ABSTRACT
Tendon-bone attachment sites, called entheses, are essential for musculoskeletal function. They are formed embryonically by Sox9+ progenitors and continue to develop postnatally, utilizing Gli1 lineage cells. Despite their importance, we lack information on the transition from embryonic to mature enthesis and on the relation between Sox9+ progenitors and the Gli1 lineage. Here, by performing a series of lineage tracing experiments in mice, we identify the onset of Gli1 lineage contribution to different entheses. We show that Gli1 expression is regulated embryonically by SHH signaling, whereas postnatally it is maintained by IHH signaling. During bone elongation, some entheses migrate along the bone shaft, whereas others remain stationary. Interestingly, in stationary entheses Sox9+ cells differentiate into the Gli1 lineage, but in migrating entheses this lineage is replaced by Gli1 lineage. These Gli1+ progenitors are defined embryonically to occupy the different domains of the mature enthesis. Overall, these findings demonstrate a developmental strategy whereby one progenitor population establishes a simple embryonic tissue, whereas another population contributes to its maturation. Moreover, they suggest that different cell populations may be considered for cell-based therapy of enthesis injuries.
KEY WORDS: Musculoskeletal development, Enthesis, Sox9, Gli1, Progenitor cell, Hedgehog signaling, R26R-Confetti, Mouse
Summary: During the development of migrating entheses, one progenitor population establishes a simple embryonic tissue, whereas another population is responsible for its postnatal maturation into a complex structure.
INTRODUCTION
The proper assembly of the musculoskeletal system is essential for the function, form and stability of the organism. During embryogenesis, an attachment between tendon and bone, known as an enthesis, is formed. Thereafter, and into the postnatal period, the rudimentary enthesis further develops into a more complex tissue. Although some knowledge on enthesis formation and maturation exists, far less is known about the processes that transform the simple embryonic enthesis into the structure of the mature enthesis.
Traditionally, entheses are divided into either fibrous or fibrocartilaginous, according to their cellular composition. Fibrous entheses form at the attachment site of a tendon that is inserted directly into the bone shaft, forming a structure that resembles a root system. This structure, which is composed of a dense connective tissue (Doschak and Zernicke, 2005; Shaw and Benjamin, 2007; Wang et al., 2013), has been shown to be regulated by parathyroid hormone-like hormone (PTHLH, also known as PTHrP) (Wang et al., 2013). Fibrocartilage entheses typically form at attachment sites of tendons to the epiphysis or to bone eminences. Relative to fibrous entheses, fibrocartilage entheses are structurally more complex, displaying a cellular gradient that is typically divided into four zones, namely tendon, fibrocartilage, mineralized fibrocartilage and bone. The graded tissue, which develops postnatally, dissipates the stress that forms at the attachment site, thereby providing the enthesis with the mechanical strength necessary to withstand compression (Benjamin et al., 2006).
Enthesis development is initiated by the specification of a specialized pool of progenitor cells that express both SRY-box 9 (Sox9), a key regulator of chondrogenesis, and the tendon marker scleraxis (Scx) (Akiyama et al., 2005; Blitz et al., 2013; Schweitzer et al., 2001; Sugimoto et al., 2013). Cell lineage studies have shown that Sox9-expressing progenitor cells contribute to the formation of the enthesis in neonatal mice (Akiyama et al., 2005; Soeda et al., 2010). Another marker for enthesis cells is GLI-Kruppel family member GLI1 (Gli1; Liu et al. 2012), a component of the hedgehog (HH) signaling pathway. Lineage studies revealed that Gli1-expressing cells act as progenitors that contribute to the formation of some adult entheses (Dyment et al., 2015; Schwartz et al., 2015). However, the relationship between progenitors of the embryonic enthesis and Gli1 lineage cells, which contribute to the mature enthesis, has not been determined.
The relative position of an enthesis along the bone directly affects its mechanical function and, subsequently, the animal's mobility (Polly, 2007; Salton and Sargis, 2009). Recently, it was shown that all entheses maintain their relative position during bone elongation (Stern et al., 2015). Unlike entheses located at the bone ends, from which the bone elongates, entheses that are located between the epiphyseal plates must maintain their positions during growth. This was shown to occur through either regulation of the relative growth rates at the two epiphyseal plates, or active migration by continuous reconstruction, a process known as bone modeling or drift (Benjamin and McGonagle, 2009; Dörfl, 1980a,b). The latter (referred to hereafter as migrating entheses) encounters a unique developmental challenge. Unlike most organs and tissues, entheses must develop into a complex graded tissue while constantly drifting. This raises the issue of whether the descendants of the embryonic enthesis progenitors continue to serve as the building blocks during maturation of migrating entheses.
In this work, we identify the embryonic stage at which Gli1 lineage is initiated and demonstrate its contribution to the postnatal enthesis. We show that embryonic Gli1 expression is initially under the regulation of sonic hedgehog (SHH). Later during postnatal development, Gli1 expression is maintained by Indian hedgehog (IHH). Moreover, we show that Sox9 lineage does not contribute to postnatal migrating entheses. Instead, Gli1 lineage cells replace the Sox9 lineage cells and populate the enthesis. Finally, we show that embryonic Gli1 lineage cells are pre-determined to contribute to the different layers of the fibrocartilaginous enthesis.
RESULTS
Some entheses undergo cellular and morphological changes during migration
The development of the rudimentary embryonic attachment site into the complex structure of a mature enthesis has received little attention (Galatz et al., 2007). As mentioned, in addition to substantial cellular and morphological changes, some entheses also migrate considerable distances along the bone during bone growth (Benjamin and McGonagle, 2009; Dörfl, 1980a,b; Stern et al., 2015). Therefore, to study the transition that migrating entheses undergo during maturation, we documented morphological and molecular changes, as well as drifting activity, in entheses from embryonic day (E) 14.5 to postnatal day (P) 14. We focused on two migrating entheses, namely the deltoid enthesis (DT), a fibrocartilaginous enthesis that forms between the deltoid tendon and the deltoid tuberosity, and the teres major enthesis (TM), a fibrous enthesis that forms between the teres major tendon and the humeral shaft.
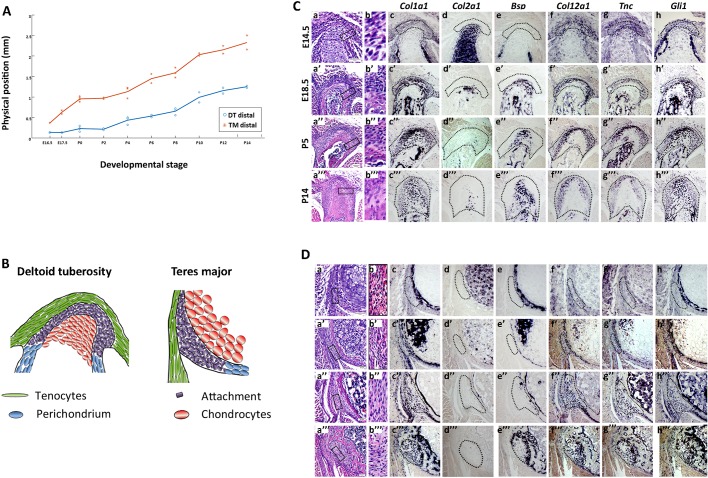
Analysis of enthesis positions during development revealed that both DT (0.778±0.026 mm, data are mean±s.d.) and TM (1.624±0.171 mm) entheses drifted considerably along the bone shaft during bone elongation (Fig. 1A). Histological sections through wild-type (WT) mouse humeri showed that both embryonic entheses displayed a simple structure of layered cells (Fig. 1B,Ca,Da). However, the overall shape of the enthesis changed dramatically throughout development, as it protruded outwards from the bone shaft and the different enthesis domains became more noticeable (Fig. 1Ca′-Ca‴,Da′-Da‴). Tissue complexity also increased, as a larger variety of cells, such as fibrocartilage cells and osteoblasts (Benjamin and McGonagle, 2009), were identified along with an increase in extracellular matrix (Fig. 1Cd,Cd′,Ch,Ch′,Dd,Dd′,Dh,Dh′). The increased complexity was also demonstrated by a change in the expression patterns of structural genes, such as bone sialoprotein (Bsp; also known as integrin binding sialoprotein, Ibsp), collagen type 2 alpha 1 (Col2a1), collagen type 12 alpha 1 (Col12a1) and tenascin C (Tnc), and regulatory genes such as Gli1. Furthermore, expression domains correlating to various structural domains emerged, namely Col12a1, Tnc, Gli1 and Col1a1 in tendon and fibrocartilage (Fig. 1Cc,Cf-Ch,Dc,Df-Dh), and Col1a1, Gli1 and Bsp in mineralized fibrocartilage and bone (Fig. 1Cc,Ce,Ch,Dc,De,Dh).
