Fig. 7.
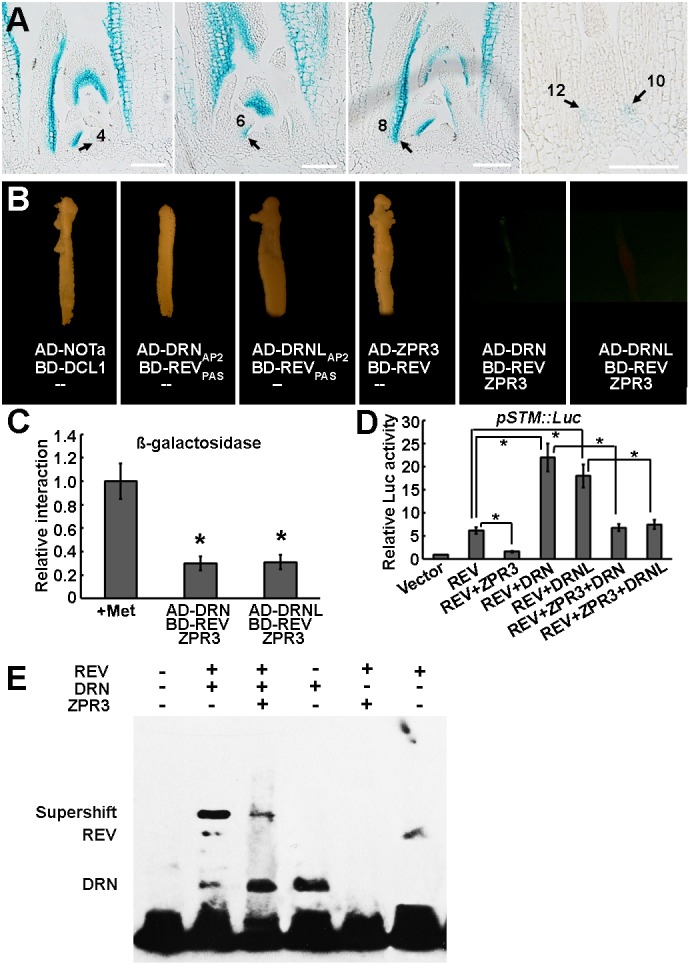
ZPR3 interferes with the DRN/DRNL-REV interaction and inhibits STM expression. (A) Patterns of ZPR3-promoter-driven GUS expression in serial longitudinal 8 μm sections through the vegetative shoot apex of a 30-day-old wild-type-like plant. Arrows indicate the leaf axils. The GUS signals are weaker in the leaf axils of P10 and older leaves. See Fig. S8D-G for additional transverse sections. Scale bars: 100 μm. (B) Y2H and Y3H assay showing the disruption of the DRN/DRNL-REV interaction by ZPR3. Yeast growth on SD-Leu-Trp-His-Ade plates showing that DRN, DRNL and ZPR3 interact with REV, respectively. The interaction of DRN or DRNL with REV was weakened after the induction of ZPR3 activity. AD-NOTa and BD-DCL1 were used as positive controls. (C) Relative β-galactosidase activity of the UAS-driven β-galactosidase reporter measured before and after ZPR3 induction in Y3H. Constructs and additional results are shown in Fig. S9. The data are mean values of three replicates±s.d. *P<0.01 (Student's t-test). (D) Relative Luc reporter gene expression in transcriptional activity assays in Arabidopsis protoplasts. The pSTM::Luc construct was co-transformed with p35S::REV alone, p35S::REV+p35S::ZPR3, p35S::REV+p35S::DRN, p35S::REV+p35S::DRNL, p35S::REV+p35S::ZPR3+p35S::DRN or p35S::REV+p35S::ZPR3+p35S::DRNL; p35S::GUS was the internal control. Data are mean±s.d. Error bars are derived from three independent biological experiments, each performed in triplicate. Note the suppression of REV activation and DRN/DRNL–REV co-activation of STM expression by ZPR3. *P<0.01 (Student's t-test). (E) Supershift in EMSA, indicating that REV and DRN interact and bind to a biotin-labeled STM promoter fragment. The addition of ZPR3 decreased the intensity of the supershift band of DRN and REV; 2 µg DRN and ZPR3, and 1 µg REV protein were used for incubation.
