ABSTRACT
Our ability to sequence genomes has vastly surpassed our ability to interpret the genetic variation we discover. This presents a major challenge in the clinical setting, where the recent application of whole-exome and whole-genome sequencing has uncovered thousands of genetic variants of uncertain significance. Here, we present a strategy for targeted human gene replacement and phenomic characterization, based on CRISPR-Cas9 genome engineering in the genetic model organism Caenorhabditis elegans, that will facilitate assessment of the functional conservation of human genes and structure-function analysis of disease-associated variants with unprecedented precision. We validate our strategy by demonstrating that direct single-copy replacement of the C. elegans ortholog (daf-18) with the critical human disease-associated gene phosphatase and tensin homolog (PTEN) is sufficient to rescue multiple phenotypic abnormalities caused by complete deletion of daf-18, including complex chemosensory and mechanosensory impairments. In addition, we used our strategy to generate animals harboring a single copy of the known pathogenic lipid phosphatase inactive PTEN variant (PTEN-G129E), and showed that our automated in vivo phenotypic assays could accurately and efficiently classify this missense variant as loss of function. The integrated nature of the human transgenes allows for analysis of both homozygous and heterozygous variants and greatly facilitates high-throughput precision medicine drug screens. By combining genome engineering with rapid and automated phenotypic characterization, our strategy streamlines the identification of novel conserved gene functions in complex sensory and learning phenotypes that can be used as in vivo functional assays to decipher variants of uncertain significance.
KEY WORDS: CRISPR-Cas9, Caenorhabditis elegans, Genome editing, Humanization, Phenomics, Variants of uncertain significance
Summary: Here, we provide a CRISPR-Cas9 human gene replacement and phenomic characterization strategy to directly replace Caenorhabditis elegans genes with their human orthologs for disease variant modeling and therapeutic screening.
INTRODUCTION
The rapid development and application of whole-exome and whole-genome sequencing technology has dramatically increased the pace at which we associate genetic variation with a particular disease (Auton et al., 2015; Bamshad et al., 2011; Gonzaga-Jauregui et al., 2012; Lek et al., 2016; Metzker, 2010; Need et al., 2012; Ng et al., 2010). However, our ability to sequence genomes has vastly surpassed our ability to interpret the clinical implications of the genetic variants we discover. The majority of genetic variants identified in clinical populations are currently classified as ‘variants of uncertain significance’, meaning that their potential role as a causative agent in the disease in question, or their pathogenicity, is unknown (Richards et al., 2015). Many variants are exceedingly rare, making it extremely difficult to designate them as pathogenic using classical genetic methods such as segregation within a pedigree, or by identifying multiple carriers of the variant. As such, it often remains challenging to predict clinical outcomes and make informed treatment decisions based on genetic data alone.
In an attempt to address this problem, several computational tools have been developed that estimate the functional consequences and pathogenicity of disease-associated variants (Richards et al., 2015). These tools use a variety of predictive features, such as evolutionary sequence conservation, protein structural and functional information, the prevalence of a variant in large putatively healthy control populations or a combination of annotations (Kircher et al., 2014; Lek et al., 2016; Richards et al., 2015). Despite extensive efforts, none of these tools used in isolation or combination can faithfully report on the functional effects of a large portion of disease-associated variation, and their accuracy is intrinsically limited to existing experimental training data (Grimm et al., 2015; Miosge et al., 2015; Starita et al., 2017). These limitations were clearly demonstrated in a recent study that showed that in vivo functional assays of 21 human genes in yeast identified pathogenic variants with significantly higher precision and specificity than computational methods (Sun et al., 2016). This means that, even for genes with well-characterized biological functions, there are often hundreds of variants of uncertain functional significance (Landrum et al., 2014; Starita et al., 2017). This creates a challenging situation that requires direct assessment of the functional effects of disease-associated variants in vivo (Starita et al., 2017).
Genetically tractable model organisms are crucial for discovering novel gene functions and the functional consequences of disease-associated genetic variants (Dunham and Fowler, 2013; Lehner, 2013; Manolio et al., 2017; Wangler et al., 2017). Governmental and private funding agencies are increasingly commissioning large-scale collaborative programs to use diverse genetic model organisms to decipher variants of uncertain significance (Chong et al., 2015; Gahl et al., 2016; Wangler et al., 2017). Among genetic model organisms, the nematode Caenorhabditis elegans has proven to be a particularly powerful animal model for the functional characterization of human genes in vivo (Kaletta and Hengartner, 2006). C. elegans is an ideal genetic model as it combines the throughput and tractability of a single-celled organism with the complex morphology and behavioral repertoire of a multi-cellular animal. In addition, ∼60-80% of human genes have an ortholog in the C. elegans genome (Kaletta and Hengartner, 2006; Lai et al., 2000; Shaye and Greenwald, 2011). Phenotypic analyses of transgenically expressed human genes are routinely done to confirm functional conservation and to observe the effects of disease-associated mutations. Notable examples of the utility of C. elegans to determine conserved human gene functions relevant to disease include the identification of presenilins as part of the gamma secretase complex, the mechanism of action of selective serotonin reuptake inhibitors, and the role of the insulin signaling pathway in normal and pathological aging (Kaletta and Hengartner, 2006; Levitan and Greenwald, 1995; Levitan et al., 1996; Murphy et al., 2003; Ranganathan et al., 2001). However, traditional methods for expression of human genes in C. elegans rely on mosaic and variable overexpression of transgenes harbored as extrachromosomal arrays or specialized genetic backgrounds that can confound phenotypic analysis. This presents several challenges that inhibit precise analysis of the often critical, but subtle, effects of missense variants and impede the use of these transgenic strains in large-scale drug screens.
The recent advent of CRISPR-Cas9-mediated genome editing has revolutionized structure-function analyses across model organisms (Cong et al., 2013; DiCarlo et al., 2013; Dickinson et al., 2013; Doudna and Charpentier, 2014; Friedland et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Jinek et al., 2012, 2013; Li et al., 2013). This system uses a single-guide RNA (sgRNA) to precisely target a nuclease (most often Cas9) to induce a DNA double-strand break at a defined location (Doudna and Charpentier, 2014). The double-strand break can then be repaired via the error-prone non-homologous end-joining pathway (often resulting in damaging frameshift mutations) or a homology repair pathway, e.g. homology-directed repair (HDR) or microhomology-mediated end joining (Nakade et al., 2014). Following a double-strand break, exogenous DNA repair templates can be used as substrates for HDR, allowing virtually any desired sequence to be inserted anywhere in the genome. Importantly, CRISPR-Cas9 genome engineering is remarkably efficient and robust in C. elegans (Dickinson and Goldstein, 2016; Norris et al., 2015).
Here, we present a broadly applicable strategy that adapts CRISPR-Cas9 genome engineering for targeted replacement of C. elegans genes with human genes. We illustrate how the library of knockout and humanized transgenics generated with this approach can be efficiently combined with automated machine vision phenotyping to rapidly discover novel gene functions, and assess the functional conservation of human genes, and how this will allow for analysis of the effects of variants of uncertain significance with unprecedented precision. It is our hope that the human gene replacement and phenomic characterization strategy delineated in this article will serve both basic and health researchers alike, by serving as an open and shareable resource that will aid any genome engineer interested in understanding the functional conservation of human genes, and the functional consequences of their variants.
RESULTS
A general genome editing strategy for direct replacement of a C. elegans gene with a single copy of its human ortholog
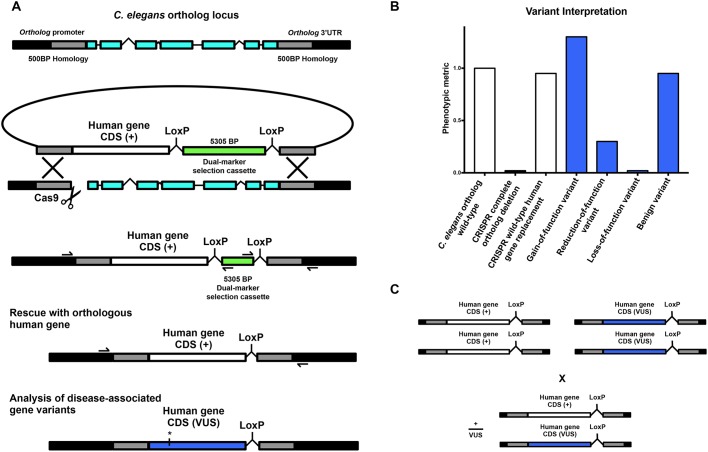
To replace the open reading frame (ORF) of an orthologous gene with a human gene, our strategy first directs an sgRNA to induce a Cas9-mediated DNA double-strand break immediately downstream of the ortholog start codon (Fig. 1A). A co-injected repair template containing ∼500 bp homology arms targeted to the regions immediately upstream and downstream of the ortholog ORF serves as a substrate for HDR. By fusing the coding DNA sequence (CDS) of a human gene of interest to the upstream homology arm, HDR integrates the human gene into the ortholog at a single copy in-frame (Fig. 1A).
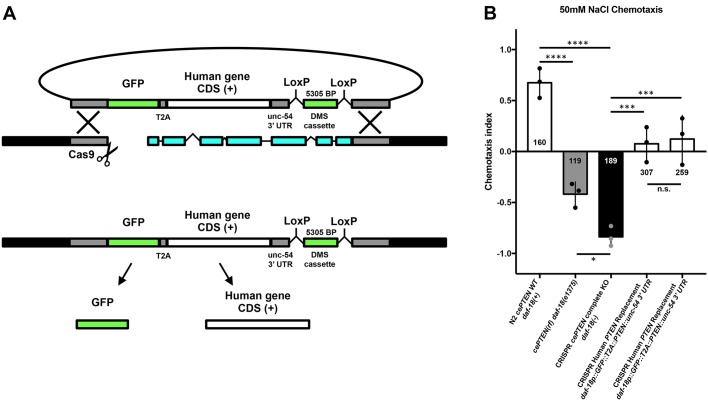
Fig. 1.