Fig. 1.
Some entheses undergo cellular and morphological changes while migrating. (A) Graph showing the physical position of the DT and TM entheses along the bone throughout development (E16.5-P14), defined as the distance of the element (mm) from a predefined longitudinal origin roughly at the center of the shaft. Positive and negative values correspond to elements that are proximal and distal to the point of origin, respectively. As indicated by the steep slope of the curves, both DT and TM migrate considerably throughout development. (B) Schematics of the embryonic DT and TM entheses. (C,D) Transverse sections through the humerus showing the DT (C) and TM (D) entheses of E14.5-P14 WT mice. (Ca,Cb,Da,Db) H&E staining. Cb and Db are magnifications of the boxed areas in Ca and Da, respectively. (Cc-Ch,Dc-Dh) ISH using DIG-labeled, anti-sense complementary RNA probes for Col1a1, Col2a1, Bsp, Col12a1, Tnc and Gli1 at E14.5 (c-h), E18.5 (c′-h′), P5 (c″-h″) and P14 (c‴-h‴). Dotted line demarcates the enthesis. Scale bars: 50 µm.
Gli1+ cell lineage contributes to the postnatal enthesis
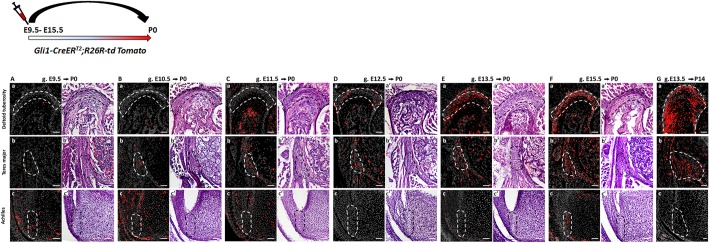
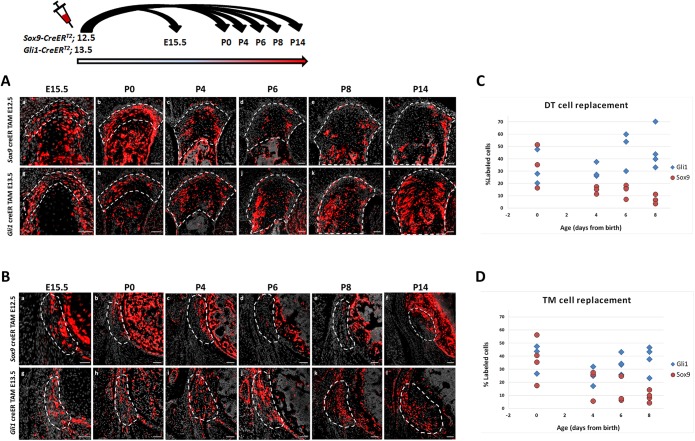
Lineage tracing experiments have shown that Gli1 lineage cells contribute to postnatal enthesis development (Dyment et al., 2015; Schwartz et al., 2015). Yet, the onset of this lineage during embryogenesis and its dynamics in different entheses have been missing. In order to identify the onset of Gli1 lineage, we performed pulse-chase experiments on Gli1-CreERT2 mice (Ahn and Joyner, 2004) crossed with R26R-tdTomato reporter mice (Madisen et al., 2010), which allowed us to mark Gli1-expressing cells at specific time points and follow their descendants. We analyzed three entheses that represented different types, namely DT (migratory-fibrocartilaginous), TM (migratory-fibrous) and Achilles (stationary-fibrocartilaginous). Examination of neonatal (P0) TM and DT entheses following tamoxifen administration at E9.5-E12.5 revealed only a few Gli1 lineage cells (Fig. 2Aa-Da,Ab-Db). However, administration at E13.5 and E15.5 resulted in extensive labeling in both migratory entheses (Fig. 2Ea,Eb,Fa,Fb). In the stationary Achilles enthesis, we observed labeled cells in tendon and cartilage at P0 following tamoxifen administration at E9.5-E10.5 (Fig. 2Ac,Bc); yet, tamoxifen administration at later time points (E11.5-E14.5) did not result in labeled cells (Fig. 2Cc,Dc,Ec). Tamoxifen administration at E15.5 labeled cells specifically in the enthesis and perichondrium and not in adjacent cartilage (Fig. 2Fc). Although we cannot rule out periodic expression of Gli1 in the Achilles enthesis cells, it is possible that the early labeling represents a direct readout of HH expression in the zone of polarizing activity (Ahn and Joyner, 2004), whereas the later expression is a specific induction of Gli1 in enthesis cells. The postnatal contribution of Gli1 lineage to migratory entheses was further established by following E13.5 lineage induction to P14 (Fig. 2Ga-Gc). Together, these results indicate that, although Gli1 lineage is not induced at the onset of enthesis formation (Blitz et al., 2013; Sugimoto et al., 2013), it contributes differentially to different entheses during embryonic development.
Fig. 2.
Gli1+ cell lineage contributes to the postnatal enthesis. (A-G) Pulse-chase cell lineage experiments using Gli1-CreERT2;R26R-tdTomato mice demonstrate the contribution of Gli1 lineage cells to various entheses. Gli1+ cells were labeled by a single tamoxifen administration at different time points (E9.5-E15.5) and their descendants were followed to P0 and P14. (Aa-Fa) The DT enthesis was extensively marked after tamoxifen administration at E13.5 and E15.5. (Ab-Fb) tdTomato+ cells were identified in the TM enthesis starting at E10.5; however, a stronger signal was seen following tamoxifen administration at E13.5 or later. (Ac-Fc) tdTomato+ cells were identified in the Achilles tendon enthesis following tamoxifen administration at E9.5 and E15.5. On the right of each image, a corresponding H&E section shows the enthesis region. (Ga-Gc) Both DT and TM, but not the Achilles entheses, were extensively marked at P14 following tamoxifen administration at E13.5. Dashed line demarcates the enthesis. Scale bars: 50 µm.
Gli1 expression in migrating entheses is initiated by SHH and maintained by IHH
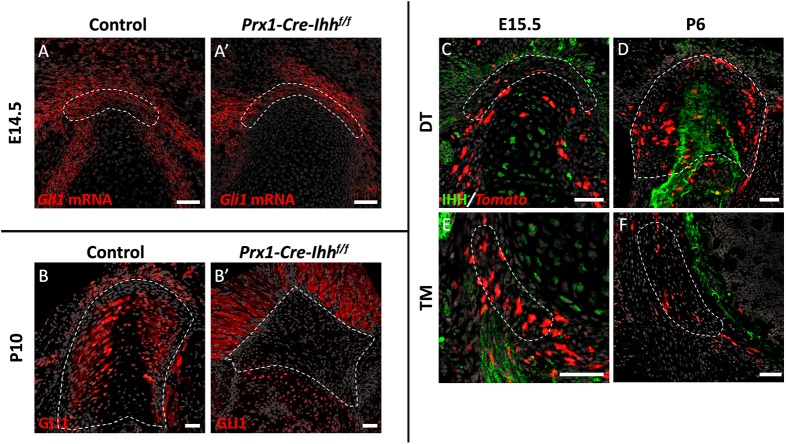
It has previously been suggested that the HH signaling pathway plays a role in regulating the activity of Gli1+ enthesis cells (Breidenbach et al., 2015; Dyment et al., 2015; Liu et al., 2013; Schwartz et al., 2015). Therefore, our finding that Gli1 expression is initiated at early stages of enthesis development (Fig. 1Ch,Dh) raised the issue of which component of the HH pathway regulates Gli1 expression in enthesis cells and whether it also affects migratory entheses. It was suggested that IHH, an effector of the HH signaling pathway, is a possible regulator of Gli1 lineage cells in adult stationary entheses. In order to examine the possible role of IHH in inducing Gli1 expression in the embryonic enthesis, we blocked Ihh expression in the limb by using Prx1-Cre-Ihh−/− mice. Interestingly, at E14.5 Gli1 was expressed in control and mutant entheses, suggesting that IHH is not necessary for Gli1 induction in embryonic enthesis progenitors (Fig. 3A,A′). We therefore examined the possible role of another regulator of HH signaling, SHH, by analyzing the Gli1 expression of embryonic enthesis cells in Shh KO embryos (Shh-GFPCre−/−). Results showed that Gli1 expression dramatically decreased in the mutant entheses compared with the WT (Fig. 4C-D′), suggesting that SHH is necessary for the induction of Gli1 in embryonic enthesis cells.
