LuxR family and its binding site was determined as 5′-AAAAGNNNNNNNNGAAAA-3′ [48]. Searching for similar motifs, I was able to identify two regions with high similarity. One of them was located 102 bp upstream of the uvrY start codon while the second was located further upstream (-415 bp) (Fig 5).
While running the virtual footprint analysis, I also noticed a high number of motifs responsible for binding of the H-NS protein. As NhaR is known to interact with H-NS protein, as in the case of nhaAR regulation, I decided to have a closer look at the location of those sites. Ten H-NS binding sites were predicted in total. Analysis of detailed location revealed that these sites form a kind of “dyad” structures with some of them oriented toward each other (H-NS 2, 3, 4) and some heading in opposite directions (H-NS1, 5) (Fig 5). I noticed that three out of five hypothetical binding sites for the H-NS regulator overlapped with the predicted NhaR sites. Also, the distal SdiA binding motif overlapped with the NhaR binding site (Fig 5), suggesting that all three regulators play a role in the regulation of uvrY expression.
NhaR interacts with H-NS and SdiA in regulation of the uvrY promoter
To analyze the effect of these three regulators, I amplified a set of nested DNA fragments with a downstream primer located at pos. -9 in relation to the uvrY start codon and different upper primers. Obtained amplicons were cloned in front of a promoterless gfp gene in a pG-GFP promoter probe vector [40] resulting in plasmids pJEK1295 (-770;-9bp in relation to uvrY start point), pJEK1270, pJEK1272, pJEK1275 (all -634;-9bp), pJEK1263 (-358;-9bp), pJEK1301 (-260;-9bp), and pJEK1295 (-196;-9bp in relation to uvrY start point) (Fig 5). All clones were sequenced to confirm DNA sequence identity. When fragment (-634,-9) was cloned, I noticed that two clones showed a stronger fluorescence than others (pJEK1272 and p1275). DNA sequence analysis revealed the presence of a single-base deletion (pJEK1272) and a single-base substitution (pJEK1275) within the H-NS5 region (Fig 5). These two, together with other plasmids, were transferred into E. coli MG1655 (JEK710) and its isogenic nhaR, H-NS, nhaRH-NS, and sdiA deletion mutants. Promoter activity was measured as fluorescence (A480-520) per culture density (A600) and expressed as relative fluorescence units (RFU). At 37°C in standard LB medium, the basic uvrY promoter activity was 25,400±4,100 RFU and was observed for pJEK1295, p1270, and p1263 in the WT, ΔnhaR, and ΔsdiA strains. As plasmid pJEK1263 did not contain NhaR1, NhaR2, SdiA1, and H-NS1 sites, it seemed that those sites are not important for the expression of uvrY promoter under these conditions. Plasmid pJEK1301 contained a 251-bp fragment that lacks H-NS2, H-NS3, as well as NhaR3 and NhaR4 binding sites and was designed specially to imitate the effect of IS5 insertion (Fig 5). Its activity in the WT strain was 1.8 times higher than longer fragments (P = 0.0012). Also, in the nhaR and sdiA mutants, the activity of the shorter fragment was 2.64 and 2.15 higher than the p1295 fragment, respectively. The shortest fragment pJEK1292 (196 upstream of the uvrY start codon) contained only H-NS5 and SdiA2 motifs. Its specific activity was again ~12% higher than pJEK1301 in the WT and sdiA mutant strains and 15% lower in the nhaR mutant (Fig 5), suggesting that the presence of NhaR and both NhaR5 and NhaR6 binding sites slightly activates expression of the uvrY promoter. In E. coli H-NS and H-NS/nhaR double-deletion mutants, the activity of all reporter plasmids was much (2.4- to 3.9-fold) higher than in the corresponding WT strain and these results were extremely statistically significant (P = 0.0007 for H-NS and P = 0.001 for the double mutant) (Fig 5B). Also, plasmids pJEK1272 and p1275, which carry single-nucleotide mutations in the H-NS5 site, showed 1.5- to 2.4-fold increases in activity, respectively, compared to the WT strain (P = 0.0013 and P = 0.0012, respectively) (Fig 5). All these data show strong statistical significance and strongly suggest that the H-NS protein is involved in inhibition of the uvrY promoter.