A general strategy for direct single-copy replacement of C. elegans genes with human genes at the ortholog’s native genomic loci. (A) A schematic of the genome editing strategy. An sgRNA targets Cas9 to induce a DNA double-strand break immediately downstream of the ortholog’s start codon. A co-injected repair template containing ∼500 bp homology arms targeted to the regions immediately upstream and downstream of the ortholog ORF serve as a substrate for HDR. By fusing the CDS of a human gene of interest to the upstream homology arm, HDR integrates the human gene in place of the ortholog at a single copy in-frame. A co-integrated DMS cassette consisting of an antibiotic resistance gene (Prps-27::neoR::unc-54 UTR) and a fluorescent marker (Pmyo-2::GFP::unc-54 UTR) greatly facilitates the identification of transgenic animals without inducing morphological or phenotypic abnormalities. Initial integration deletes the entire ORF of the C. elegans ortholog while separating the human gene from the ortholog’s transcriptional terminator to inhibit expression, creating an ortholog deletion allele for phenotypic analysis (note that the cassette is not shown to scale for most human gene CDSs). Subsequent injection of Cre recombinase excises the selection cassette and connects the human gene to the orthologous transcriptional termination sequence, such that a single copy of the human gene will now be expressed under the control of all the ortholog’s 5′ and 3′ cis- and trans-regulatory machinery. Validation of the desired edit is performed using standard amplification and Sanger sequencing of the target region (primer binding locations represented by half arrows in the schematic). For analysis of a human gene variant of uncertain significance (VUS), the variant of interest is incorporated into the HDR plasmid using standard in vitro methods, such as site-directed mutagenesis, and the same genome editing process is repeated using the same validated sgRNA and homology arms. (B) Human gene replacement allows for straightforward interpretation of variant functional effect. This process allows for (1) initial generation and phenotypic analysis of a complete null allele in the C. elegans orthologous gene; (2) direct integration of the human gene to determine whether the human gene can compensate for loss of the orthologous gene, measuring functional conservation; and (3) structure-function analysis of the effects of variants of uncertain significance on wild-type gene function. (C) This strategy allows for straightforward assessment of heterozygous alleles using standard genetic crosses.
To streamline genome editing we have based our method on a recently described dual-marker-selection (DMS) cassette screening protocol (Norris et al., 2015). The DMS cassette consists of an antibiotic resistance gene (Prps-27::neoR::unc-54 UTR) and a fluorescent marker (Pmyo-2::GFP::unc-54 UTR) that greatly facilitates the identification of transgenic animals (Norris et al., 2015). We chose this cassette over similar methods as it can be used in any wild-type or mutant strain amenable to transgenesis and does not require any specialized genetic backgrounds, and avoids the use of morphology and/or behavior altering selection markers that necessitate cassette excision prior to phenotypic analysis (Bend et al., 2016; Dickinson et al., 2013, 2015). In our strategy, the DMS cassette is placed between the human gene of interest and the downstream homology arm (Fig. 1A). This deletes the entire ORF of the ortholog and separates the human gene from the ortholog’s transcriptional terminator upon initial integration. In many cases, this efficiently creates a useful deletion allele of the ortholog with no human gene expression (this can be confirmed via inclusion of an epitope tag if immunological reagents are unavailable). In the unlikely event that human gene expression does occur without a transcription terminator, a second repair template using the same validated homology arms and sgRNA – and no human gene – can be integrated to create an ortholog null. A deletion allele can also be ordered from the Caenorhabditis Genetics Center where available. Importantly, the DMS cassette is flanked by two LoxP sites housed within synthetic introns that allow subsequent excision of the selection cassette via transient expression of Cre recombinase (Norris et al., 2015). DMS cassette excision connects the human gene to the endogenous C. elegans ortholog’s transcriptional termination sequence, such that a single copy of the human gene will now be expressed under the control of all of the ortholog’s 5′ and 3′ cis- and trans-regulatory machinery (Fig. 1B). Validation of the desired edit is performed using standard PCR amplification and Sanger sequencing of both the 5′ and 3′ junctions of the target region (Fig. 1A; Au et al., 2018).
For structure-function analysis of a human gene variant of uncertain significance, the variant of interest is incorporated into the HDR plasmid using standard in vitro methods such as site-directed mutagenesis, and the same genome editing process is repeated using the same validated sgRNA and homology arms. This process allows for (1) the initial generation and phenotypic analysis of a complete deletion allele in the C. elegans orthologous gene; (2) direct integration of the human gene to determine whether the human gene can compensate for loss of the orthologous gene, measuring functional conservation; and (3) structure-function analysis of the effects of variants of uncertain significance on wild-type gene function (Fig. 1B). Importantly, the vast majority of variants of uncertain significance identified in patients are heterozygous, and the integrated nature of the transgenes generated with this strategy allows for straightforward assessment of heterozygous alleles using standard genetic crosses (Fig. 1C).
PTEN as a prototypic disease-associated gene for targeted human gene replacement
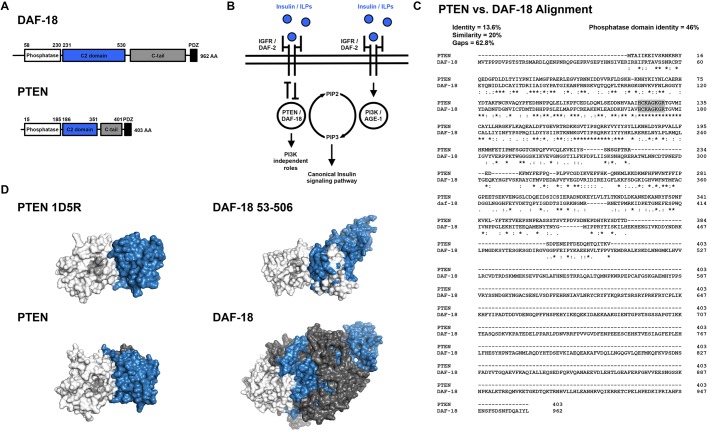
As a proof of principle, we demonstrated the utility of our strategy by focusing on the critical disease-associated gene phosphatase and tensin homolog (PTEN). PTEN is a lipid phosphatase that antagonizes the phosphoinositide 3-kinase (PI3K) signaling pathway by dephosphorylating phosphatidylinositol (3,4,5)-trisphosphate (PIP3) (Li et al., 1997; Maehama and Dixon, 1998). Heterozygous germline PTEN mutations are associated with diverse clinical outcomes, including several tumor predisposition phenotypes (collectively called PTEN harmatoma tumor syndrome), intellectual disability and autism spectrum disorders (Hobert et al., 2014; Li et al., 1997; Liaw et al., 1997; McBride et al., 2010; O'Roak et al., 2012; Orrico et al., 2009; Sanders et al., 2015; Varga et al., 2009). Despite extensive study, it is currently impossible to predict the clinical outcome of a PTEN mutation carrier using sequence data alone (Matreyek et al., 2018; Mighell et al., 2018). PTEN also has several technical advantages that make it an ideal test case: (1) PTEN functions in the highly conserved insulin signaling pathway that is well characterized in C. elegans (Ozes et al., 2001; Ogg and Ruvkun, 1998; Mihaylova et al., 1999; Fig. 2B); (2) C. elegans has a single PTEN ortholog called daf-18 (Fig. 2A-D), and transgenic overexpression of human PTEN using extrachromosomal arrays has been shown to rescue reduced longevity and dauer-defective phenotypes induced by mutations in daf-18 (Liu and Chin-Sang, 2015; Solari et al., 2005); and (3) C. elegans harboring homozygous daf-18 null alleles are viable and display superficially normal morphology and spontaneous locomotor behavior (Mihaylova et al., 1999; Fig. S1).
Fig. 2.
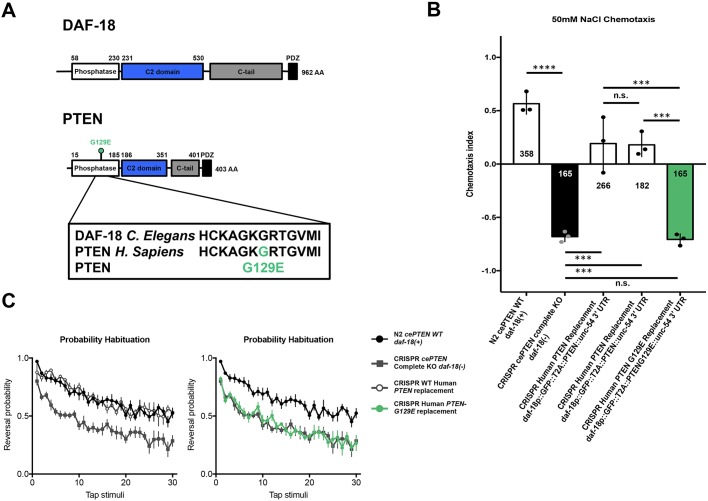
Functional and structural similarity of C. elegans DAF-18 and human PTEN. (A) Protein domain annotations for DAF-18 and PTEN. The canonical DAF-18 amino acid (AA) sequence is more than twice as long as the canonical PTEN AA sequence, primarily due to elongation of the C2 membrane targeting and C-tail domains. (B) Both DAF-18 and PTEN function as lipid and protein phosphatases to antagonize the highly conserved canonical insulin signaling pathway. (C) Clustal alignment of DAF-18 and PTEN. DAF-18 and PTEN share a highly conserved phosphatase domain (46% identity) and fully conserved catalytic site (residues highlighted in gray). DAF-18 has a markedly longer and less conserved C-terminal region than human PTEN, resulting in low overall AA similarity (20%) and identity (13.6%). Note that although the C-terminal region is much longer, there are small conserved motifs spread throughout that are not illustrated by this alignment (Liu et al., 2014). (D) Comparison of DAF-18 and PTEN structural models. (Top left) Solved crystal structure of human PTEN (1D5R reference structure; Lee et al., 1999). (Top right) Predicted structure of DAF-18 AA 53-506, indicating similar 3D structure to human PTEN. (Bottom left) Predicted structure of full-length PTEN. (Bottom right) DAF-18, illustrating the increased size of DAF-18. Note that the full-length DAF-18 model is likely to be inaccurate due to poor homology-based modeling of the non-conserved C-terminal region. Domain color mapping matches that in A, except that the catalytic site within the phosphatase domain is shaded light gray.