Fig. 3.
Gli1 expression in the enthesis is maintained by IHH. (A,A′) Fluorescent ISH for Gli1 on sections through the DT of E14.5 control (A) and Prx1-Cre-Ihhf/f embryos (A′) demonstrates that, in the absence of IHH signaling, Gli1 expression in the enthesis remains similar. (B,B′) Immunostaining for GLI1 protein in P10 control (B) and Prx1-Cre-Ihhf/f (B′) DT sections shows that, in the absence of IHH, GLI1 protein expression is lost from the DT enthesis. (C-F) Immunostaining for IHH protein in Gli1-CreERT2;R26R-tdTomato DT (C,D) and TM (E,F) entheses. Tamoxifen was administered at E13.5 and mice were sacrificed at E15.5 (C,E) or P6 (D,F). IHH expression is seen in hypertrophic chondrocytes at E15.5 and in the mineralizing part of the enthesis at P6. IHH is also expressed in surrounding muscle at E15.5. Dashed line demarcates the enthesis. Scale bars: 50 µm.
Fig. 4.
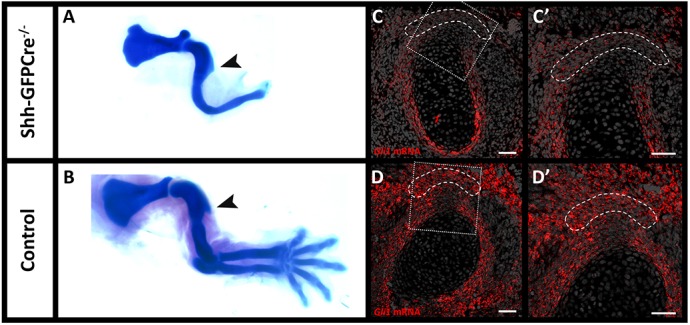
Gli1 expression in the enthesis is initiated by SHH. (A,B) Skeletal preparations from control and Shh-GFPCre−/− embryos at E14.5 demonstrate that, although limb morphology is greatly impaired in the mutant, proximal humerus morphology remains similar and the DT is formed. Arrowheads indicate the deltoid tuberosity. (C-D′) Fluorescent ISH for Gli1 mRNA on sections through the DT of E14.5 Shh-GFPCre−/− (C-C′) and control (D-D′) embryos show that, in the absence of SHH signaling, Gli1 mRNA and expression in the DT enthesis is lost. C′ and D′ are magnifications of the boxed areas in C and D, respectively. Dashed line demarcates the enthesis. Scale bars: 50 µm.
That result was intriguing, because we observed that Gli1 was constantly expressed by both embryonic and postnatal enthesis cells (Fig. 1Ah-Ah‴,Bh-Bh‴), whereas Shh is expressed in the limb only during embryogenesis (Harfe et al., 2004). This raised the issue of how Gli1 expression in the enthesis is maintained after the loss of Shh expression. To address the possibility that Ihh controls Gli1 expression in the postnatal enthesis, even though it is not involved in the induction of Gli1 expression in embryonic enthesis progenitors, we analyzed Prx1-Cre-Ihh−/− mice at P10. Results showed that, although Gli1 expression was maintained in mutant muscle and bone, in the enthesis it was completely lost (Fig. 3B,B′), which indicates that Ihh is indeed required for the maintenance of Gli1 expression in enthesis cells. To identify the source of Ihh in the enthesis, we performed immunofluorescence staining for IHH protein in P6 enthesis sections. As seen in Fig. 3 (C-F), IHH was highly expressed in mineralized fibrocartilage and bone regions at the enthesis center, along with its expression in surrounding muscles. Taken together, these results suggest that Gli1 expression by embryonic enthesis progenitors is induced by SHH and later, in the maturing enthesis, maintained by IHH that originates in mineralized fibrocartilage and bone.
Sox9 lineage cells of the embryonic enthesis do not contribute to postnatal migrating entheses
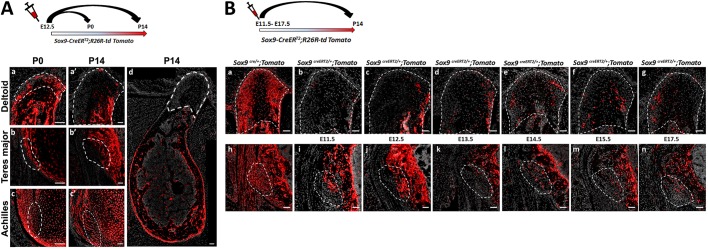
The embryonic enthesis originates from Scx+/Sox9+ progenitor cells (Blitz et al., 2013; Sugimoto et al., 2013). Yet, the contribution of these progenitors to the postnatal enthesis and their relation to the Gli1 lineage cells have never been studied. To fill this gap, we first examined the contribution of Sox9 lineage to the postnatal enthesis. To that end, we performed a pulse-chase cell lineage experiment using mice that express Cre-ER under control of the Sox9 promoter (Sox9-CreERT2) crossed with R26R-tdTomato reporter mice (Soeda et al., 2010). It has previously been demonstrated that tamoxifen administration at E12.5 effectively labels embryonic enthesis cells (Blitz et al., 2013; Soeda et al., 2010). Indeed, examination at P0 following tamoxifen administration at E12.5 showed that the DT, TM and Achilles entheses were populated by tdTomato+ cells, which suggests that, at that stage, the enthesis is populated by Sox9 lineage cells (Fig. 5Aa-Ac). Yet, surprisingly, at P14 we observed a dramatic decrease in the contribution of tdTomato-expressing cells to the two migrating entheses (Fig. 5Aa′,Ab′,Ad), although the cells in the stationary enthesis and in the humerus were still extensively labeled (Fig. 5Ac′,Ad). These results suggest that postnatal stationary enthesis cells were descendants of the Sox9+ embryonic lineage, whereas in migrating entheses this lineage was lost.
Fig. 5.
Sox9 lineage cells of the embryonic enthesis do not contribute to postnatal migrating entheses. (A) Pulse-chase cell lineage experiment using Sox9-CreERT2;R26R-td Tomato mice demonstrates that Sox9 lineage contributes differently to migrating and to stationary entheses. Sox9+ cells were marked at E12.5 by a single tamoxifen administration. At P0, tdTomato+ cells were identified in migrating DT and TM entheses (Aa,Ab) as well as in stationary Achilles entheses (Ac). At P14, only a few tdTomato+ cells were identified in migrating entheses (Aa′,Ab′,Ad), whereas extensive staining was still seen in stationary entheses (Ac′) and in other areas of the bone, as seen in a section through the humerus in Ad. (B) The contribution of Sox9 lineage to the DT and TM entheses was evaluated by crossing Sox9-Cre or Sox9-CreERT2 mice with R26R-tdTomato reporter mice. (Ba,Bh) The total contribution of the lineage was evaluated by examination of Sox9-Cre;R26R-tdTomato mice at P14. (Bb-Bg,Bi-Bn) The relative contribution of Sox9 lineage cells specified at different time points was evaluated by pulse-chase experiments on Sox9-CreERT2;R26R-tdTomato mice, in which a single dose of tamoxifen was administered at various stages from E11.5 through E17.5. Dashed line demarcates the enthesis. Scale bars: 50 µm.
The labeled stationary entheses could serve as a positive internal control for the effectiveness of labeling. Nevertheless, to rule out the possibility that tamoxifen was administered at the wrong time point, we repeated the experiment while administering tamoxifen at different time points from E11.5 to E17.5 (Fig. 5B). Examination at P14 showed minimal contribution of Sox9 lineage cells to the postnatal entheses, similar to the results obtained following pulsing at E12.5. Taken together, these results indicate that the embryonic Sox9 lineage contributes poorly to postnatal migrating entheses, suggesting that these entheses are populated by another cell lineage postnatally.
Gli1 lineage cells replace the embryonic Sox9 lineage during enthesis maturation
Our finding that both embryonic Gli1 and Sox9 lineages contribute to the stationary postnatal enthesis suggests that these two genes mark a common cell lineage. Conversely, the finding that embryonic Sox9 lineage contributes to embryonic but not to postnatal migrating enthesis implies that, postnatally, embryonic Gli1 lineage replaces the Sox9 lineage to form the mature enthesis.