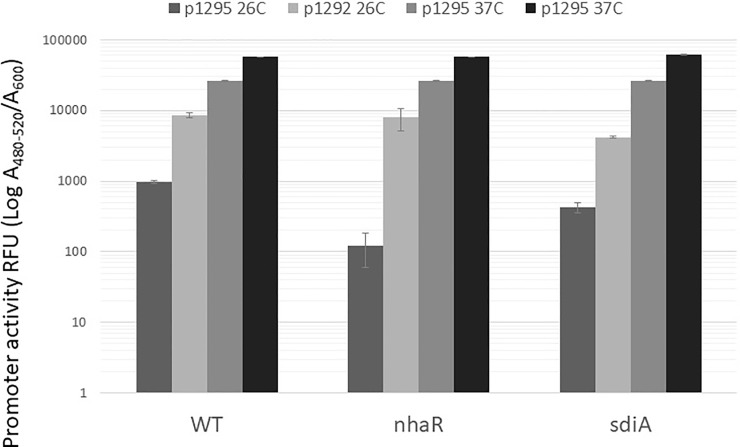
I also analyzed uvrY promoter activity under the conditions where I saw the biggest effect of nhaR mutation on the csrA system (18 h, 26°C LB pH 8.4 TAPS). Results showed that under these conditions the activity of the uvrY promoter was much lower than at 37°C (Fig 6). For the longest fragment, which contained the whole intergenic region (pJEK1295), that difference was more than 26-fold, while for the shortest one (pJEK1292), that difference was 6.6-fold, in the WT strain. The major observation was that in both nhaR and sdiA mutants, the activities of the longer fragment were reduced 8 and 2.3 times compared to the WT strain (Fig 6). Both results were statistically significant with P = 0.0004 and P = 0.0024, respectively. That 8-fold difference in uvrY promoter activity between the WT and nhaR mutant was closer to the effect observed in NhaR Western blot analysis where the difference was much higher (7-fold) than between the corresponding uvrY’-‘lacZ fusions (Fig 4). In the case of the shorter fragment, mutation in the nhaR gene did not affect its activity (P = 0.25) while sdiA mutation reduced its activity 2-fold (P = 0.0048). As that fragment contains the second SdiA binding site (SdiA2, Fig 5), it was not surprising that the lack of that regulator reduced its expression activity. Unfortunately, I was not able to analyze uvrY promoter activity in the H-NS mutant strain under these conditions using the pG-GFP constructs. Antibiotic selection and cost of plasmid-expressing GFP protein together with the H-NS mutation made the strain unable to grow in LB pH 8.4 TAPS at 26°C.
Fig 6. Transcriptional activity of selected fragments from the uvrY promoter region in stress conditions.
E. coli MG1655 (JEK710) and its isogenic nhaR and sdiA deletion mutants with reporter plasmids were grown for 18 h at 26°C in LB pH8.4 TAPS. Fluorescence and cell density were measured to calculate promoter activity in RFU. Data shows representative results from 3 experiments.