An automated chemotaxis paradigm reveals a conserved nervous system role for PTEN in controlling NaCl preference
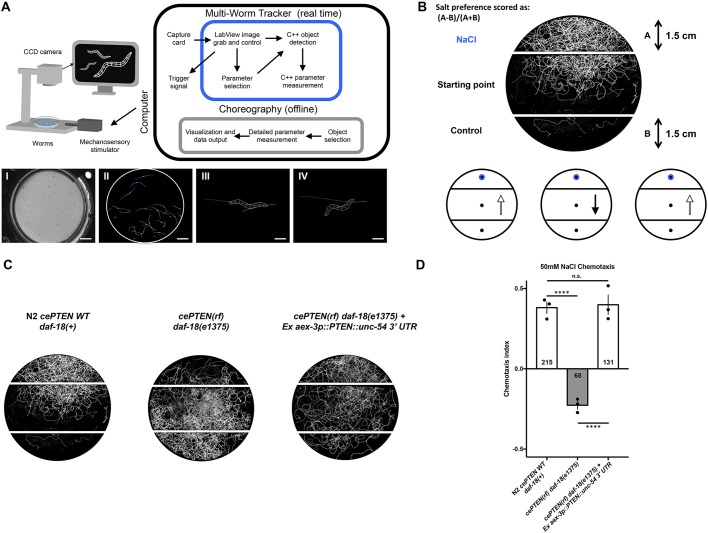
Structure-function analyses of PTEN variants necessitate an in vivo phenotypic assay that reports on PTEN function. When wild-type worms are grown in the presence of NaCl and their food source (Escherichia coli), they display naïve attractive navigation behavior toward NaCl. This behavior is called NaCl chemotaxis and can be quantified by measuring the navigation behavior of a population of animals up a controlled NaCl concentration gradient (Ward, 1973). Previous work has shown that daf-18 is required for attractive navigation behavior up a concentration gradient of NaCl, such that animals with deletion or reduction-of-function mutations in daf-18 display innate aversion to NaCl (Tomioka et al., 2006). Interestingly, insulin/PI3K signaling normally actively promotes salt avoidance under naïve conditions and daf-18 functions in a single chemosensory neuron (ASER) to antagonize the insulin/PI3K pathway and promote salt attraction (Adachi et al., 2010; Tomioka et al., 2006). Using our machine vision system, the Multi-Worm Tracker, we developed an automated high-throughput NaCl chemotaxis paradigm (Fig. 3A,B) and replicated the finding that daf-18(e1375) reduction-of-function mutants display strong aversion to NaCl (Swierczek et al., 2011; Fig. 3C,D). We then generated a transgenic line using traditional extrachromomosal array technology that directed pan-neuronal expression of wild-type human PTEN using the aex-3 promoter (Kuroyanagi et al., 2010). Pan-neuronal expression of human PTEN is able to rescue the daf-18 reduction-of-function phenotype and restore attractive NaCl chemotaxis to wild-type levels (Fig. 3C,D). This work establishes NaCl chemotaxis as an in vivo behavioral assay of conserved nervous system PTEN functions.
Fig. 3.
A conserved neuronal role for PTEN in NaCl preference revealed by a novel automated chemotaxis paradigm. (A) (Top) All phenotypic analysis was conducted using our machine vision system, the Multi-Worm Tracker. The Multi-Worm Tracker delivers stimuli and performs image acquisition, object selection and parameter selection in real time, while Choreography software extracts detailed phenotypic information offline. (Bottom) (I) A Petri plate of C. elegans; (II) a Petri plate of C. elegans selected for analysis by the Multi-Worm Tracker; (III and IV) Multi-Worm Tracker digital representations showing the degree of phenotypic detail. An example behavior scored by the Multi-Worm Tracker: the C. elegans response to a mechanosensory tap to the side of the Petri plate is brief backwards locomotion (from III to IV). Scale bars: 1 cm (I and II), 0.25 mm (III and IV). (B,C) Behavioral track tracing of a plate of worms from a novel automated Multi-Worm Tracker NaCl chemotaxis paradigm, illustrating attractive navigation behavior of wild-type animals toward a point source of NaCl. (B) Circles and arrows (bottom, left to right) and (C) worm tracks (left to right) represent navigation trajectories of wild-type attraction to a point source of NaCl, a daf-18(e1375) reduction-of-function decrease in NaCl chemotaxis, and a transgenic rescue of NaCl preference via pan-neuronal overexpression of wild-type human PTEN in daf-18(e1375) reduction-of-function mutants. (D) Quantitative chemotaxis index scores across genotypes. Pan-neuronal expression of human PTEN rescues the reversed NaCl preference of daf-18(e1375) mutants to wild-type levels. Circles represent plate replicates run on the same day; numbers in bars represent the number of individual animals registered by the tracker and located outside the center starting region (i.e. included in the preference score) across the three plate replicates for each genotype. Error bars represent s.d., using the number of plates as n (n=3). ****P<0.0001; n.s., not significant; one-way ANOVA and Tukey's post hoc test.
Complete deletion of the daf-18 ORF causes strong NaCl avoidance that is not rescued by direct single-copy replacement with the canonical human PTEN CDS
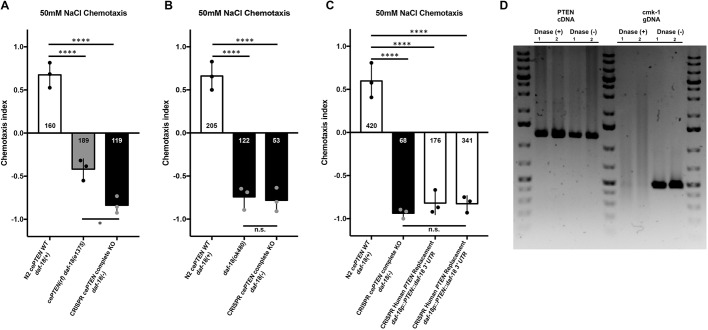
Next, we used our strategy to create a daf-18 deletion allele and single-copy replacement daf-18 with the 1212 bp canonical human PTEN CDS. Complete deletion of the 4723 bp daf-18 ORF resulted in strong aversion to NaCl and chemotaxis down the salt gradient (Fig. 4A). We achieved genome editing efficiency for human gene replacement similar to what has been previously reported for plasmid-based CRISPR-Cas9-mediated DMS cassette integration alone (Au et al., 2018; Norris et al., 2015, 2017). Chemotaxis avoidance of worms harboring the daf-18 complete deletion allele generated using CRISPR-Cas9 was not significantly different from that of worms carrying the previously characterized large deletion allele daf-18(ok480), confirming effective inactivation of daf-18 (Fig. 4B). DMS cassette excision and expression of a single copy of human PTEN was unable to substitute for daf-18 and did not rescue attractive NaCl chemotaxis behavior (Fig. 4C). This result was observed in two independent single-copy human PTEN knock-in lines on several independent experimental replications (Fig. 4C). Transcription of human PTEN was confirmed using reverse transcription PCR in both knock-in lines (Fig. 4D). Sanger sequencing confirmed error-free insertion of transgenes at base-pair resolution before and after cassette excision. There are several potential reasons why expression of human PTEN using extrachromasomal arrays rescued daf-18 mutant phenotypes (Solari et al., 2005; Fig. 3C,D), whereas targeted single-copy replacement with PTEN did not (Fig. 4C). The two most prominent differences between these two technologies are the expression levels of the transgenes and the use of the endogenous 3′ UTR in the CRISPR-Cas9 knock-in versus the unc-54 myosin 3′ UTR used in most C. elegans transgenes, to ensure proper processing of transcripts in all tissues, including all constructs previously shown to rescue daf-18 phenotypes with human PTEN (Merritt and Seydoux, 2010; Solari et al., 2005; Fig. 3C,D).
Fig. 4.
Complete daf-18 ORF deletion causes strong NaCl avoidance that is not rescued by direct replacement with human PTEN. (A) NaCl chemotaxis preference scores of wild-type, daf-18(e1375) reduction-of-function and CRISPR daf-18 complete deletion mutants. daf-18 complete deletion mutants show significantly stronger NaCl avoidance than daf-18(e1375) reduction-of-function mutants. (B) CRISPR daf-18 complete deletion mutants are not significantly different from animals harboring the putative null daf-18(ok480) deletion allele. (C) Expression of a single-copy PTEN transgene from the native daf-18 locus is insufficient to significantly rescue NaCl chemotaxis toward wild-type levels. (D) Reverse transcription PCR, confirming expression of full-length PTEN mRNA in the two independent knock-in lines used for behavioral analysis. Previously validated primers that target cmk-1 intronic regions of genomic DNA do not produce products following DNase treatment, confirming purity of the cDNA. Circles represent plate replicates run on the same day; numbers in bars represent the number of individual animals registered by the tracker and located outside the center starting region (i.e. included in preference score) across the three plate replicates for each genotype. Error bars represent s.d., using the number of plates as n (n=3). ****P<0.0001, *P<0.05; n.s., not significant; one-way ANOVA and Tukey's post hoc test.
A streamlined human gene replacement strategy functionally replaces daf-18 with human PTEN
In order to address these limitations and increase the speed of transgenesis, we created an alternative repair template strategy that includes a transcriptional termination sequence so that expression of the human gene begins immediately upon integration (Fig. 5A). We included the validated unc-54 3′ UTR sequence fused to the 3′ end of human PTEN (Fig. 5A). Expression of wild-type human PTEN using this genome editing strategy significantly rescued NaCl chemotaxis, indicating production of functional PTEN immediately upon genomic integration prior to cassette excision (Fig. 5B). Again, for this editing strategy, we achieved efficiency similar to what has been previously reported for DMS cassette integration alone (Au et al., 2018; Norris et al., 2015, 2017). This alternative approach also offers the added benefits of increased throughput (as a second injection step to excise the cassette was not required for human gene expression as it is in the first strategy) and the option for retained visual transgenic markers (either within the selection cassette or by adding a 2A sequence to drive reporter expression from the same promoter), which simplifies the generation and phenotypic analysis of heterozygotes and double mutants (Fig. 5A; Ahier and Jarriault, 2014; Calarco and Norris, 2018; Norris et al., 2017). Given the demonstrated versatility of CRISPR-Cas9-mediated genome editing in C. elegans, these results suggest that our strategy should be broadly applicable for in vivo analysis of diverse human disease-associated genes.
Fig. 5.
A streamlined human gene replacement strategy functionally replaces daf-18 with human PTEN. (A) Streamlined CRISPR-Cas9 gene replacement strategy. Inclusion of the validated unc-54 3′ UTR in the upstream homology arm increases the speed of transgenesis by removing the need for cassette excision to induce transgene expression. This alternative approach also offers the option for retained visual transgenic markers, which simplifies the generation and phenotypic analysis of heterozygotes and double mutants. The inclusion of a GFP::T2A cassette is an optional addition to allow for confirmation of transgene expression without altering human gene function (Ahier and Jarriault, 2014). (B) Expression of wild-type human PTEN using this genome editing strategy significantly rescued NaCl chemotaxis toward wild-type levels. Circles represent plate replicates run on the same day; numbers in and below bars represent the number of individual animals registered by the tracker and located outside the center starting region (i.e. included in preference score) across the three plate replicates for each genotype. Error bars represent s.d., using the number of plates as n (n=3). ****P<0.0001, ***P<0.001, *P<0.05; n.s., not significant; one-way ANOVA and Tukey's post hoc test.
daf-18 deletion causes mechanosensory hyporesponsivity that is rescued by targeted replacement with human PTEN
A long-standing goal has been to understand how human disease-associated variants alter normal gene function to produce sensory and learning abnormalities characteristic of diverse neurogenetic disorders. The massive number variants of uncertain significance recently implicated in the etiology neurogenetic disorders necessitates a dramatic increase in throughput of both transgenic construction and behavioral phenotyping if this goal is to be achieved (Ben-Shalom et al., 2017; Geschwind and Flint, 2015; Lim et al., 2017; Sanders et al., 2015; Starita et al., 2017; Wangler et al., 2017). By combing streamlined human gene integration with rapid machine vision phenotypic analysis of C. elegans, our strategy greatly simplifies the identification of novel conserved gene functions in complex sensory and learning phenotypes.