To study the process of lineage replacement and to follow its dynamics, we traced both lineages throughout enthesis development, from E15.5 to maturation at P14, by pulse-chase experiments. As seen in Fig. 6, at E15.5 and P0 in both the DT and TM, Sox9 lineage cells populated the embryonic enthesis. However, from P0 their number decreased dramatically and, by P8, the enthesis contained only a limited number of these cells. Concurrently, the number of cells of the Gli1 lineage gradually increased in both entheses and, by P8, Gli1+ cells inhabited most of the enthesis structure. Interestingly, the gradual population of the DT and TM entheses by Gli1 lineage cells correlated with the temporal dynamics of enthesis migration (Fig. 1A). These results support our hypothesis that, during early postnatal development of migrating entheses, a new population that is derived from Gli1+ progenitors substitutes the Sox9 lineage cells of the embryonic enthesis.
Fig. 6.
Gli1 lineage cells replace Sox9 lineage cells during enthesis maturation. (A,B) Sox9-CreERT2;R26R-tdTomato and Gli1-CreERT2;R26R-tdTomato mice were labeled by tamoxifen administration at E12.5 and E13.5, respectively, and sacrificed at E15.5-P14. Transverse sections through the DT (A) and TM (B) show an increase in Sox9 lineage cells at E15.5-P0. Between P0 and P14, a continuous decrease in Sox9 lineage cells is seen. Concurrently, Gli1 lineage cell number gradually increased in both DT and TM entheses. By P8, both entheses were populated by Gli1 lineage cells. (C,D) The percentages of tdTomato+ cells in Sox9-CreERT2;R26R-tdTomato and Gli1-CreERT2;R26R-tdTomato limbs were significantly different in the DT (C, P=0.0010) and TM (D, P=0.0019). Additionally, there was a significant decrease in the percentage of tdTomato+ cells in Sox9-CreERT2;R26R-tdTomato limbs over time (P=0.020). Dashed line demarcates the enthesis. Scale bars: 50 µm.
Cellular mechanism that underlies the elimination of Sox9 lineage cells in migrating entheses
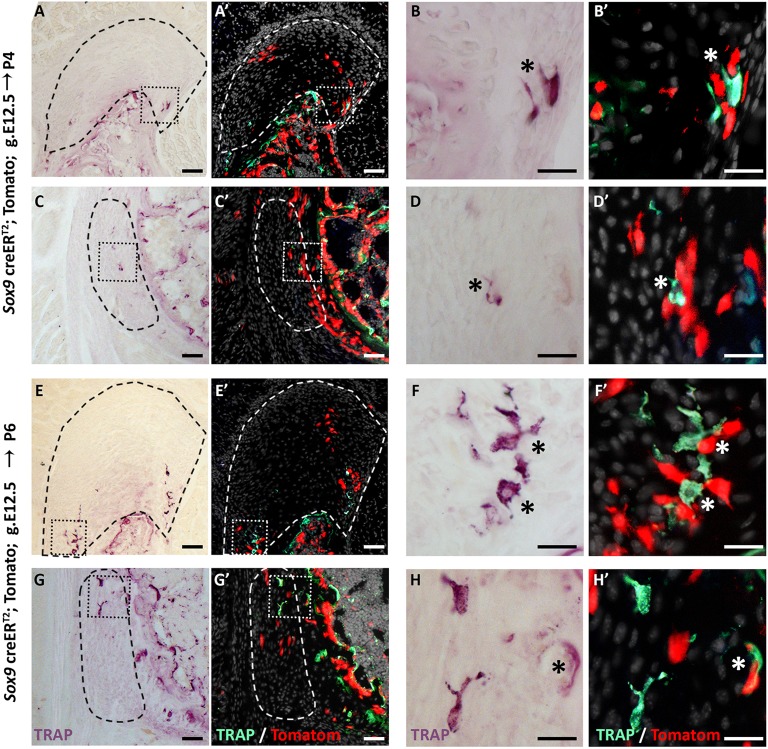
The loss of Sox9+ lineage cells during the replacement process raised the issue of the mechanism that underlies their disappearance from the enthesis. Several mechanisms could account for the elimination of these cells, including bone modeling by osteoclasts, cell death and phagocytosis. To decide between these options, we analyzed P6 and P4 Sox9-CreER;tdTomato mouse humeri following tamoxifen administration at E12.5. We first traced the presence of osteoclasts using TRAP staining. As seen in Fig. 7, we identified TRAP+ cells at the enthesis in proximity to Sox9+ lineage cells, which suggests that Sox9-lineage elimination is performed, at least in part, by bone modeling. Next, we addressed the possibility of apoptosis by TUNEL assay. As seen in Fig. 8, we identified TUNEL+ cells in both entheses, which suggests that cell death might contribute to the cell lineages replacement process. Finally, we examined the presence of phagocytic cells that could eliminate Sox9+ lineage cells using F4/80 antibody staining. Results showed a few F4/80+ cells in proximity to Sox9+ lineage cells, suggesting that they too participate in Sox9+ lineage elimination (Fig. 9). Altogether, these results suggest that Sox9+ cells are eliminated by a combination of different mechanisms, including bone modeling, cell death and phagocytosis.
Fig. 7.
Close spatial proximity between Sox9+ lineage cells and osteoclasts. (A-H′) TRAP staining of P4 and P6 humeri shows osteoclasts (asterisk) in proximity to Sox9+ lineage cells in the DT (A-B′,E-F′) and in the TM (C-D′,G-H′). In A′-H′, TRAP expression images (A,C,E,G) were inverted and overlaid with tdTomato signal from Sox9-CreER;tdTomato lineage cells. B,B′,D,D′,F,F′,H,H′ are magnifications of the boxed areas in A,A′,C,C′,E,E′,G,G′, respectively. Dashed line demarcates the enthesis. Scale bars: 50 µm in A,C,E,G; 20 µm in B,D,F,H.
Fig. 8.
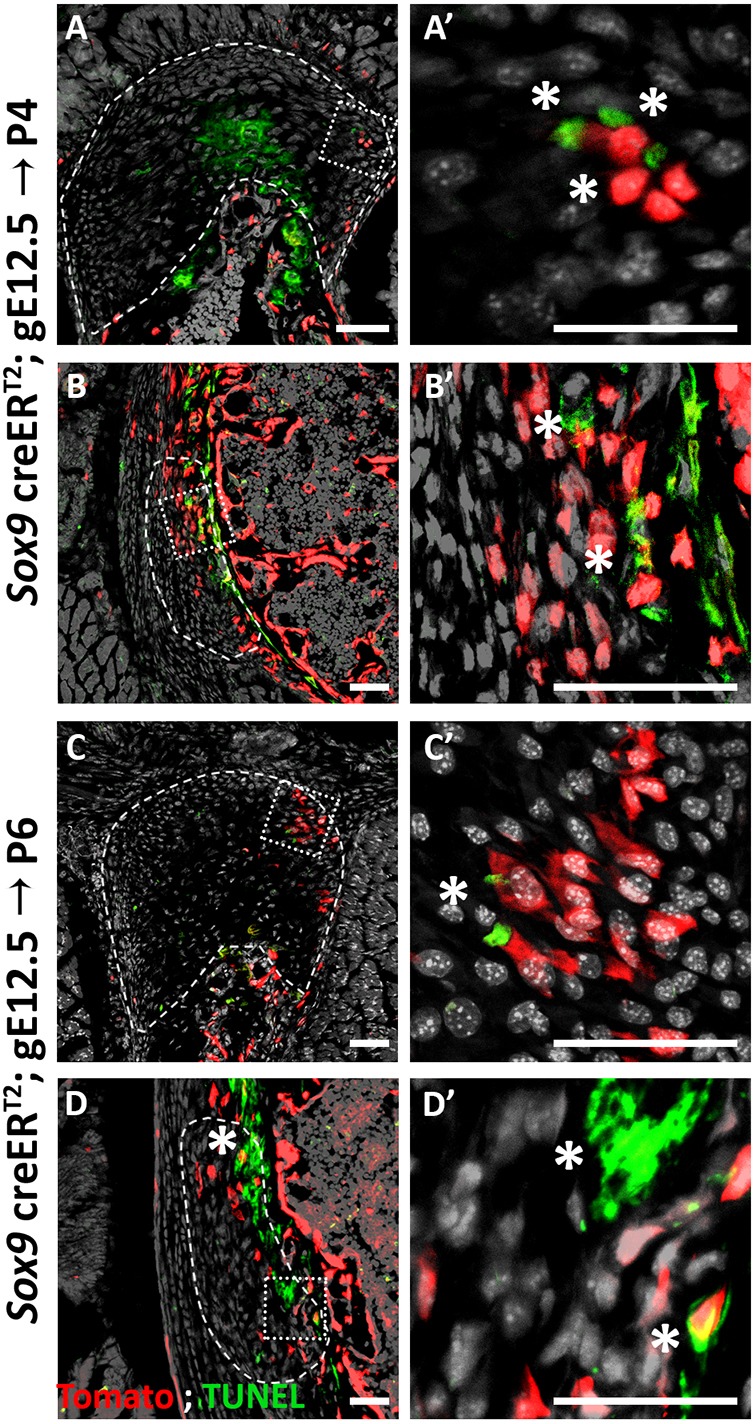
Some enthesis cells undergo cell death. (A-D′) TUNEL staining of P4 (A,B) and P6 (C,D) humeri show apoptotic cells in the DT (A,C) and in the TM (B,D). E13.5 autopod was used as a control (see Fig. S1). Dashed line demarcates the enthesis; asterisks indicate TUNEL-positive cells. A′,B′,C′,D′ are magnifications of the boxed areas in A,B,C,D, respectively. Scale bars: 50 µm.