Mapping of the sdiA-uvrY intergenic region containing the regulatory signals recognized by NhaR
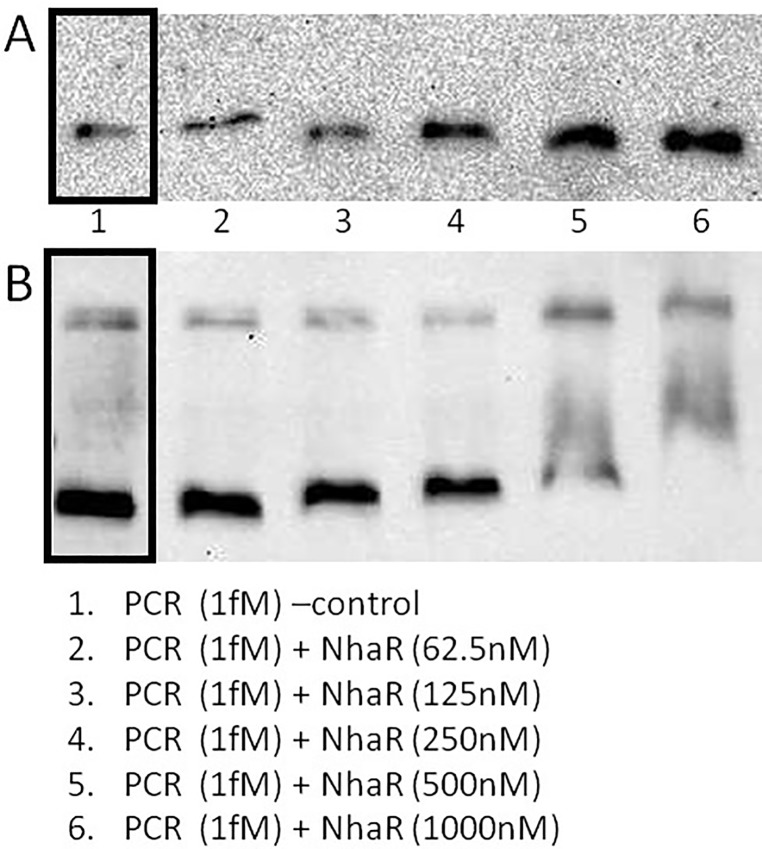
Two fragments pJEK1295 and p1292 showed different sensitivity to nhaR mutation (Fig 6). Although in silico analysis showed that all hypothetical NhaR binding sites were located further upstream, I decided to confirm if that DNA region contains the cis-elements recognized by NhaR. I PCR-amplified sequences from pJEK1292 and p1263 that contained four predicted NhaR sites (NhaR3-6, Fig 5) using a DIG-labeled primer. Each fragment was tested for binding to the purified NhaR-His tagged protein in a DNA gel retardation assay (Fig 7). It was previously shown that the purified protein binds specifically to the pgaA gene promoter [22]. My data showed that in fact the smaller PCR fragment did not react with increased NhaR protein concentrations (Fig 7A), while the longer amplicon was shifted when 125 and 250 nM NhaR was added, with even bigger shifts at higher protein concentrations (Fig 7B). That experiment confirmed that in fact there were no sites recognized by the NhaR protein in pJEK1292 and predicted sites located further upstream (pJEK1263) were responsible for NhaR regulation.
Fig 7. Mapping of the DNA region containing the cis-regulatory elements of uvrY recognized by NhaR.
PCR- amplified fragments from pJEK1292 (A) and p1263 (B) were incubated with increasing amounts of NhaR protein for 20 min. at room temperature in a buffer containing 10mM Tris pH7.5, 50mM KCl, 1mM DTT, 5mM MgCl2 and 2.5% glycerol and detected by immunoblotting. In “B” lower concentrations of NhaR were removed and lines 2–6 were moved left next to the control line 1. Boxed lines–controls without NhaR.