We explored whether daf-18 mutants displayed behavioral deficits in mechanosensory responding and/or habituation, a conserved form of non-associative learning that is altered in several neurodevelopmental and neuropsychiatric disorders (McDiarmid et al., 2017, 2018; Rankin et al., 2009; Stessman et al., 2017; Timbers et al., 2017; van der Voet et al., 2014). When a non-localized mechanosensory stimulus is delivered to the side of the Petri plate C. elegans are cultured on, they respond by eliciting a brief reversal before resuming forward locomotion. Wild-type C. elegans habituate to repeated stimuli by learning to decrease the probability of eliciting a reversal (Rankin et al., 1990). To determine whether daf-18 is important for mechanoresponding and/or non-associative learning, we examined habituation of the daf-18(e1375) and daf-18 complete deletion mutants. Compared with wild-type animals, both daf-18(e1375) reduction-of-function and daf-18 complete deletion mutants exhibited significantly reduced probability of eliciting a reversal response throughout the habituation training session, indicating mechanosensory hyporesponsivity (Fig. 6A,B). Despite this hyporesponsivity, the plasticity of responses, or the pattern of the gradual decrement in the probability of emitting of a reversal response throughout the training session, was not significantly altered in daf-18 mutants (Fig. 6A,B,E). Importantly, targeted single-copy replacement of daf-18 with human PTEN was sufficient to rescue the mechanosensory hyporesponsivity phenotype across the training session toward wild-type levels (Fig. 6D). These results identify a novel conserved role for PTEN in mechanosensory responding, a fundamental biological process disrupted in diverse genetic disorders (Badr et al., 1987; McDiarmid et al., 2017; Orefice et al., 2016). More broadly, they illustrate how the library of transgenic animals generated with our strategy can be used to rapidly characterize the role of diverse human genes in complex sensory and learning behaviors. These novel phenotypes can then be used to investigate the functional consequences of disease-associated variants in intact and freely behaving animals, and to screen for therapeutics that reverse the effects of a particular patient’s missense variant.
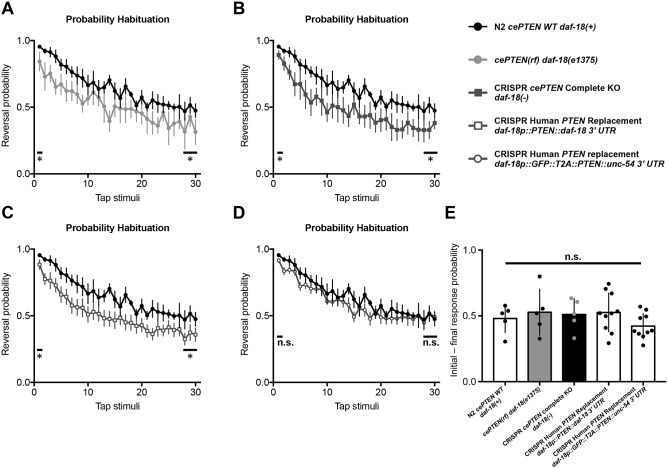
Fig. 6.
daf-18 deletion causes mechanosensory hyporesponsivity that is rescued by targeted replacement with human PTEN. (A-D) Average probability of eliciting a reversal response to 30 consecutive tap stimuli delivered at a 10 s ISI. (A) daf-18(e1375) reduction-of-function and (B) daf-18 complete deletion mutants exhibit significantly reduced probability of eliciting a reversal response throughout the habituation training session, indicating mechanosensory hyporesponsivity. (C) Replacement of daf-18 with human PTEN using the original strategy does not rescue mechanosensory hyporesponsivity. (D) Expression of human PTEN using the streamlined replacement strategy rescues mechanosensory responding to wild-type levels. Error bars represent s.e.m. (E) Habituation, or the ability to learn to decrease the probability of eliciting of a reversal response, throughout the training session was not significantly altered in daf-18 mutants. Circles represent plate replicates run on the same day. Error bars represent s.d., using the number of plates as n (n=5 or 10). *P<0.05; n.s., not significant; one-way ANOVA and Tukey's post hoc test.
Human gene replacement and in vivo phenotypic assessment accurately identifies the functional consequences of the pathogenic PTEN-G129E variant
To demonstrate the feasibility of our human gene replacement strategy for assessing variants of uncertain significance, we set out to determine whether our in vivo functional assays could discern known pathogenic variants. Early studies characterizing the role of PTEN as a tumor suppressor suggested that impaired protein phosphatase activity was key to the etiology of PTEN disorders (Tamura et al., 1998). However, this notion was challenged by the identification of cancer patients harboring a missense mutation that changes a glycine residue in the catalytic signature motif to a glutamate, which was predicted to abolish the lipid phosphatase activity of PTEN while leaving the protein phosphatase activity intact (Liaw et al., 1997; Myers et al., 1997). Subsequent biochemical analyses supported the now widely accepted view that the lipid phosphatase activity of PTEN is critical to its tumor suppressor activity (Myers et al., 1998).
We used site-directed mutagenesis to incorporate the PTEN-G129E (PTEN, c386G>A) missense variant into our repair template and used our human gene replacement strategy to replace daf-18 with a single copy of human PTEN-G129E (Fig. 7A). Animals harboring the PTEN-G129E variant displayed strong NaCl avoidance, equivalent to that of animals carrying the complete daf-18 deletion allele, indicating loss of function (Fig. 7B). Similarly, PTEN-G129E mutants also displayed mechanosensory hyporesponsivity that was not significantly different from that of daf-18 deletion carriers (Fig. 7C). These in vivo phenotypic results accurately classify the pathogenic PTEN-G129E as a strong loss-of-function variant. In addition, by taking advantage of a pathogenic variant with well-characterized biochemical effects, these results identify a necessary role for PTEN lipid phosphatase activity in both chemotaxis and mechanosensory responding, providing novel insight into the molecular mechanisms underlying these forms of sensory processing. Taken together, these results demonstrate the potential of human gene replacement and phenomic characterization to rapidly identify the functional consequences variants of uncertain significance.
Fig. 7.
Human gene replacement and in vivo phenotypic assessment accurately identifies functional consequences of the pathogenic PTEN-G129E variant. (A) Location (top) and conserved amino acid sequence change (bottom) of the pathogenic lipid phosphatase inactive PTEN-G129E variant within the PTEN phosphatase domain. (B) Animals harboring the PTEN G129E variant displayed strong NaCl avoidance, equivalent to that of animals carrying the complete daf-18 deletion allele, indicating loss of function. Circles represent plate replicates run on the same day; numbers in and below bars represent the number of individual animals registered by the tracker and located outside the center starting region (i.e. included in preference score) across the three plate replicates for each genotype. Error bars represent s.d., using the number of plates as n (n=3). (C) Similarly, PTEN-G129E mutants also displayed mechanosensory hyporesponsivity that was not significantly different from that of daf-18 deletion mutants. Error bars represent s.e.m. ****P<0.0001, ***P<0.001; n.s., not significant; one-way ANOVA and Tukey's post hoc test.
DISCUSSION
We have developed and validated a broadly applicable strategy for targeted human gene replacement and phenomic characterization in C. elegans that will facilitate assessment of the functional conservation of human genes and structure-function analysis of variants of uncertain significance with unprecedented precision. We established an automated NaCl chemotaxis paradigm and demonstrate that pan-neuronal overexpression or direct replacement of daf-18 with its human ortholog PTEN using CRISPR-Cas9 is sufficient to rescue reversed NaCl chemotaxis preference induced by complete daf-18 deletion. We further identified a novel mechanosensory hyporesponsive phenotype for daf-18 mutants that could also be rescued by targeted replacement with human PTEN. In vivo characterization of mutants harboring a single copy of the known lipid phosphatase inactive G129E variant accurately classified this variant as pathogenic, and revealed a crucial role for PTEN lipid phosphatase activity in NaCl chemotaxis and mechanosensory responding. We provide novel high-throughput in vivo functional assays for PTEN, as well as validated strains, reagents, sgRNA and repair template constructs to catalyze further analysis of this critical human disease-associated gene. More broadly, we provide a conceptual framework that illustrates how genome engineering and automated machine vision phenotyping can be combined to efficiently generate and characterize a library of knockout and humanized transgenic strains that will allow for straightforward and precise analysis of human genes and disease-associated variants in vivo (Fig. 8).
Fig. 8.
A conceptual framework for in vivo functional analysis of human genetic variation using C. elegans. (A) (Clockwise from top) A human gene and/or variant of uncertain significance is implicated in disease etiology through clinical sequencing. Targeted CRISPR-Cas9 human gene replacement or analogous methods are used to generate a library of knockout, human wild-type and variant transgenic strains. Large isogenic synchronous colonies of these transgenic worms are grown, and their morphology, baseline locomotion and sensory phenotypes are rapidly characterized using machine vision to establish novel functional assays and interpret variant effects. In vivo functional data can be used to probe epistatic network disruptions and cluster variants based on multi-parametric phenotypic profiles. The integrated humanized transgenic lines and functional assays greatly facilitate downstream applications, including precision medicine drug screens designed to identify compounds that reverse the effects of a particular patient’s missense variant. (B) Advantages of targeted human gene replacement using C. elegans.