Fig. 9.
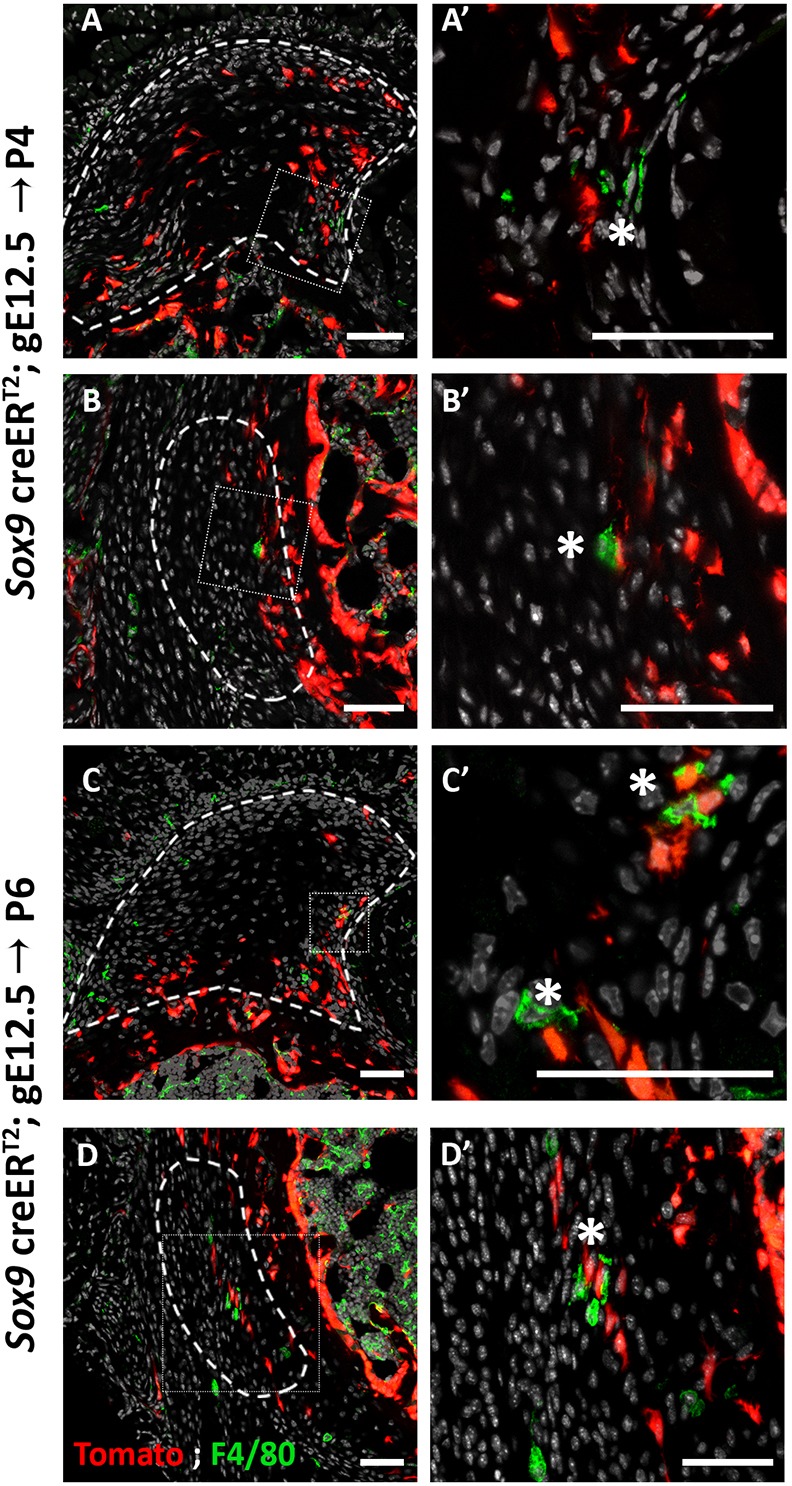
Sox9+ lineage cells are in close proximity to phagocytes. (A-D′) F4/80 staining, marking phagocytes, of P4 (A,B) and P6 (C,D) humeri show phagocytic cells (asterisk) proximal to Sox9+ lineage cells in the DT (A,A′,C,C′) and in the TM (B,B′,D,D′). A′-D′ are magnifications of the boxed areas in A-D, respectively. Dashed line demarcates the enthesis. Scale bars: 50 µm.
Gli1+ progenitors are compartmentalized during embryonic enthesis development
The increasing cellular complexity of the developing enthesis, and our finding that progenitors of the Gli1 lineage that will contribute to the postnatal enthesis are already present during embryonic development, led us to ask whether these progenitors are already assigned to a specific domain within the forming enthesis; for example, fibrocartilage or mineralized fibrocartilage. To study this possibility, we conducted pulse-chase experiments on Gli1-CreERT2 mice crossed with R26R-Confetti mice (Snippert et al., 2010) that, upon recombination, stochastically express GFP, RFP, CFP or YFP, allowing for the identification of different cell clones. Finding clones that cross domain boundaries would indicate that there is no compartmentalization of the Gli1 progenitors. Conversely, finding restricted clones would support the existence of embryonic compartmentalization of these cells.
We focused on the two main domains of the DT enthesis, namely mineralized and non-mineralized fibrocartilage, and on the border between entheseal mesenchymal cells and bone in the TM enthesis. Examination of P14 limbs following tamoxifen administration at E13.5 revealed multiple clones in both entheses, most of which were restricted to a specific domain (Fig. 10). These results suggest that at E13.5, the Gli1 lineage progenitors are already assigned a specific enthesis domain.
Fig. 10.
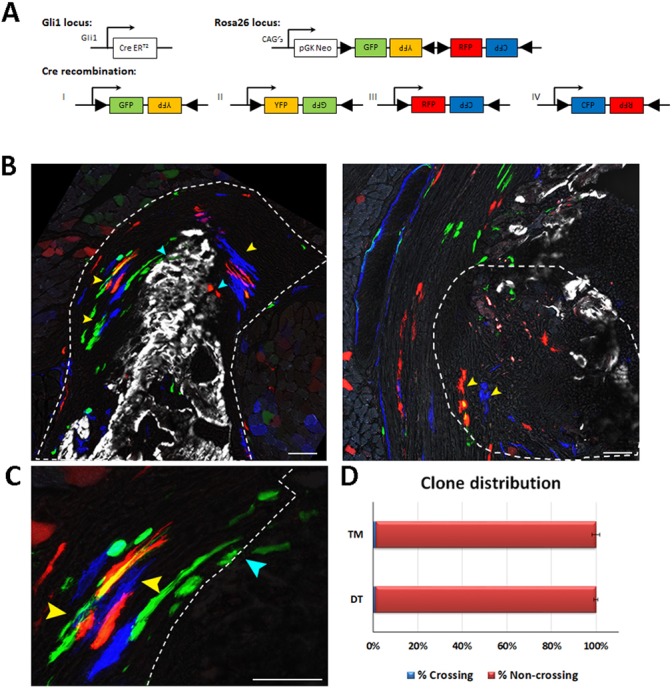
Gli1+ enthesis progenitors are compartmentalized during embryogenesis. Pulse-chase cell lineage experiments using Gli1-CreERT2;R26R-Confetti mice, in which Gli1+ progenitor cell clones were labeled. Gli1+ cells were labeled by a single tamoxifen administration at E13.5 and their descendants were followed to P14. One hour before sacrifice, mice were injected with Calcein Blue to mark mineralized tissue. (A) Schematic of possible Cre recombination outcomes. (B) Multiple clones were identified in the DT enthesis; however, the clones were restricted to either fibrocartilage or mineralized fibrocartilage and did not cross the border between the layers (yellow arrowheads). Cyan arrowheads indicate clones that crossed the border between the layers. The clones identified in the TM enthesis were restricted to the fibrous part and did not penetrate the bone. Dashed line demarcates the enthesis. (C) Magnification of DT clones in B showing crossing (cyan arrowheads) and non-crossing clones (yellow arrowheads). Dashed line marks the mineralization border. (D) Only a small percentage of identified Gli1+ clones crossed the mineralization border in the DT (1.09%±0.9) and in the TM (1.22%±1.725). Scale bars: 50 µm.