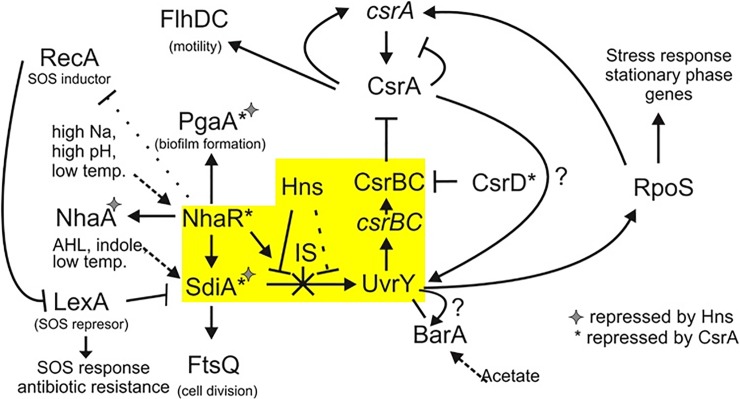
Based on my experimental data as well as available literature, I drew a broader model of interactions involving NhaR, H-NS, and SdiA that affect the Csr regulatory system (Fig 8). As the UvrY/CsrA branch of my model has been recently reviewed [49], here I focus only on the part involving NhaR regulation. The NhaR has been shown previously to regulate its own operon (nhaAR) [13], the pgaABCD operon [22], as well as the osmC gene [20] (omitted in my model). Here I showed that NhaR under specific stress conditions is necessary for expression of the uvrY gene. It has been shown previously that UvrY activity was increased under the alkaline conditions [34]; however, a detailed mechanism of that regulation was not revealed. Here I showed that that mechanism depended on NhaR and involved interaction with H-NS and SdiA, as I showed that regulation is extremely important at low temperature when both H-NS and SdiA are induced [24]. In addition, under alkaline conditions, the activity of amino acid deaminases including TnpA is induced resulting in indole production [1] which can act as a quorum-sensing particle activating SdiA [50]. The effect of indole and SdiA on biofilm formation has been published [50]; however, I could not confirm those data using my E. coli strains (data not shown). I noticed that in some laboratory strains the regulation of the uvrY gene was completely destroyed by the presence of the IS5 insertion sequence in the uvrY promoter. That randomly occurring transposition into a nhaR “hot spot” can explain observed differences in response to indole.
Fig 8. Model of interactions between NhaR, SdiA and H-NS affecting the Csr system activity.
Possible indirect effect of NhaR on RecA is shown as a dotted line. Transcriptional and posttranscriptional gene inductors are shown in the case of nhaR, sdiA and BarA. A dotted line in the case of H-NS represents a possible weak repression by reaction with the H-NS5 binding site. The yellow box highlights the interactions analyzed in this work.
However, the NhaR is necessary for the regulation of uvrY, it seems that the H-NS protein is the major player in that system. An effect of H-NS mutation on β-1,6-GlcNAc (PGA) synthesis and biofilm formation has been checked in our lab previously, but only weak changes were observed [22]. As the H-NS protein shows a domain structure, it has been shown that some mutations do not affect its activity [51]. I noticed that in the previous mutant a Tn10 insertion was located near the 3’end of the gene and could explain the data. When a complete deletion H-NS mutant was tested in a standard 96-well plate experiment at 26°C, an almost 4.7-fold increase in biofilm formation with respect to the WT strain was observed (data not shown).
In the “omics” era, it is relatively easy to study gene expression in different environmental-stress conditions. Recent work using an Integrative FourD-omic approach (INFO) [52] and the next generation sequencing of immunoprecipitated DNA fragments (CLIP-seq) [53] revealed and confirmed a global role of the CsrA system in gene regulation. Despite that, detailed mechanisms of regulation and interactions affecting expression of specific genes or pathways still have to be studied at the molecular level. My current work may serve as an introduction to more detailed molecular studies of interactions among the NhaR, H-NS, and SdiA regulators.
Supporting information
DNA sequence upstream the uvrY gene (C); primers are underlined, primer extension products starts are marked as capital letters, predicted binding sites for Hns underlined italics with arrows showing orientation, SdiA and NhaR binding sites are bold and bold underlined respectively; IS5 insertion site is marked by a triangle.