Comparing CRISPR-Cas9 targeted human gene replacement with orthology-based variant assessment methods
To date, the most widely used genome editing-based human disease variant assessment strategy in C. elegans uses sequence alignments to identify and engineer the corresponding amino acid change into the orthologous C. elegans gene (Bend et al., 2016; Bulger et al., 2017; Canning et al., 2018; Pierce et al., 2018; Prior et al., 2017; Sorkaç et al., 2016; Troulinaki et al., 2018). A major advantage of this approach is that, by using the C. elegans gene, intronic regulation, protein-protein interactions, subcellular localization and biochemical activity of the protein of interest are, by design, perfectly modeled by C. elegans. Even when expressed at physiologically relevant levels directly from the ortholog’s native loci, and with evidence of phenotypic rescue, it is not guaranteed that a transgenic human protein will recapitulate all functions and interactions of the orthologous C. elegans protein. This presents an important consideration when attempting to replace C. elegans proteins that must interact in extremely precise heteromeric complexes to perform their molecular functions (e.g. certain ion channel subunits) (Bend et al., 2016; Prior et al., 2017). The most obvious limitation of this approach is that it can only be used to study orthologous amino acids. Amino acids that have been conserved throughout evolution are, by definition, the least tolerant to mutation and are thus far more likely to be detrimental to protein function when mutated (Starita et al., 2017; Weile et al., 2017). Indeed, many variant effect prediction algorithms rely on sequence conservation as the main predictor that a variant will be deleterious (Richards et al., 2015; Starita et al., 2017). It is also important to note that a large portion of amino acids will not be conserved to humans (e.g. >50% of amino acids are not conserved from DAF-18 to PTEN, Fig. 2C) and alignment algorithms that identify orthologous amino acids are imperfect. Many current implementations of orthologous amino acid engineering also require constant generation of completely new sgRNAs and repair templates for subsequent edits. Human gene replacement, in contrast, allows any coding variant of uncertain significance to be studied in vivo using the same validated sgRNA and homology arms.
One potential limitation of human gene replacement is that it relies on a C. elegans ortholog to replace. Estimates suggest that there are C. elegans orthologs for 60-80% of human disease-associated genes. This means there will not be an ortholog available for a minority of human genes (Kaletta and Hengartner, 2006; Lai et al., 2000; Shaye and Greenwald, 2011). This is even less likely to be a problem for disease variant modeling, as disease-associated genes are more likely to be highly conserved (Aerts et al., 2006; López-Bigas and Ouzounis, 2004). A recent study also showed that ∼10% of human disease-associated genes are able to functionally substitute for their yeast paralogs (in addition to orthologs), further increasing the number of human genes that can be studied by replacement (Yang et al., 2017). Still, in the event that there is no suitable C. elegans ortholog or paralog available for a human gene of interest, the human gene can simply be integrated at a putatively neutral genomic location using CRISPR-Cas9 or transposon mobilization and expressed from a heterologous promoter (Frøkjær-Jensen et al., 2008, 2012, 2014). This approach can be used to screen for phenotypes induced by expression of any human gene, and to determine whether a variant exacerbates or eliminates these effects (Baruah et al., 2017).
With the rapidly expanding set of precise genome editing techniques available to C. elegans, researchers interested in variants of uncertain significance now have the freedom to choose the approach that best suits their particular needs and interests. The approaches described here provide a diverse collection of methods that can be sequentially tested in a pragmatic hierarchy of precision, beginning with direct replacement and working down until phenotypic rescue is achieved. Regardless, it will always be ideal to have corroborating evidence of variant effect from multiple techniques, indeed multiple model systems, to best inform clinical decisions.
Combining human gene replacement and automated phenomic characterization to discover conserved gene functions and establish variant functional assays
A necessary step in the establishment of human gene functional assays is the identification of phenotypes that are rescued by or induced upon expression of the reference (wild-type) human gene, as we have done here for human PTEN and NaCl chemotaxis. Indeed, the establishment of functional assays remains a major bottleneck for variant assessment across species (Starita et al., 2017; Weile et al., 2017). Although traditional extrachromosomal array transgenes offer a quick way to establish such assays, several aspects of these transgenes can severely impede this process. These include, but are not limited to, (1) variable overexpression of transgenes, which can lead to silencing of transgenes in certain tissues, complicating phenotypic analysis (e.g. multi-copy transgenes are expressed in the soma but silenced in the germline, while low- and single-copy transgenes are expressed in both) (Kelly et al., 1997; Merritt and Seydoux, 2010); and (2) variably mosaic expression, which can make it extremely difficult to assess the rescue of partially penetrant and subtle complex phenotypes, as an animal must simultaneously carry the extrachromosomal array and be one of the members of the population that displays the partially penetrant phenotype that can often be difficult to score.
The human gene replacement approach described here allows for generation of ortholog deletion alleles directly in any wild-type or mutant strain amenable to transgenesis using the same reagents designed for replacement, thereby reducing the confounding effects of background mutations on phenotype discovery. The use of excisable selectable markers that do not severely alter morphology, baseline locomotion and several evoked sensory behaviors further simplifies phenotypic analysis and removes the need for any specialized genetic backgrounds (Norris et al., 2015; Figs 4-6; Fig. S1). Using this approach in combination with machine vision, we provide two in vivo functional assays for human PTEN, NaCl chemotaxis and mechanosensory responsivity. In particular, NaCl chemotaxis possesses several characteristics that make it an ideal functional assay: (1) a large functional range between deletion and human gene rescue to discern potentially subtle functional differences among missense variants; (2) scalability, as many plates can be run simultaneously; and (3) straightforward analysis using an automatically calculated preference index (alternatively, many laboratories score chemotaxis manually by simple blinded counting). Importantly, these reagents and functional assays can now be used in precision medicine drug screens aimed at identifying compounds that counteract the effect of a particular patient's missense variant. Further broad-scale phenomic characterization of targeted knockouts and mutant libraries, combined with new databases that curate ortholog functional annotation across model organisms, should expedite the process of in vivo functional assay development (Lee et al., 2018a; McDiarmid et al., 2018; Thompson et al., 2013; Wang et al., 2017b; Yemini et al., 2013).
Further applications of targeted human gene replacement
One of the goals of this article is to illustrate how Cas9-triggered homologous recombination can be adapted to directly replace C. elegans genes with human genes. An exciting adaptation of this approach will be to combine targeted human gene insertions with bi-partite systems for precise spatial-temporal control of human transgene expression, as has recently been done to overexpress human genes as UAS-complementary DNA (cDNA) constructs in Drosophila (Luo et al., 2017; Lee et al., 2018b; Wangler et al., 2017). The recent and long-awaited development of the cGAL4-UAS system should allow a similar approach to be developed for C. elegans, currently the only organism for which the complete cell lineage and neuronal wiring diagram is known (Sulston and Horvitz, 1977; Sulston et al., 1983; Wang et al., 2016; White et al., 1986).
While one of the clearest uses for targeted replacement is precise structure-function analysis of variants of uncertain significance, there are several exciting applications beyond modeling disease-associated alleles. Targeted human gene replacement is also particularly well suited for investigation of the evolutionary principles that determine the replaceability of genes. By allowing for human gene expression immediately upon genomic integration, this approach should also be adaptable to essential genes. A homozygous integrant will only be obtained if the human gene can substitute for the orthologous essential gene, creating a complementation test out of the transgenesis process. This will allow for systematic and precise interrogation of the sequence characteristics and functional properties required for successful human gene replacement (Kachroo et al., 2015). A library of humanized worms would also open the door to the rich resources of tools available to visualize and manipulate human genes and proteins that are often unavailable for C. elegans researchers (e.g. high-quality antibodies, biochemically characterized or known pathogenic control variants, and experimentally determined crystal structures, Figs 2 and 7; Berman et al., 2000). Given the throughput that has been achieved for reporter gene analysis (several thousand genes) and genome editing in C. elegans, it should be possible to generate a humanized C. elegans library of similar size to those recently created in yeast (Dickinson and Goldstein, 2016; Dupuy et al., 2007; Hamza et al., 2015; Hunt-Newbury et al., 2007; Kachroo et al., 2015; Norris et al., 2017; Sun et al., 2016; Yang et al., 2017). Integrated transgenes would offer the possibility of humanizing entire cellular processes for detailed in vivo analysis in a relatively complex, yet tractable, metazoan with increasingly powerful tools for spatial-temporal control of transgene expression and protein degradation (Armenti et al., 2014; Wang et al., 2016, 2017a; Zhang et al., 2015).
Deep mutational scanning and related technologies have recently made it feasible to characterize the functional effects of virtually every possible amino acid change of a protein on a particular cellular phenotype (Fowler and Fields, 2014; Fowler et al., 2010). Several of such exhaustive sequence-function maps have been recently been generated in yeast and human cell culture systems (Findlay et al., 2014, 2018; Majithia et al., 2016; Matreyek et al., 2018; Mighell et al., 2018; Weile et al., 2017). These tools offer amazing resources that serve as ‘lookup tables’ of functional missense variation in human genes, to enable experimentally confirmed variant interpretation immediately upon first clinical presentation (Starita et al., 2017; Weile et al., 2017). An ambitious and exciting goal for the C. elegans community will be to further streamline genome engineering and high-throughput phenotyping to achieve the first comprehensive sequence-function map in a metazoan.
MATERIALS AND METHODS
Strains and culture
Worms were cultured on nematode growth medium (NGM) seeded with E. coli (OP50) as described previously (Brenner, 1974). N2 Bristol and CB1375 daf-18(e1375) strains were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA). daf-18(e1375) harbors a 30 bp insertion in the fourth exon and is predicted to insert six amino acids before introducing an early stop codon that truncates the C-terminal half of the protein while leaving the phosphatase domain intact (Ogg and Ruvkun, 1998).
The following strains were created for this work:
VG674, daf-18(e1375); yvEx674[paex-3::PTEN::unc-54; pmyo-2::mCherry::unc-54 UTR];
VG810-813, daf-18(e1375); yvEx810-813[paex-3::PTEN::unc-54 UTR; pmyo-2::mCherry::unc-54 UTR];
VG712, daf-18(yv3[daf-18p::PTEN+LoxP pmyo-2::GFP::unc-54 UTR prps-27::NeoR::unc-54 UTR LoxP+daf-18 UTR]);
VG713, daf-18(yv4[daf-18p::PTEN+LoxP pmyo-2::GFP::unc-54 UTR prps-27::NeoR::unc-54 UTR LoxP+daf-18 UTR]);
VG714, daf-18(yv5[daf-18p::PTEN+LoxP+daf-18 UTR]);
VG715, daf-18(yv5[daf-18p::PTEN+LoxP+daf-18 UTR]);
VG817, daf-18(yv7[daf-18p::GFP::T2A::PTEN::unc-54 UTR+LoxP pmyo-2::GFP::unc-54 UTR prps-27::NeoR::unc-54 UTR LoxP+daf-18 UTR]);
VG818, daf-18(yv8[daf-18p::GFP::T2A::PTEN::unc-54 UTR+LoxP pmyo-2::GFP::unc-54 UTR prps-27::NeoR::unc-54 UTR LoxP+daf-18 UTR]);
VG867, daf-18(yv14[daf-18p::GFP::T2A::PTEN-G129E::unc-54 UTR+LoxP pmyo-2::GFP::unc-54 UTR prps-27::NeoR::unc-54 UTR LoxP+daf-18 UTR]).