DISCUSSION
The transition from the embryonic to the mature enthesis has been understudied. Here, we use genetic lineage tracing to unravel the cellular developmental sequence of fibrous and fibrocartilaginous entheses. We show that, although Gli1+ progenitors contribute to entheses of both types, their contribution differs greatly between stationary and migratory entheses. In stationary entheses, Gli1+ progenitors are descendants of Sox9+ progenitors that have established the embryonic enthesis. However, in migrating entheses, a separate, compartmentalized population of Gli1+ progenitors is recruited to the embryonic enthesis, co-populates it alongside Sox9+ progenitors and, eventually, replaces them during postnatal development.
As mentioned, fibrous and fibrocartilaginous entheses exhibit marked differences in structure and composition (Benjamin et al., 2002). Notwithstanding the importance of this distinction, the observed differences in cellular origin between stationary and migrating entheses calls for a revision of the traditional classification and suggests that it should be taken into account whether an enthesis is ‘migratory’ or ‘stationary’.
Organ development can be mediated by several cellular mechanisms. In a linear mechanism, an embryonic set of progenitors forms a primordium and then continues to proliferate and differentiate to form the mature organ. Another mechanism is cell recruitment, during which cells are supplied to the forming organ after primordium establishment. A third mechanism involves template replacement, in which an initial template is formed by one type of cells to be later replaced by another cell population, which will form the mature organ. Interestingly, in the musculoskeletal system there are examples of all these modes of development. Muscles and joints develop through cell recruitment (Buckingham et al., 2003; Shwartz et al., 2016), whereas most of the skeleton develops through template replacement, namely by endochondral ossification (Kronenberg, 2003). Here, we show that stationary entheses develop linearly by embryonic Sox9+ progenitors that form the postnatal enthesis and later upregulate the expression of Gli1. However, we show that migrating entheses develop through template replacement, as Gli1+ progenitors replace the Sox9+ embryonic enthesis cells and form the mature enthesis.
Previous studies have shown not only that Gli1 is a marker for the forming enthesis, but that the HH pathway plays an active role in regulating enthesis maturation and regeneration (Breidenbach et al., 2015; Dyment et al., 2015; Liu et al., 2013; Schwartz et al., 2015, 2017). Ihh expression was identified in proximity to the Gli1+ progenitor population in stationary entheses (Dyment et al., 2015; Liu et al., 2013; Schwartz et al., 2015). Moreover, loss-of-function of Smo, a component of the HH pathway, in Scx-expressing cells resulted in reduced mineralization of the enthesis (Breidenbach et al., 2015; Dyment et al., 2015; Liu et al., 2013; Schwartz et al., 2015). Yet, the question of which ligand of the HH pathway regulates the specification of Gli1+ cells in the embryo and, specifically, in migrating enthuses, has remained unanswered. Our results clearly suggest that Gli1 expression is regulated by both SHH and IHH during enthesis development. In the embryo, we show that SHH, but not IHH, signaling is essential for Gli1 expression in the enthesis, implicating SHH in Gli1+ progenitor specification. However, during postnatal development, Ihh expression is vital for maintaining Gli1 enthesis expression.
An obvious concern regarding this conclusion relates to the mispatterning of the Shh mutant limb, which might lead to aberrant Gli1 expression. Although one cannot completely rule out this possibility, the patterning of the mutant's proximal humerus was comparable with that of WT. Thus, it is reasonable to assume that the loss of Gli1 expression was a direct effect of SHH loss. This suggests that SHH has a specific role in regulating Gli1 expression in the enthesis.
During endochondral ossification, IHH regulates hypertrophic chondrocyte differentiation and, thereby, bone elongation (Vortkamp et al., 1996). It is therefore possible that, by controlling both these processes, IHH coordinates enthesis migration with the concurrent bone elongation to preserve enthesis position along the shaft. However, the signals that govern enthesis positioning and migration along the bone, including the possible effect of IHH on these processes, are yet to be elucidated.
Another key issue surrounds the mechanism that facilitates the replacement between Sox9+ and Gli1+ cell populations. Finding evidence for the existence of osteoclasts, phagocytes and cells that are undergoing apoptosis in proximity to Sox9+ cells during the replacement process suggests that all of these mechanisms might contribute to Sox9+ lineage cell elimination. Yet, none of these findings was compelling enough to provide an explanation for the massive loss of Sox9+ cells from the attachment site. Therefore, our results do not rule out other mechanisms for the elimination of Sox9 lineage cells, including the migration of these cells to other regions in the bone. Moreover, whether the replacement process is the mechanism that allows migrating entheses to maintain their relative position along the bone is yet to be determined.
Finally, our finding that Gli1-expressing cells have already been assigned domains in the embryo suggests the existence of an earlier, yet unknown, mechanism that regulates the compartmentalization of these progenitors. It would be interesting to determine whether, besides being compartmentalized, the fate of these cells is also pre-determined or, similar to the neural crest cells (Trainor and Krumlauf, 2000), these cells are multipotent and their fate is determined later by cell-community effect. Resolving these issues may hold therapeutic potential.
To conclude, our findings shed light on developmental linearity in organogenesis. Using the enthesis as a model system, we identified two different strategies of development (Fig. 11). The first, found in stationary entheses, is linear development, in which embryonic progenitors and their descendants contribute to the entire process of enthesis development, from embryogenesis to maturity. Conversely, in migrating entheses, another developmental strategy was identified in which one type of progenitor cells form an embryonic template, only to be later replaced by another cell lineage that contributes to the mature organ.
Fig. 11.
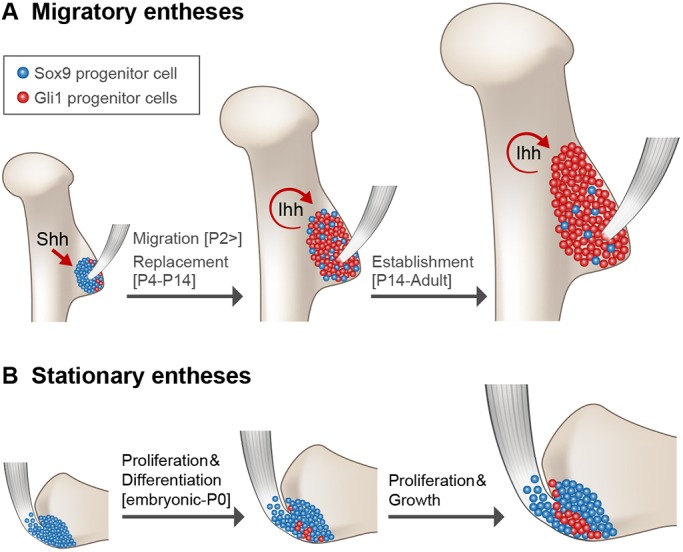
Migrating entheses develop through template replacement. (A-B) Schematic of the developmental sequence of migratory and stationary entheses. During embryogenesis, Sox9 lineage cells form an enthesis template. (A) In migrating entheses, Sox9 lineage cells are replaced by a second population of Gli1 lineage cells, which eventually form the mature enthesis. Specification of Gli1 lineage is regulated by SHH, whereas maintenance of this population is regulated by IHH. (B) In stationary entheses, embryonic Sox9 lineage cells proliferate and differentiate and, eventually, populate the postnatal enthesis structure. During late embryonic development, some Sox9 lineage cells start expressing Gli1.
MATERIALS AND METHODS
Animals
All experiments involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the Weizmann Institute. Histology was performed on BL6 mice.
For lineage tracing experiments, Sox9-Cre (Akiyama et al., 2005), Sox9-CreER (Soeda et al., 2010) and Gli1-CreERT2 (Ahn and Joyner, 2004; Jackson Laboratory) mice were crossed with R26R-tdTomato mice [B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; Madisen et al., 2010] or with R26R-Confetti mice [B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J; Snippert et al., 2010].
To create Shh KO mice, mice heterozygous for a mutation in Shh [B6.Cg-Shhtm1(EGFP/cre)Cjt/J; Jackson Laboratory] were intercrossed; heterozygotes or WT littermates were used as a control. The generation of Prx1-Cre (Logan et al., 2002) has been previously described. To generate Prx1-Cre-Ihh mutant mice, Ihh-floxed mice (B6N;129S4-Ihhtm1Blan/J; Jackson Laboratory) (Razzaque et al., 2005) were mated with Prx1-Cre mice. As a control, Prx1-Cre– animals were used.
For cell lineage experiments, Sox9-CreERT2/+ or Gli1-CreERT2/+ males were crossed with R26R-tdTomato females to produce embryos carrying both the relevant CreERT2 and R26R-tdTomato alleles. For fate mapping, Gli1-CreERT2/+ males were crossed with R26R-Confetti females to produce embryos carrying both the Gli1-CreERT2 and Confetti alleles.
In all experiments, n≥3 embryos or mice from different litters. In all timed pregnancies, plug date was defined as E0.5. For harvesting of embryos, timed-pregnant females were sacrificed by cervical dislocation. The gravid uterus was dissected out and suspended in a bath of ice-cold PBS and the embryos were harvested after removal of amnion and placenta. Tail genomic DNA was used for genotyping.
Histological analysis, in situ hybridization and immunofluorescence
For histology and in situ hybridization (ISH), embryos were harvested at various ages, dissected and fixed in 4% paraformaldehyde (PFA)/PBS at 4°C overnight. After fixation, tissues were dehydrated to 70% EtOH and embedded in paraffin. The embedded tissues were cut to 7-µm-thick sections and mounted onto slides. Hematoxylin and Eosin (H&E) staining was performed following standard protocols. Non-fluorescent and fluorescent ISH was performed as has been previously described using digoxigenin (DIG)-labeled probes (Shwartz and Zelzer, 2014). Probes are listed in Table S1. For analysis of Shh-GFPCre−/− mice, only mutants in which enthesis integrity was preserved were used.
For immunofluorescence staining, 10-µm-thick cryosections were air-dried for 1 h before staining. For IHH staining, sections were washed 2× in PBS+0.05% Tween-100 (PBST) for 5 min and blocked to prevent non-specific binding with 7% goat serum and 1% bovine serum albumin (BSA) dissolved in PBST. Then, sections were incubated with rabbit anti-IHH antibody (Abcam, AB39364, 1:50) at 4°C overnight. The next day, sections were washed 2× in PBST and incubated for 1 h with Cy2-conjugated fluorescent antibody (Jackson Laboratory, 711-225-152, 1:100). Slides were mounted with Immu-mount aqueous-based mounting medium (Thermo Fisher Scientific).
For Gli1 staining, 10-µm-thick cryosections were air dried for 1 h and fixed in 4% PFA for 10 min. Then, sections were washed 2× in PBST and endogenous peroxidase was quenched using 3% H2O2 in PBS. Next, antigen retrieval was performed using 0.3% Triton in PBS. Non-specific binding was blocked using 7% horse serum and 1% BSA dissolved in PBST for 1 h. Then, sections were incubated with Goat anti-GLI1 antibody (R&D Systems, AF3455, 1:100) overnight at room temperature. The next day, sections were washed 2× in PBST and incubated with Biotin anti-goat (Jackson Laboratory, 705065147, 1:100) for 1 h and then with streptavidin-HRP (Perkin Elmer, NEL750001EA, 1:200) for 1 h. HRP was developed using a TSA amplification kit (Perkin Elmer, 1:100) for 20 min, counterstained with DAPI and mounted with Immu-mount aqueous- based mounting medium.
Physical position of entheses
For identification of enthesis physical position, 2-4 limb samples at stages E16.5-P14 were scanned ex vivo using iodine contrast agent to allow visualization of the soft tissue. DT and TM positions were identified manually, and their physical position along the bone was calculated as described previously (Stern et al., 2015).
Cell lineage analysis
Tamoxifen (Sigma-Aldrich, T-5648) was dissolved in corn oil (Sigma-Aldrich, C-8267) at a final concentration of 5 mg/ml. Time-mated R26R-tdTomato females were administered 1 mg of tamoxifen by oral gavage (Fine Science Tools) at designated time points as indicated.
Cell count
For each age, two to four limbs from different litters were harvested, embedded in optimal cutting temperature compound (OCT) and sectioned at a thickness of 10 µm. Sections were imaged using a Zeiss LSM 780 microscope approximately every 80 µm, capturing red and DAPI channels. Images were then processed in ImageJ according to the following protocol. Each image was converted to an RGB stack and the region of interest was manually identified and cropped. Then, image levels were adjusted to improve separation between nuclei. A binary threshold was set automatically and conjoined nuclei were automatically separated using the binary watershed function. Using a home-made Matlab script, the number of nuclei was counted. Each nucleus that was co-localized with a red channel signal was counted as a tdTomato+ cell. For each section, the percentage of tdTomato+ cells was calculated and the average percentage of these cells was calculated for each bone. From each genotype and for each age group, at least three individual bones were sampled, and nine to 17 sections per bone were analyzed, depending on bone length. Statistical significance, defined as P<0.05, was determined using two-way ANOVA.
Fate mapping Confetti experiment
Tamoxifen was dissolved in corn oil at a final concentration of 20 mg/ml. Time-mated R26R-Confetti females were administered 4 mg of tamoxifen by oral gavage at E13.5. Cre+ pups were sacrificed at P14. Each pup was injected with Calcein Blue (Sigma, #m1255; 30 mg/kg) 1 h before sacrifice. Limbs were harvested, fixed for 30 min in 4% PFA at 4°C, embedded in OCT and sectioned at a thickness of 10 µm. Sections were imaged using a Zeiss LSM 780 microscope approximately every 80 µm.
Microscope settings and image analysis
At least 1024×1024 pixel, 8-bit images were acquired using the ×20 lens. A z-stack of two to four images was taken from each section. To detect GFP and YFP, the argon laser 488 nm was used. For RFP detection, a red diode laser emitting at 561 nm was used, and blue mCFP was excited using a laser line at 458 nm. Calcein Blue staining was detected using the 405 nm laser line. GFP fluorescence was collected between ∼500-598 nm, Airy 1; RFP fluorescence was collected between ∼606-654 nm, Airy 1; mCFP fluorescence was collected between ∼464-500 nm, Airy 1 and Calcein Blue was collected between ∼410-451 nm. For each image, a corresponding bright-field image was captured. The acquired images were processed using Photoshop and ImageJ.
To calculate the percentage of Gli1+ clones found in both mineralized and non-mineralized fibrocartilage, the border between the zones was identified using the Calcein Blue signal. Clones that crossed the mineral border were manually counted and their percentage was calculated first for each section, then for each limb and, finally, for each type of enthesis. Data are presented as mean±s.d.
Supplementary Material
Acknowledgements
We thank Nitzan Konstantin for expert editorial assistance, Dr Ron Rotkoff for his help with statistical analysis and Dr Patrick Tschopp from the Clifford J. Tabin Lab at the Harvard Medical School for his assistance in generating Shh KO mice. Special thanks to all members of the Zelzer Laboratory for encouragement and advice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.F., E.Z.; Software: T.S.; Formal analysis: N.F., T.S.; Investigation: N.F.; Resources: S.R., S.K., D.P., B.A.P., R.S.; Writing - original draft: N.F., E.Z.; Writing - review & editing: N.F., E.Z.; Visualization: N.F.; Supervision: E.Z.; Project administration: E.Z.; Funding acquisition: E.Z.
Funding
This study was supported by grants from the National Institutes of Health (R01 AR055580) and the European Research Council (310098, and Proof of Concept grant 737473). It was also supported by the Weizmann Institute of Science, through the following funders: the Jeanne and Joseph Nissim Foundation for Life Sciences Research, the Y. Leon Benoziyo Institute for Molecular Medicine, Beth Rom-Rymer, the Estate of David Levinson, the Bernard M. and Audrey Jaffe Foundation, the Georges Lustgarten Cancer Research Fund, the David and Fela Shapell Family Center for Genetic Disorders, the David and Fela Shapell Family Foundation INCPM Fund for Preclinical Studies, and the Estate of Bernard Bishin for the Weizmann Institute of Science-Clalit Program. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.165381.supplemental
References
- Ahn S. and Joyner A. L. (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505-516. 10.1016/j.cell.2004.07.023 [DOI] [PubMed] [Google Scholar]
- Akiyama H., Kim J.-E., Nakashima K., Balmes G., Iwai N., Deng J. M., Zhang Z., Martin J. F., Behringer R. R., Nakamura T. et al. (2005). Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. USA 102, 14665-14670. 10.1073/pnas.0504750102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin M. and McGonagle D. (2009). Entheses: tendon and ligament attachment sites. Scand. J. Med. Sci. Sports 19, 520-527. 10.1111/j.1600-0838.2009.00906.x [DOI] [PubMed] [Google Scholar]
- Benjamin M., Kumai T., Milz S., Boszczyk B. M., Boszczyk A. A. and Ralphs J. R. (2002). The skeletal attachment of tendons--tendon “entheses”. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 133, 931-945. 10.1016/S1095-6433(02)00138-1 [DOI] [PubMed] [Google Scholar]
- Benjamin M., Toumi H., Ralphs J. R., Bydder G., Best T. M. and Milz S. (2006). Where tendons and ligaments meet bone: attachment sites (‘entheses’) in relation to exercise and/or mechanical load. J. Anat. 208, 471-490. 10.1111/j.1469-7580.2006.00540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz E., Sharir A., Akiyama H. and Zelzer E. (2013). Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 140, 2680-2690. 10.1242/dev.093906 [DOI] [PubMed] [Google Scholar]
- Breidenbach A. P., Aschbacher-Smith L., Lu Y., Dyment N. A., Liu C.-F., Liu H., Wylie C., Rao M., Shearn J. T., Rowe D. W. et al. (2015). Ablating Hedgehog signaling in tenocytes during development impairs biomechanics and matrix organization of the adult murine patellar tendon enthesis. J. Orthop. Res. 33, 1142-1151. 10.1002/jor.22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D. and Relaix F. (2003). The formation of skeletal muscle: from somite to limb. J. Anat. 202, 59-68. 10.1046/j.1469-7580.2003.00139.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfl J. (1980a). Migration of tendinous insertions. I. Cause and mechanism. J. Anat. 131, 179-195. 10.1002/jor.22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfl J. (1980b). Migration of tendinous insertions. II. Experimental modifications. J. Anat. 131, 229-237. [PMC free article] [PubMed] [Google Scholar]
- Doschak M. R. and Zernicke R. F. (2005). Structure, function and adaptation of bone-tendon and bone-ligament complexes. J. Musculoskelet. Neuronal Interact. 5, 35-40. [PubMed] [Google Scholar]
- Dyment N. A., Breidenbach A. P., Schwartz A. G., Russell R. P., Aschbacher-Smith L., Liu H., Hagiwara Y., Jiang R., Thomopoulos S., Butler D. L. et al. (2015). Gdf5 progenitors give rise to fibrocartilage cells that mineralize via hedgehog signaling to form the zonal enthesis. Dev. Biol. 405, 1-12. 10.1016/j.ydbio.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatz L., Rothermich S., VanderPloeg K., Petersen B., Sandell L. and Thomopoulos S. (2007). Development of the supraspinatus tendon-to-bone insertion: localized expression of extracellular matrix and growth factor genes. J. Orthop. Res. 25, 1621-1628. 10.1002/jor.20441 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Kronenberg H. M. (2003). Developmental regulation of the growth plate. Nature 423, 332-336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Liu C.-F., Aschbacher-Smith L., Barthelery N. J., Dyment N., Butler D. and Wylie C. (2012). Spatial and temporal expression of molecular markers and cell signals during normal development of the mouse patellar tendon. Tissue Eng. Part A 18, 598-608. 10.1089/ten.tea.2011.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-F., Breidenbach A., Aschbacher-Smith L., Butler D. and Wylie C. (2013). A role for Hedgehog signaling in the differentiation of the insertion site of the patellar tendon in the mouse. PLoS ONE 8, e65411 10.1371/journal.pone.0065411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N. and Tabin C. J. (2002). Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis33, 77-80 10.1002/gene.10092 [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polly P. D. (2007). Limbs in mammalian evolution. In Fins into Limbs: Evolution, Development, and Transformation (ed B. K. Hall), pp. 245-268. University of Chicago Press. [Google Scholar]
- Razzaque M. S., Soegiarto D. W., Chang D., Long F. and Lanske B. (2005). Conditional deletion of Indian hedgehog from collagen type 2α1-expressing cells results in abnormal endochondral bone formation. J. Pathol. 207, 453-461. 10.1002/path.1870 [DOI] [PubMed] [Google Scholar]
- Salton J. A. and Sargis E. J. (2009). Evolutionary morphology of the tenrecoidea (Mammalia) hindlimb skeleton. J. Morphol. 270, 367-387. 10.1002/jmor.10697 [DOI] [PubMed] [Google Scholar]
- Schwartz A. G., Long F. and Thomopoulos S. (2015). Enthesis fibrocartilage cells originate from a population of Hedgehog-responsive cells modulated by the loading environment. Development 142, 196-206. 10.1242/dev.112714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A. G., Galatz L. M. and Thomopoulos S. (2017). Enthesis regeneration: a role for Gli1+ progenitor cells. Development 144, 1159-1164. 10.1242/dev.139303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R., Chyung J. H., Murtaugh L. C., Brent A. E., Rosen V., Olson E. N., Lassar A. and Tabin C. J. (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855-3866. [DOI] [PubMed] [Google Scholar]
- Shaw H. M. and Benjamin M. (2007). Structure-function relationships of entheses in relation to mechanical load and exercise. Scand. J. Med. Sci. Sports 17, 303-315. 10.1111/j.1600-0838.2007.00689.x [DOI] [PubMed] [Google Scholar]
- Shwartz Y. and Zelzer E. (2014). Nonradioactive in situ hybridization on skeletal tissue sections. Methods Mol. Biol. 1130, 203-215. 10.1007/978-1-62703-989-5_15 [DOI] [PubMed] [Google Scholar]
- Shwartz Y., Viukov S., Krief S. and Zelzer E. (2016). Joint development involves a continuous influx of Gdf5-positive cells. Cell Rep. 15, 2577-2587. 10.1016/j.celrep.2016.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H. J., van der Flier L. G., Sato T., van Es J. H., van den Born M., Kroon-Veenboer C., Barker N., Klein A. M., van Rheenen J., Simons B. D. et al. (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134-144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Soeda T., Deng J. M., de Crombrugghe B., Behringer R. R., Nakamura T. and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635-644. 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern T., Aviram R., Rot C., Galili T., Sharir A., Kalish Achrai N., Keller Y., Shahar R. and Zelzer E. (2015). Isometric scaling in developing long bones is achieved by an optimal epiphyseal growth balance. PLoS Biol. 13, e1002212 10.1371/journal.pbio.1002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y., Takimoto A., Akiyama H., Kist R., Scherer G., Nakamura T., Hiraki Y. and Shukunami C. (2013). Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140, 2280-2288. 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- Trainor P. and Krumlauf R. (2000). Plasticity in mouse neural crest cells reveals a new patterning rolefor cranial mesoderm. Nat. Cell Biol. 2, 96-102. 10.1038/35000051 [DOI] [PubMed] [Google Scholar]
- Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M. and Tabin C. J. (1996). Regulation of rate of cartilage differentiation by Indian Hedgehog and PTH-related protein. Science 273, 613-622. 10.1126/science.273.5275.613 [DOI] [PubMed] [Google Scholar]
- Wang M., Vanhouten J. N., Nasiri A. R., Johnson R. L. and Broadus A. E. (2013). PTHrP regulates the modeling of cortical bone surfaces at fibrous insertion sites during growth. J. Bone Miner. Res. 28, 598-607. 10.1002/jbmr.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.