(TIFF)
(TIFF)
Acknowledgments
This work was supported by Dr. Tony Romeo NIH grant GM066794 and grant FLA-MCS-004949 from the University of Florida CRIS project. The author thanks Dr. Garth Ehrlich, FAAAS, Professor of Microbiology and Immunology, Professor of Otolaryngology-Head and Neck Surgery, Executive Director for the Center for Advanced Microbial Processing (CAMP), Center for Genomic Sciences, Genomics Core Facility, Clinical and Translational Research Institute, Drexel College of Medicine, for the financial support in publishing this paper. The author would like to thank Drs. Archana Pannuri for help with Western blotting, Christopher Vakulskas for supplying the uvrY-Flag construct, and Manish Kumar for help in biofilm experiments. I thank Jocelyn Hammond for English Language editing and comments.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Dr. Tony Romeo NIH grant GM066794 and grant FLA-MCS-004949 from the University of Florida CRIS project.
References
- 1.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9(5):330–43. Epub 2011/04/06. nrmicro2549 [pii] 10.1038/nrmicro2549 ; PubMed Central PMCID: PMC3247762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotter PD, Hill C. Surviving the acid test: Responses of Gram-Positive bacteria to pow pH. Microbiology and Molecular Biology Reviews. 2003;67(3):429–53. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beales N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: A Review. Comprehensive Reviews in Food Science and Food Safety. 2004;3(1):1–20. 10.1111/j.1541-4337.2004.tb00057.x [DOI] [PubMed] [Google Scholar]
- 4.Bearson S, Bearson B, Foster JW. Acid stress responses in enterobacteria. FEMS Microbiology Letters. 1997;147(2):173–80. 10.1111/j.1574-6968.1997.tb10238.x [DOI] [PubMed] [Google Scholar]
- 5.Richard HT, Foster JW. Acid resistance in Escherichia coli Advances in Applied Microbiology. Volume 52: Academic Press; 2003. p. 167–86. [DOI] [PubMed] [Google Scholar]
- 6.Proksch E. pH in nature, humans and skin. The Journal of Dermatology. 2018;0(0). 10.1111/1346-8138.14489 [DOI] [PubMed] [Google Scholar]
- 7.Fuchs R, Schmid S, Mellman I. A possible role for Na+,K+-ATPase in regulating ATP-dependent endosome acidification. Proceedings of the National Academy of Sciences. 1989;86(2):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: New insights. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2005;1717(2):67–88. 10.1016/j.bbamem.2005.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gianella R, Broitman S, Zamcheck N. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Annual Internal Medicine. 1973;78(2):271–6. [DOI] [PubMed] [Google Scholar]
- 10.Padan E, Zilberstein D, Schuldiner S. pH homeostasis in bacteria. Biochim Biophys Acta. 1981;650(2–3):151–66. Epub 1981/12/01. . [DOI] [PubMed] [Google Scholar]
- 11.Padan E, Tzubery T, Herz K, Kozachkov L, Rimon A, Galili L. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter. Biochim Biophys Acta. 2004;1658(1–2):2–13. Epub 2004/07/30. 10.1016/j.bbabio.2004.04.018 S0005272804001264 [pii]. . [DOI] [PubMed] [Google Scholar]
- 12.Pinner E, Kotler Y, Padan E, Schuldiner S. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1993;268(3):1729–34. Epub 1993/01/25. . [PubMed] [Google Scholar]
- 13.Rahav-Manor O, Carmel O, Karpel R, Taglicht D, Glaser G, Schuldiner S, et al. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J Biol Chem. 1992;267(15):10433–8. Epub 1992/05/25. . [PubMed] [Google Scholar]
- 14.Padan E, Gerchman Y, Rimon A, Rothman A, Dover N, Carmel-Harel O. The molecular mechanism of regulation of the NhaA Na+/H+ antiporter of Escherichia coli, a key transporter in the adaptation to Na+ and H+. Novartis Found Symp. 1999;221:183–96; discussion 96–9. Epub 1999/04/20. . [DOI] [PubMed] [Google Scholar]
- 15.White-Ziegler CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S. Low temperature (23 °C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology. 2008;154(1):148–66. 10.1099/mic.0.2007/012021-0 [DOI] [PubMed] [Google Scholar]