Strain and plasmid generation
The reference PTEN CDS (UniProt consensus, identifier P60484-1) was obtained from a pCMV-PTEN plasmid (Addgene plasmid #28298) and cloned into a TOPO gateway entry clone (Invitrogen), according to the manufacturer’s instructions. The PTEN entry clone was recombined with an pDEST-aex-3p destination vector (obtained from Dr Hidehito Kuroyanagi; Kuroyanagi et al., 2010) to generate the aex-3p::PTEN::unc-54 UTR rescue construct using gateway cloning (Invitrogen), according to the manufacturer’s instructions.
The Moerman laboratory guide selection tool (http://genome.sfu.ca/crispr/) was used to identify the daf-18-targeting sgRNA. The daf-18 sgRNA sequence GGAGGAGGAGTAACCATTGG was cloned into the pU6::klp-12 sgRNA vector (obtained from the Calarco laboratory, University of Toronto, Ontario, Canada) using site-directed mutagenesis and used for all editing experiments. The daf-18p::PTEN CDS and daf-18p::GFP::T2A::PTEN::unc-54 UTR upstream homology arms were synthesized by Integrated DNA Technologies and cloned into the loxP_myo2_neoR repair construct (obtained from the Calarco laboratory) using Gibson Assembly.
C. elegans wild-type N2 strain was used for all CRISPR-Cas9 editing experiments. Genome edits were created as previously described (Norris et al., 2015). In brief, plasmids encoding sgRNA, Cas9 co-transformation markers pCFJ90 and pCFJ104 (Jorgensen laboratory, Addgene) and the selection cassette flanked by homology arms (∼500 bp) containing PTEN were injected into wild-type worms. Animals containing the desired insertions were identified by G418 resistance, loss of extrachromosomal array markers and uniform dim fluorescence of the inserted GFP.
Genotype confirmation
Correct replacement of the daf-18 ORF with human PTEN was confirmed by amplifying the two regions spanning the upstream and downstream insertion borders using PCR followed by Sanger sequencing (primer binding locations depicted in Fig. 1). The genotyping strategy is essentially as described for deletion allele generation via DMS cassette insertion in Norris et al. (2015).
The forward and reverse primers used to amplify the upstream insertion region were 5′-TGCCGTTTGAATTAGCGTGC-3′ (located within the daf-18 genomic promoter region) and 5′-CCCTCAATGTCTCTACTTGT-3′ (located within the myo-2 promoter of the selection cassette), respectively.
The forward and reverse primers used to amplify the downstream insertion region were 5′-TTCCTCGTGCTTTACGGTATCG-3′ (located within the Neomycin resistance gene) and 5′-CTCAACACGTTCGGAGGGTAAA-3′ (located downstream of the daf-18 genomic coding region), respectively.
Following cassette excision via injection of Cre recombinase, the daf-18 promoter (5′-TGCCGTTTGAATTAGCGTGC-3′) and daf-18 downstream (5′-CTCAACACGTTCGGAGGGTAAA-3′) primers were used to amplify human PTEN and confirm error-free insertion at the daf-18 locus via Sanger sequencing (Fig. 1).
RNA extraction, library preparation and cDNA amplification
Total RNA was isolated from mixed-stage VG714 and VG715 PTEN knock-in animals using a GeneJET RNA Purification Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Total RNA was treated with DNase (New England Biolabs) and purified with an RNeasy MinElute spin column (Qiagen), according to the manufacturer’s instructions. cDNA libraries were prepared from crude and purified total RNA using Superscript III (Invitrogen). All genes were amplified from cDNA libraries with Platinum Taq DNA Polymerase (Thermo Fisher Scientific) and gene-specific primer sets.
The forward and reverse primers used to amplify the PTEN CDS were 5′-ATGACAGCCATCATCAAAGA-3′ and 5′-TCAGACTTTTGTAATTTGTG-3′, respectively. The forward and reverse cmk-1 intronic control primers (Ardiel et al., 2018) were 5′-AGGGTAGGCTAGAGTCTGGGATAGAT-3′ and 5′-ACGACTCCGTTGTCGTGCATAAAC-3′, respectively.
Protein structure modeling and visualization
The PTEN 1D5R reference structure (Berman et al., 2000; Lee et al., 1999) was visualized using PyMOL software (DeLano, 2002). Structural models for full-length human PTEN, DAF-18 53-506 and full-length DAF-18 were predicted using Phyre2 (Kelley et al., 2015) and visualized using PyMOL.
NaCl chemotaxis behavioral assays
The chemotaxis behavioral assay was conducted on a 6-cm assay plate (2% agar), in which a salt gradient was formed overnight by inserting a 2% agar plug containing 50 mM NaCl (∼5 mm in diameter) 1 cm from the edge of the plate. A control 2% agar plug without NaCl was inserted 1 cm from the opposite edge of the plate. Strains were grown on NGM plates seeded with E. coli (OP50) for 3 or 4 days. Worms on the plates were collected and washed three times using M9 buffer, before being pipetted onto an unseeded NGM plate to remove excess buffer and select animals carrying transformation markers. Adult worms were transferred and placed at the center of the assay plates and were tracked for 40 min on the Multi-Worm Tracker (Swierczek et al., 2011). After the tracking period, the chemotaxis index was calculated as (A–B)/(A+B), where A was the number of animals that were located in a 1.5-cm-wide region on the side of the assay plate containing the 2% agar plug with 50 mM NaCl, and B was the number of animals that were located in a 1.5-cm-wide region on the side of the assay plate containing the 2% agar plug without NaCl (Fig. 3B). Animals not located in either region (i.e. in the middle section of the assay plate) were not counted toward the chemotaxis index. One hundred to two hundred animals were used per plate, and two or three plate replicates were used for each line in each experiment. Any statistical comparisons were carried out on plates assayed concurrently (i.e. on the same day).
Mechanosensory habituation behavioral assays
Worms were synchronized for behavioral testing on Petri plates containing NGM seeded with 50 µl OP50 liquid culture 12-24 h before use. Five gravid adults were picked to plates and allowed to lay eggs for 3-4 h before removal. The animals were maintained in a 20°C incubator for 72 h. Plates of worms were placed into the tapping apparatus and, after a 100 s acclimatization period, 30 taps were administered at a 10 s interstimulus interval (ISI). Comparisons of ‘final response’ comprised the average of the final three stimuli. Any statistical comparisons were carried out on plates assayed concurrently (i.e. on the same day).
Multi-Worm Tracker behavioral analysis and statistics
Multi-Worm Tracker software (version 1.2.0.2) was used for stimulus delivery and image acquisition (Swierczek et al., 2011). Behavioral quantification with Choreography software (version 1.3.0_r103552; Swierczek et al., 2011) used ‘--shadowless’, ‘--minimum-move-body 2’ and ‘--minimum-time 20’ filters to restrict the analysis to animals that moved at least two body lengths and were tracked for at least 20 s. The MeasureReversal plugin was used to identify reversals occurring within 1 s (dt=1) of the mechanosensory stimulus onset. Custom MATLAB (MathWorks) and R (https://www.r-project.org) scripts (available on request) organized and summarized Choreography output files. Final figures were generated using GraphPad Prism version 7.00 for Mac OS X. Each experiment was independently replicated at least twice. No blinding was necessary because the Multi-Worm Tracker scores behavior objectively. Morphology metrics, baseline locomotion metrics, initial and final reversal responses, habituation difference scores or chemotaxis indices from all plates were pooled, and metrics were compared across strains with ANOVA and Tukey’s honest significant difference (HSD) tests. For all statistical tests, an alpha value of 0.05 was used to determine significance.
Supplementary Material
Acknowledgements
We thank Dr Evan L. Ardiel for useful comments and discussion regarding the manuscript; Dr Kurt Haas for motivating discussions to write the manuscript; Erica Li-Leger and the Moerman laboratory for assistance with experiments; Dr John Calarco, Dr Erik Jorgensen and Dr Hidehito Kuroyanagi and their laboratories for sharing their constructs and protocols or making them publicly available; and Warren M. Meyers and Christine R. Ackerley for useful advice and discussions regarding figure design and/or protein structural modeling. We also thank the Caenorhabditis Genetic Center for strains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.A.M., V.A., K.M., D.G.M., C.H.R.; Methodology: T.A.M., V.A., K.M., D.G.M.; Software: T.A.M.; Validation: T.A.M., V.A.; Formal analysis: T.A.M.; Investigation: T.A.M., V.A., A.D.L., J.L., K.M., D.G.M., C.H.R.; Resources: T.A.M., V.A., K.M., D.G.M., C.H.R.; Data curation: T.A.M.; Writing - original draft: T.A.M.; Writing - review & editing: T.A.M., V.A., K.M., D.G.M., C.H.R.; Visualization: T.A.M.; Supervision: K.M., D.G.M., C.H.R.; Project administration: D.G.M., C.H.R.; Funding acquisition: C.H.R.
Funding
This work was supported by the Canadian Institutes of Health Research (Doctoral Research Award to T.A.M.; MOP 130287 to C.H.R.).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.036517.supplemental
References
- Adachi T., Kunitomo H., Tomioka M., Ohno H., Okochi Y., Mori I. and Iino Y. (2010). Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans. Genetics 186, 1309-1319. 10.1534/genetics.110.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., De Smet F., Tranchevent L.-C., De Moor B., Marynen P., Hassan B. et al. (2006). Gene prioritization through genomic data fusion. Nat. Biotechnol. 24, 537-544. 10.1038/nbt1203 [DOI] [PubMed] [Google Scholar]
- Ahier A. and Jarriault S. (2014). Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics 196, 605-613. 10.1534/genetics.113.160846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardiel E. L., McDiarmid T. A., Timbers T. A., Lee K. C. Y., Safaei J., Pelech S. L. and Rankin C. H. (2018). Insights into the roles of CMK-1 and OGT-1 in interstimulus interval dependent habituation in Caenorhabditis elegans. Proc. Royal Sci. B 285, 20182084 10.1098/rspb.2018.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti S. T., Lohmer L. L., Sherwood D. R. and Nance J. (2014). Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141, 4640-4647. 10.1242/dev.115048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au V., Li-Leger E., Raymant G., Flibotte S., Chen G., Martin K., Fernando L., Doell C., Rosell F. L., Wang S. et al. (2018). CRISPR/Cas9 Methodology for the Generation of Knockout Deletions in Caenorhabditis elegans.. G3: Genes, Genomes, Genetics 8, g3-200778 10.1534/g3.118.200778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., Abecasis G. R., Altshuler D. M., Durbin R. M., Abecasis G. R., Bentley D. R., Chakravarti A., Clark A. G., Donnelly P., Eichler E. E. et al. (2015). A global reference for human genetic variation. Nature 526, 68-74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr G. G., Witt-Engerström I. and Hagberg B. (1987). Brain stem and spinal cord impairment in Rett syndrome: somatosensory and auditory evoked responses investigations. Brain Dev. 9, 517-522. 10.1016/S0387-7604(87)80076-1 [DOI] [PubMed] [Google Scholar]
- Bamshad M. J., Ng S. B., Bigham A. W., Tabor H. K., Emond M. J., Nickerson D. A. and Shendure J. (2011). Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745-755. 10.1038/nrg3031 [DOI] [PubMed] [Google Scholar]
- Baruah P. S., Beauchemin M., Parker J. A. and Bertrand R. (2017). Expression of human Bcl-xL (Ser49) and (Ser62) mutants in Caenorhabditis elegans causes germline defects and aneuploidy. PLoS ONE 12, e0177413 10.1371/journal.pone.0177413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shalom R., Keeshen C. M., Berrios K. N., An J. Y., Sanders S. J. and Bender K. J. (2017). Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol. Psychiatry 82, 224-232. 10.1016/j.biopsych.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bend E. G., Si Y., Stevenson D. A., Bayrak-Toydemir P., Newcomb T. M., Jorgensen E. M. and Swoboda K. J. (2016). NALCN channelopathies. Neurology 87, 1131-1139. 10.1212/WNL.0000000000003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N. and Bourne P. E. (2000). The protein data bank. Nucleic Acids Res. 28, 235-242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger D. A., Fukushige T., Yun S., Semple R. K., Hanover J. A. and Krause M. W. (2017). Caenorhabditis elegans DAF-2 as a model for human insulin receptoropathies. G3: Genes, Genomes, Genetics 7, 257-268. 10.1534/g3.116.037184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J. A. and Norris A. D. (2018). Synthetic genetic interaction (CRISPR-SGI) profiling in caenorhabditis elegans. Bio Protoc. 8 10.21769/BioProtoc.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P., Park K., Gonçalves J., Li C., Howard C. J., Sharpe T. D., Holt L. J., Pelletier L., Bullock A. N. and Leroux M. R. (2018). CDKL family kinases have evolved distinct structural features and ciliary function. Cell Rep. 22, 885-894. 10.1016/j.celrep.2017.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J. X., Buckingham K. J., Jhangiani S. N., Boehm C., Sobreira N., Smith J. D., Harrell T. M., McMillin M. J., Wiszniewski W., Gambin T. et al. (2015). The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 97, 199-215. 10.1016/j.ajhg.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). The PyMOL molecular graphics system. http://pymol.org.
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J. and Church G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336-4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J. and Goldstein B. (2016). CRISPR-based methods for caenorhabditis elegans genome engineering. Genetics 202, 885-901. 10.1534/genetics.115.182162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J. and Goldstein B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D. and Goldstein B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A. and Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096-1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Dunham M. J. and Fowler D. M. (2013). Contemporary, yeast-based approaches to understanding human genetic variation. Curr. Opin. Genet. Dev. 23, 658-664. 10.1016/j.gde.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D., Bertin N., Hidalgo C. A., Venkatesan K., Tu D., Lee D., Rosenberg J., Svrzikapa N., Blanc A., Carnec A. et al. (2007). Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25, 663-668. 10.1038/nbt1305 [DOI] [PubMed] [Google Scholar]
- Findlay G. M., Boyle E. A., Hause R. J., Klein J. C. and Shendure J. (2014). Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120-123. 10.1038/nature13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay G. M., Daza R. M., Martin B., Zhang M. D., Leith A. P., Gasperini M., Janizek J. D., Huang X., Starita L. M. and Shendure J. (2018). Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217-222. 10.1038/s41586-018-0461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M. and Fields S. (2014). Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801-807. 10.1038/nmeth.3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Fleishman S. J., Kellogg E. H., Stephany J. J., Baker D. and Fields S. (2010). High-resolution mapping of protein sequence-function relationships. Nat. Methods 7, 741-746. 10.1038/nmeth.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M. and Calarco J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741-743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Wayne Davis M., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S.-P., Grunnet M. and Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M. and Jorgensen E. M. (2012). Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117-118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., LaBella M., Pozniakovsky A., Moerman D. G. and Jorgensen E. M. (2014). Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11, 529-534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl W. A., Mulvihill J. J., Toro C., Markello T. C., Wise A. L., Ramoni R. B., Adams D. R. and Tifft C. J. (2016). The NIH undiagnosed diseases program and network: applications to modern medicine. Mol. Genet. Metab. 117, 393-400. 10.1016/j.ymgme.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind D. H. and Flint J. (2015). Genetics and genomics of psychiatric disease. Science 349, 1489-1494. 10.1126/science.aaa8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C., Lupski J. R. and Gibbs R. A. (2012). Human genome sequencing in health and disease. Annu. Rev. Med. 63, 35-61. 10.1146/annurev-med-051010-162644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., Wildonger J. and O'Connor-Giles K. M. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029-1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. G., Azencott C.-A., Aicheler F., Gieraths U., MacArthur D. G., Samocha K. E., Cooper D. N., Stenson P. D., Daly M. J., Smoller J. W. et al. (2015). The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 36, 513-523. 10.1002/humu.22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza A., Tammpere E., Kofoed M., Keong C., Chiang J., Giaever G., Nislow C. and Hieter P. (2015). Complementation of yeast genes with human genes as an experimental platform for functional testing of human genetic variants. Genetics 201, 1263-1274. 10.1534/genetics.115.181099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert J. A., Embacher R., Mester J. L., Frazier T. W. and Eng C. (2014). Biochemical screening and PTEN mutation analysis in individuals with autism spectrum disorders and macrocephaly. Eur. J. Hum. Genet. 22, 273-276. 10.1038/ejhg.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A. et al. (2007). High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5, e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A. and Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E. and Doudna J. (2013). RNA-programmed genome editing in human cells. Elife 2, e00471 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A. H., Laurent J. M., Yellman C. M., Meyer A. G., Wilke C. O. and Marcotte E. M. (2015). Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921-925. 10.1126/science.aaa0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T. and Hengartner M. O. (2006). Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5, 387-399. 10.1038/nrd2031 [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N. and Sternberg M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845-858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K. and Fire A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O'Roak B. J., Cooper G. M. and Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310-315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., Ohno G., Sakane H., Maruoka H. and Hagiwara M. (2010). Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat. Protoc. 5, 1495-1517. 10.1038/nprot.2010.107 [DOI] [PubMed] [Google Scholar]
- Lai C.-H., Chou C. Y., Ch'ang L. Y., Liu C. S. and Lin W. (2000). Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10, 703-713. 10.1101/gr.10.5.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Riley G. R., Jang W., Rubinstein W. S., Church D. M. and Maglott D. R. (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980-D985. 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-O., Yang H., Georgescu M.-M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P. and Pavletich N. P. (1999). Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323-334. 10.1016/S0092-8674(00)81663-3 [DOI] [PubMed] [Google Scholar]
- Lee R. Y. N., Howe K. L., Harris T. W., Arnaboldi V., Cain S., Chan J., Chen W. J., Davis P., Gao S., Grove C. et al. (2018a). WormBase 2017: molting into a new stage. Nucleic Acids Res. 46, D869-D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.-T., Zirin J., Kanca O., Lin W.-W., Schulze K. L., Li-Kroeger D., Tao R., Devereaux C., Hu Y., Chung V. et al. (2018b). A gene-specificT2A-GAL4library for Drosophila. Elife 7, e35575 10.7554/eLife.35574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. (2013). Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 14, 168-178. 10.1038/nrg3404 [DOI] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B. et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285-291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D. and Greenwald I. (1995). Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature 377, 351-354. 10.1038/377351a0 [DOI] [PubMed] [Google Scholar]
- Levitan D., Doyle T. G., Brousseau D., Lee M. K., Thinakaran G., Slunt H. H., Sisodia S. S. and Greenwald I. (1996). Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 93, 14940-14944. 10.1073/pnas.93.25.14940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R. et al. (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943-1947. 10.1126/science.275.5308.1943 [DOI] [PubMed] [Google Scholar]
- Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., Li Y., Gao N., Wang L., Lu X. et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681-683. 10.1038/nbt.2661 [DOI] [PubMed] [Google Scholar]
- Liaw D., Marsh D. J., Li J., Dahia P. L. M., Wang S. I., Zheng Z., Bose S., Call K. M., Tsou H. C., Peacoke M. et al. (1997). Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16, 64-67. 10.1038/ng0597-64 [DOI] [PubMed] [Google Scholar]
- Lim C.-S., Kang X., Mirabella V., Zhang H., Bu Q., Araki Y., Hoang E. T., Wang S., Shen Y., Choi S. et al. (2017). BRaf signaling principles unveiled by large-scale human mutation analysis with a rapid lentivirus-based gene replacement method. Genes Dev. 31, 537-552. 10.1101/gad.294413.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Chin-Sang I. D. (2015). C. elegans as a model to study PTEN's regulation and function. Methods 77-78, 180-190. 10.1016/j.ymeth.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Liu J., Visser-Grieve S., Boudreau J., Yeung B., Lo S., Chamberlain G., Yu F., Sun T., Papanicolaou T., Lam A. et al. (2014). Insulin activates the insulin receptor to downregulate the PTEN tumour suppressor. Oncogene33, 3878-3885 10.1038/onc.2013.347 [DOI] [PubMed] [Google Scholar]
- López-Bigas N. and Ouzounis C. A. (2004). Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 32, 3108-3114. 10.1093/nar/gkh605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Rosenfeld J. A., Yamamoto S., Harel T., Zuo Z., Hall M., Wierenga K. J., Pastore M. T., Bartholomew D., Delgado M. R. et al. (2017). Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLoS Genet. 13, e1006905 10.1371/journal.pgen.1006905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T. and Dixon J. E. (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375-13378. 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Majithia A. R., Tsuda B., Agostini M., Gnanapradeepan K., Rice R., Peloso G., Patel K. A., Zhang X., Broekema M. F., Patterson N. et al. (2016). Prospective functional classification of all possible missense variants in PPARG. Nat. Genet. 48, 1570-1575. 10.1038/ng.3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Fowler D. M., Starita L. M., Haendel M. A., MacArthur D. G., Biesecker L. G., Worthey E., Chisholm R. L., Green E. D., Jacob H. J. et al. (2017). Bedside back to bench: building bridges between basic and clinical genomic research. Cell 169, 6-12. 10.1016/j.cell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek K. A., Starita L. M., Stephany J. J., Martin B., Chiasson M. A., Gray V. E., Kircher M., Khechaduri A., Dines J. N., Hause R. J. et al. (2018). Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 50, 874-882. 10.1038/s41588-018-0122-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K. L., Varga E. A., Pastore M. T., Prior T. W., Manickam K., Atkin J. F. and Herman G. E. (2010). Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 3, 137-141. 10.1002/aur.132 [DOI] [PubMed] [Google Scholar]
- McDiarmid T. A., Bernardos A. C. and Rankin C. H. (2017). Habituation is altered in neuropsychiatric disorders-A comprehensive review with recommendations for experimental design and analysis. Neurosci. Biobehav. Rev. 80, 286-305. 10.1016/j.neubiorev.2017.05.028 [DOI] [PubMed] [Google Scholar]
- McDiarmid T. A., Yu A. J. and Rankin C. H. (2018). Beyond the response-High throughput behavioral analyses to link genome to phenome in Caenorhabditis elegans. Genes, Brain Behav. e12437 10.1111/gbb.12437 [DOI] [PubMed] [Google Scholar]
- Merritt C. and Seydoux G. (2010). Transgenic solutions for the germline. WormBook 1-21. 10.1895/wormbook.1.148.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M. L. (2010). Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31-46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- Mighell T. L., Evans-Dutson S. and O'Roak B. J. (2018). A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotypes relationships. Am. J. Hum. Genet. 102, 943-955. 10.1016/j.ajhg.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova V. T., Borland C. Z., Manjarrez L., Stern M. J. and Sun H. (1999). The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. USA 96, 7427-7432. 10.1073/pnas.96.13.7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miosge L. A., Field M. A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S. et al. (2015). Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. USA 112, E5189-E5198. 10.1073/pnas.1511585112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H. and Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Myers M. P., Stolarov J. P., Eng C., Li J., Wang S. I., Wigler M. H., Parsons R. and Tonks N. K. (1997). P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc. Natl. Acad. Sci. USA 94, 9052-9057. 10.1073/pnas.94.17.9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. P., Pass I., Batty I. H., Van der Kaay J., Stolarov J. P., Hemmings B. A., Wigler M. H., Downes C. P. and Tonks N. K. (1998). The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA 95, 13513-13518. 10.1073/pnas.95.23.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T. et al. (2014). Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5, 5560 10.1038/ncomms6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need A. C., Shashi V., Hitomi Y., Schoch K., Shianna K. V., McDonald M. T., Meisler M. H. and Goldstein D. B. (2012). Clinical application of exome sequencing in undiagnosed genetic conditions. J. Med. Genet. 49, 353-361. 10.1136/jmedgenet-2012-100819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. B., Buckingham K. J., Lee C., Bigham A. W., Tabor H. K., Dent K. M., Huff C. D., Shannon P. T., Jabs E. W., Nickerson D. A. et al. (2010). Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42, 30-35. 10.1038/ng.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Kim H.-M., Colaiácovo M. P. and Calarco J. A. (2015). Efficient genome editing in caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201 10.1534/genetics.115.180679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Gracida X. and Calarco J. A. (2017). CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. Elife 6, e28129 10.7554/eLife.28129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S. and Ruvkun G. (1998). The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2, 887-893. 10.1016/S1097-2765(00)80303-2 [DOI] [PubMed] [Google Scholar]
- Orefice L. L., Zimmerman A. L., Chirila A. M., Sleboda S. J., Head J. P. and Ginty D. D. (2016). Peripheral mechanosensory neuron dysfunction underlies tactile and behavioral deficits in mouse models of ASDs. Cell 166, 299-313. 10.1016/j.cell.2016.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak B. J., Vives L., Fu W., Egertson J. D., Stanaway I. B., Phelps I. G., Carvill G., Kumar A., Lee C., Ankenman K. et al. (2012). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619-1622. 10.1126/science.1227764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrico A., Galli L., Buoni S., Orsi A., Vonella G. and Sorrentino V. (2009). Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin. Genet. 75, 195-198. 10.1111/j.1399-0004.2008.01074.x [DOI] [PubMed] [Google Scholar]
- Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E. and Donner D. B. (2001). A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA 98, 4640-4645. 10.1073/pnas.051042298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Stewart M. D., Gulsuner S., Walsh T., Dhall A., McClellan J. M., Klevit R. E. and King M.-C. (2018). De novo mutation inRING1with epigenetic effects on neurodevelopment. Proc. Natl. Acad. Sci. USA 115, 1558-1563. 10.1073/pnas.1721290115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior H., Jawad A. K., MacConnachie L. and Beg A. A. (2017). Highly efficient, rapid and Co-CRISPR independent genome editing in Caenorhabditis elegans. G3 (Bethesda) 7, 3693-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R., Sawin E. R., Trent C. and Horvitz H. R. (2001). Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J. Neurosci. 21, 5871-5884. 10.1523/JNEUROSCI.21-16-05871.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin C. H., Beck C. D. O. and Chiba C. M. (1990). Caenorhabditis elegans: a new model system for the study of learning and memory. Behav. Brain Res. 37, 89-92. 10.1016/0166-4328(90)90074-O [DOI] [PubMed] [Google Scholar]
- Rankin C. H., Abrams T., Barry R. J., Bhatnagar S., Clayton D. F., Colombo J., Coppola G., Geyer M. A., Glanzman D. L., Marsland S. et al. (2009). Habituation revisited: an updated and revised description of the behavioral characteristics of habituation. Neurobiol. Learn. Mem. 92, 135-138. 10.1016/j.nlm.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W. W., Hegde M., Lyon E., Spector E. et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405-423. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., He X., Willsey A. J., Ercan-Sencicek A. G., Samocha K. E., Cicek A. E., Murtha M. T., Bal V. H., Bishop S. L., Dong S. et al. (2015). Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215-1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D. and Greenwald I. (2011). OrthoList: a compendium of c. elegans genes with human orthologs. PLoS ONE 6, e20085 10.1371/journal.pone.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F., Bourbon-Piffaut A., Masse I., Payrastre B., Chan A. M.-L. and Billaud M. (2005). The human tumour suppressor PTEN regulates longevity and dauer formation in Caenorhabditis elegans. Oncogene 24, 20-27. 10.1038/sj.onc.1207978 [DOI] [PubMed] [Google Scholar]
- Sorkaç A., Alcantara I. C. and Hart A. C. (2016). In vivo modelling of ATP1A3 G316S-induced ataxia in c. elegans using crispr/cas9-mediated homologous recombination reveals dominant loss of function defects. PLoS ONE 11, e0167963 10.1371/journal.pone.0167963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Ahituv N., Dunham M. J., Kitzman J. O., Roth F. P., Seelig G., Shendure J. and Fowler D. M. (2017). Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 101, 315-325. 10.1016/j.ajhg.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman H. A. F., Xiong B., Coe B. P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N. et al. (2017). Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515-526. 10.1038/ng.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E. and Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G. and Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C. L., Bansal P., Sahni N., Yi S. et al. (2016). An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 26, 670-680. 10.1101/gr.192526.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczek N. A., Giles A. C., Rankin C. H. and Kerr R. A. (2011). High-throughput behavioral analysis in C. elegans. Nat. Methods 8, 592-598. 10.1038/nmeth.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M., Gu J., Matsumoto K., Aota S., Parsons R. and Yamada K. M. (1998). Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 280, 1614-1617. 10.1126/science.280.5369.1614 [DOI] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., Adair R., Au V., Chaudhry I., Fernando L., Hutter H. et al. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749-1762. 10.1101/gr.157651.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R. and Iino Y. (2006). The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51, 613-625. 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Troulinaki K., Büttner S., Marsal Cots A., Maida S., Meyer K., Bertan F., Gioran A., Piazzesi A., Fornarelli A., Nicotera P. et al. (2018). WAH-1/AIF regulates mitochondrial oxidative phosphorylation in the nematodeCaenorhabditis elegans. Cell Death Discov. 4, 2 10.1038/s41420-017-0005-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M., Nijhof B., Oortveld M. A. W. and Schenck A. (2014). Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci. Biobehav. Rev. 46, 326-342. 10.1016/j.neubiorev.2014.01.013 [DOI] [PubMed] [Google Scholar]
- Varga E. A., Pastore M., Prior T., Herman G. E. and McBride K. L. (2009). The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 11, 111-117. 10.1097/GIM.0b013e31818fd762 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu J., Gharib S., Chai C. M., Schwarz E. M., Pokala N. and Sternberg P. W. (2016). cGAL, a temperature-robust GAL4–UAS system for Caenorhabditis elegans. Nat. Methods 14, 145-148. 10.1038/nmeth.4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tang N. H., Lara-Gonzalez P., Zhao Z., Cheerambathur D. K., Prevo B., Chisholm A. D., Desai A. and Oegema K. (2017a). A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development 144, 2694-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Al-Ouran R., Hu Y., Kim S.-Y., Wan Y.-W., Wangler M. F., Yamamoto S., Chao H.-T., Comjean A., Mohr S. E. et al. (2017b). MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 100, 843-853. 10.1016/j.ajhg.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M. F., Yamamoto S., Chao H.-T., Posey J. E., Westerfield M., Postlethwait J., Hieter P., Boycott K. M., Campeau P. M., Bellen H. J. et al. (2017). Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207, 9-27. 10.1534/genetics.117.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. (1973). Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. USA 70, 817-821. 10.1073/pnas.70.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weile J., Sun S., Cote A. G., Knapp J., Verby M., Mellor J. C., Wu Y., Pons C., Wong C., van Lieshout N. et al. (2017). A framework for exhaustively mapping functional missense variants. Mol. Syst. Biol. 13, 957 10.15252/msb.20177908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N. and Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 314, 1-340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Yang F., Sun S., Tan G., Costanzo M., Hill D. E., Vidal M., Andrews B. J., Boone C. and Roth F. P. (2017). Identifying pathogenicity of human variants via paralog-based yeast complementation. PLoS Genet. 13, e1006779 10.1371/journal.pgen.1006779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E., Jucikas T., Grundy L. J., Brown A. E. X. and Schafer W. R. (2013). A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods 10, 877-879. 10.1038/nmeth.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z. and Dernburg A. F. (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374-4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.