- 16.Dover N, Padan E. Transcription of nhaA, the main Na(+)/H(+) antiporter of Escherichia coli, is regulated by Na(+) and growth phase. J Bacteriol. 2001;183(2):644–53. Epub 2001/01/03. 10.1128/JB.183.2.644-653.2001 ; PubMed Central PMCID: PMC94921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dover N, Higgins CF, Carmel O, Rimon A, Pinner E, Padan E. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol. 1996;178(22):6508–17. Epub 1996/11/01. ; PubMed Central PMCID: PMC178537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pannuri A, Yakhnin H, Vakulskas CA, Edwards AN, Babitzke P, Romeo T. Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J Bacteriol. 2012;194(1):79–89. Epub 2011/11/01. JB.06209-11 [pii] 10.1128/JB.06209-11 ; PubMed Central PMCID: PMC3256615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toesca I, Perard C, Bouvier J, Gutierrez C, Conter A. The transcriptional activator NhaR is responsible for the osmotic induction of osmC(p1), a promoter of the stress-inducible gene osmC in Escherichia coli. Microbiology. 2001;147(Pt 10):2795–803. Epub 2001/09/29. 10.1099/00221287-147-10-2795 . [DOI] [PubMed] [Google Scholar]
- 20.Sturny R, Cam K, Gutierrez C, Conter A. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. Journal of Bacteriology. 2003;185(15):4298–304. 10.1128/JB.185.15.4298-4304.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186(9):2724–34. Epub 2004/04/20. 10.1128/JB.186.9.2724-2734.2004 ; PubMed Central PMCID: PMC387819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goller C, Wang X, Itoh Y, Romeo T. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2006;188(23):8022–32. Epub 2006/09/26. JB.01106-06 [pii] 10.1128/JB.01106-06 ; PubMed Central PMCID: PMC1698181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Micro. 2004;2(5):391–400. [DOI] [PubMed] [Google Scholar]
- 24.White-Ziegler CA, Davis TR. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. Journal of Bacteriology. 2009;191(3):1106–10. 10.1128/JB.00599-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms In: Romeo T, editor. Bacterial Biofilms. Current Topics in Microbiology and Immunology. 322: Springer; Berlin Heidelberg; 2008. p. 249–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67(17):2897–908. Epub 2010/05/07. 10.1007/s00018-010-0381-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56(6):1648–63. Epub 2005/05/27. MMI4648 [pii] 10.1111/j.1365-2958.2005.04648.x . [DOI] [PubMed] [Google Scholar]
- 28.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40(1):245–56. Epub 2001/04/12. mmi2380 [pii]. . [DOI] [PubMed] [Google Scholar]
- 29.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, et al. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Esigma and Esigma(S), and indirect transcriptional activation by CsrA. Mol Microbiol. 2011;81(3):689–704. Epub 2011/06/24. 10.1111/j.1365-2958.2011.07723.x ; PubMed Central PMCID: PMC3189700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yuksel U, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272(28):17502–10. Epub 1997/07/11. . [DOI] [PubMed] [Google Scholar]
- 31.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48(3):657–70. Epub 2003/04/16. 3459 [pii]. . [DOI] [PubMed] [Google Scholar]
- 32.Yang TY, Sung YM, Lei GS, Romeo T, Chak KF. Posttranscriptional repression of the cel gene of the ColE7 operon by the RNA-binding protein CsrA of Escherichia coli. Nucleic Acids Res. 2010;38(12):3936–51. Epub 2010/04/10. gkq177 [pii] 10.1093/nar/gkq177 ; PubMed Central PMCID: PMC2896534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, et al. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184(18):5130–40. Epub 2002/08/24. 10.1128/JB.184.18.5130-5140.2002 ; PubMed Central PMCID: PMC135316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mondragon V, Franco B, Jonas K, Suzuki K, Romeo T, Melefors O, et al. pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J Bacteriol. 2006;188(23):8303–6. Epub 2006/09/19. JB.01052-06 [pii] 10.1128/JB.01052-06 ; PubMed Central PMCID: PMC1698187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, et al. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J Bacteriol. 2011;193(22):6162–70. Epub 2011/09/13. JB.05975-11 [pii] 10.1128/JB.05975-11 ; PubMed Central PMCID: PMC3209218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho MI, Alvarez AF, Gonzalez Chavez R, Romeo T, Merino E, Georgellis D. Effects of the Global Regulator CsrA on the BarA/UvrY Two-Component Signaling System. Journal of Bacteriology. 2015;197(5):983–91. 10.1128/JB.02325-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183(20):6017–27. Epub 2001/09/22. 10.1128/JB.183.20.6017-6027.2001 ; PubMed Central PMCID: PMC99681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184(1):290–301. Epub 2001/12/14. 10.1128/JB.184.1.290-301.2002 ; PubMed Central PMCID: PMC134780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krol JE, Nguyen HD, Rogers LM, Beyenal H, Krone SM, Top EM. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl Environ Microbiol. 2011;77(15):5079–88. Epub 2011/06/07. AEM.00090-11 [pii] 10.1128/AEM.00090-11 ; PubMed Central PMCID: PMC3147451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romeo T, Preiss J. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5'-diphosphate 3'-diphosphate and analysis of in vivo transcripts. J Bacteriol. 1989;171(5):2773–82. Epub 1989/05/01. ; PubMed Central PMCID: PMC209963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20(18):2605–17. Epub 2006/09/19. 20/18/2605 [pii] 10.1101/gad.1461606 ; PubMed Central PMCID: PMC1578682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Stark TF, Beattie WG, Moses RE. Multiple control elements for the uvrC gene unit of Escherichia coli. Nucleic acids research. 1986;14(5):2301–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, et al. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21(22):4187–9. 10.1093/bioinformatics/bti635 [DOI] [PubMed] [Google Scholar]
- 45.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annual Review of Microbiology. 1993;47(1):597–626. 10.1146/annurev.mi.47.100193.003121 [DOI] [PubMed] [Google Scholar]
- 46.Sen N, Mishra M, Khan F, Meena A, Sharma A. D-MATRIX: a web tool for constructing weight matrix of conserved DNA motifs. Bioinformation. 2009;3(10):415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, et al. The Complete Genome Sequence of <em>Escherichia coli</em> K-12. Science. 1997;277(5331):1453 [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto K, Yata K, Fujita N, Ishihama A. Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Molecular Microbiology. 2001;41(5):1187–98. 10.1046/j.1365-2958.2001.02585.x [DOI] [PubMed] [Google Scholar]
- 49.Romeo T, Vakulskas CA, Babitzke P. Posttranscriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ Microbiol. 2013;15(2):313–24. 10.1111/j.1462-2920.2012.02794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Jayaraman A, Wood T. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiology. 2007;7(1):42 10.1186/1471-2180-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donato GM, Kawula TH. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. Journal of Bacteriology. 1999;181(3):941–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sowa SW, Gelderman G, Leistra AN, Buvanendiran A, Lipp S, Pitaktong A, et al. Integrative FourD omics approach profiles the target network of the carbon storage regulatory system. Nucleic Acids Research. 2017;45(4):1673–86. 10.1093/nar/gkx048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potts AH, Vakulskas CA, Pannuri A, Yakhnin H, Babitzke P, Romeo T. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nature Communications. 2017;8(1):1596 10.1038/s41467-017-01613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequence upstream the uvrY gene (C); primers are underlined, primer extension products starts are marked as capital letters, predicted binding sites for Hns underlined italics with arrows showing orientation, SdiA and NhaR binding sites are bold and bold underlined respectively; IS5 insertion site is marked by a triangle.
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper.