Abstract
Mitochondrial respiratory deficiencies have been observed in numerous neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases. For decades, these reductions in oxidative phosphorylation (OxPhos) have been presumed to trigger an overall bioenergetic crisis in the neuron, resulting in cell death. While the connection between respiratory defects and neuronal death has never been proven, this hypothesis has been supported by the detection of nonspecific mitochondrial DNA mutations in these disorders. These findings led to the notion that mitochondrial respiratory defects could be initiators of these common neurodegenerative disorders, instead of being consequences of a prior insult, a theory we believe to be misconstrued. Herein, we review the roots of this mitochondrial hypothesis and offer a new perspective wherein mitochondria are analyzed not only from the OxPhos point of view, but also as a complex organelle residing at the epicenter of many metabolic pathways.
Introduction
The accumulation of mutations in mitochondrial DNA (mtDNA) and subsequent reductions in mitochondrial oxidative phosphorylation (OxPhos) are thought to occur during the course of neurodegenerative disorders such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (1). Since we are often looking for the answer where we see clues, it should not come as a surprise that over the years a large body of literature has led to the controversial, but nevertheless popular, hypothesis that mitochondrial defects are instrumental in provoking neuronal death in common adult-onset neurodegenerative disorders.
When mitochondria are studied in isolation from the rest of the cellular machinery, and from the traditional bioenergetic point of view, their role in the regulation of overall cellular homeostasis is usually overlooked. Mitochondria are not only “powerhouses” that generate energy via OxPhos but also integrate numerous cellular signaling and metabolic pathways (2) that have only recently been considered in the study of disease. Alterations in these pathways can result in mitochondrial dysfunction that, in turn, can affect OxPhos output, rather than vice versa.
Thus, while the alterations in mitochondrial respirationreported in many neurological conditions (Tables 1–3) may truly reflect a primary OxPhos defect, they might also be mere bystander consequences of non–OxPhos-related cellular problems. Furthermore, detection of an OxPhos defect often leads the disorder to be labeled a mitochondrial disease that is conceptually no different from a classic OxPhos disease (see below). Yet, could it be that altered OxPhos is a metabolic feature of many disorders without necessarily being etiologically or pathogenically significant? The question, incidentally, applies not only to common neurodegenerative disorders (Table 1) such as AD and PD, but also to other neurological conditions (Tables 2 and 3) such as very common peripheral neuropathies (Table 3), including those induced by chemotherapeutic agents and HIV as well as those associated with diabetes that are reported to comprise a mitochondrial component (3). The task of deciphering the role of mitochondria in neurodegeneration is further complicated by the fact that enhancing mitochondrial respiration within compromised neurons may well be beneficial even if the actual disease mechanism is not OxPhos-dependent.
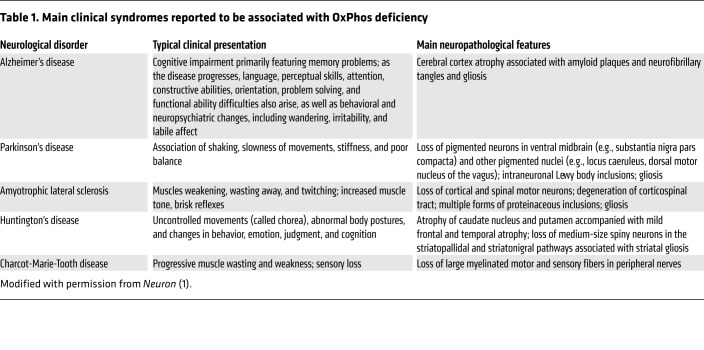
Table 1. Main clinical syndromes reported to be associated with OxPhos deficiency.
Table 3. Mitochondria-related neuropathies.
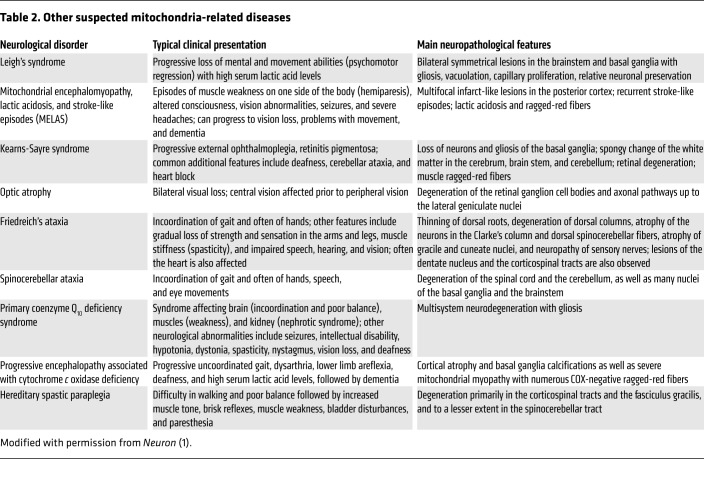
Table 2. Other suspected mitochondria-related diseases.
In this Review, we first provide a brief historical context for how the mitochondrial OxPhos hypothesis of neurodegeneration emerged. We then take the provocative position that the links between adult-onset neurodegenerative disorders and OxPhos have been misconstrued. Finally, in light of newly published data, we offer an alternative explanation as to how mitochondrial deficits may participate in the neurodegenerative process.
Reduced OxPhos and ATP production in neurological conditions
Before discussing potential links between mitochondria and neurodegeneration, we need to clarify what is an “authentic” mitochondrial disease due to OxPhos deficiency. Primary mitochondrial disorders result from mutations in nuclear DNA or mtDNA that encode subunits of the OxPhos system or factors involved in its expression and assembly (4). Many primary mitochondrial disorders present with a range of neurological manifestations, and, conversely, neurological conditions highly suggestive of being due to a mitochondrial problem typically include a relapsing-remitting pattern, with incremental worsening and partial recovery; an age at onset prior to 40 (generally during childhood); a maternal pattern of inheritance; a multisystem presentation, reflecting the involvement of different parts of the nervous system and frequently affecting other organs; and the presence of lactic acidosis in blood and cerebrospinal fluid as well as mitochondrial proliferation in skeletal muscle (ragged-red fibers). Furthermore, even if cognitive decline, parkinsonism, ataxia, or other common signs of neurodegenerative disorders occur in authentic mitochondrial diseases, they are almost never observed in isolation, in contrast to prototypical neurodegenerative disorders. In rare instances in which parkinsonism was indeed observed in patients with mtDNA mutations, atypical PD features, such as deafness and peripheral neuropathy, were also noted (5).
It is increasingly recognized that identical molecular abnormalities may give rise to heterogeneous clinical and neuropathological phenotypes, as illustrated by the tauopathies (6) and leucine-rich repeat kinase 2 (LRRK2) mutations (7). This raises the possibility that adult-onset neurodegenerative disorders might indeed be authentic mitochondrial disorders that simply do not fit into the canonical mold of mitochondrial disease depicted above. Therefore, if the clinical presentation is of little help in clarifying the link between mitochondria and neurodegeneration, can genetics be a more fruitful approach?
Efforts to find specific mtDNA alterations in PD yielded confusing results, as some groups demonstrated the existence of mtDNA mutations, including partial deletions, in patients with parkinsonism (8–13) whereas others found few or none (14–17). Sequencing of aged healthy individuals showed the accumulation of somatic mtDNA mutations, leading to the proposal that mtDNA point mutations in PD patients rendered their mitochondria susceptible to failure (18, 19). Data from other groups, however, challenged these results (20). Likewise, the quest for mtDNA candidate genes in PD revealed a significant risk associated with the A4336G mutation in mt-tRNAGlu (21), which was challenged by others (22, 23). Lastly, specific mtDNA haplotypes conferring susceptibility to PD were reported (24), but how much of a role these gene variants play in the occurrence of PD remains to be established.
A search for AD-specific mtDNA mutations was equally inconclusive (25–27). An early report identified a point mutation in the NADH dehydrogenase 2 (ND2) subunit of complex I (CI) in brain tissue of 19 AD patients (28), despite the lack of evidence for maternal inheritance in familial AD (29). These results were not replicated by others (30), nor were reports of mtDNA mutations (31, 32) in mt-tRNAGln (33) and in subunits I and II (COX I/II) of complex IV (CIV; also known as cytochrome c oxidase) (34). Also, the ND2 mutation is not present solely in AD (28), implying that it is likely a neutral polymorphism (35). Some groups found elevated levels of the relatively common mtDNA 4977-bp deletion (36) in AD brain compared with controls (37, 38), but this deletion has also been associated with aging (39) and other neurodegenerations, such as amyotrophic lateral sclerosis (ALS) (40). Similarly, reports of reduced mRNA levels for mitochondrial CI and CIV subunits in AD brains (41–44) were not correlated with the appearance of disease phenotypes (45). More recent studies have corroborated the increased presence of mtDNA partial deletions (46, 47) and point mutations (48), altered mtDNA methylation (49), and reduced expression and activity of respiratory chain complexes (50) in AD.
It may thus be concluded from the above that even if future studies unequivocally link mutations in mtDNA to neurodegenerative disorders such as AD and PD, they do not appear to be primary causes and may even reflect nonspecific changes in dying neurons or result from technical artifacts (51). Another possibility as to why findings of mtDNA mutations in neurodegenerative disorders have been inconsistent is that they may be acquired in the course of the disease as a consequence of a prior specific insult. For example, an initial error might cause defects in the replication and repair of mtDNA (52), resulting in genome instability and the appearance mtDNA defects in the course of neurodegeneration (53, 54). As in the aging process (27, 55–57), these mutations would accumulate in cells until they crossed a threshold beyond which decline in bioenergetics emerges (58, 59). Thus, given that different tissues and cells have different thresholds and compensatory mechanisms (58, 60), understanding mitochondrial defects in neurodegenerative disorders as consequences instead of as initiators would help explain not only some of the contradictory results obtained by different groups (61), but also the differences in the clinical presentations as compared with authentic OxPhos disorders (62).
Common neurodegenerative disorders and the mitochondrial hypothesis
If common neurodegenerative disorders depart clinically from authentic mitochondrial diseases, how did the hypothesis arise that a causal link might exist between adult-onset neurodegeneration and mitochondria? Historically it is likely that the first proposal of such a connection involved PD. In the early 1980s, contemporaneously with the identification of a number of mitochondrial syndromes, it was discovered that exposure to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) via illicit drug use led to an acute parkinsonian syndrome that was clinically indistinguishable from PD (63). This PD-like condition was shown to result from blockade of mitochondrial electron flow at the level of CI (64, 65). Subsequently, reports that MPTP (66) and other CI inhibitors (67–69) produced some of the features of PD in rodents strengthened the idea that CI inhibition can cause PD-like neurodegeneration. The similarity between MPTP-induced parkinsonism and PD led investigators to assess mitochondrial respiration in biospecimens from PD patients. They found significant reductions in respiratory chain activity in PD tissues (70), particularly in CI in PD brain (71, 72) and platelets (73, 74), as well as structural alterations in CI subunits (75), leading to the conclusion that deficient CI function was key to PD pathogenesis. However, other results cast doubt on this view, as some groups were unable to confirm CI deficiencies in PD muscle (76). Moreover, the chronic use of levodopa, a widely used anti-PD therapy, was found to alter OxPhos activity in rodent brains (77, 78). Finally, the cellular origin of reported reductions in CI activity was called into question, as postmortem brains from PD patients are mostly devoid of dopaminergic neurons, a main target of PD neurodegeneration (66).
Almost contemporaneously with the discovery of MPTP’s parkinsonian effect, the connection between mitochondria and AD emerged from reports of mitochondrial morphological alterations in postmortem brain sections (79, 80) and metabolic alterations in fibroblasts from patients, such as reduced glucose and deficits in calcium homeostasis (81). Given the neuronal vulnerability to these insults, it was hypothesized that mitochondrial dysfunction underlay the behavioral deficits in AD. Furthermore, cytoplasmic hybrids (cybrids) repopulated with mitochondria from AD patients reportedly displayed mitochondrial alterations (82) resembling those found in the disease (83–86). And, reported reductions in CIV activity (87) and CI and CIV deficiencies in AD platelets and brain tissue (88–90) led to the idea that CIV deficiency could be behind AD pathogenesis (91, 92). However, detractors pointed out not only that the reported CIV reductions in AD are below the threshold for dysfunction (93, 94), but that similarly small reductions in CIV subunits were observed in other neurodegenerative diseases (95), implying that the CIV deficiencies likely reflected non–AD-specific changes. Moreover, as in PD, additional studies challenged the aforementioned findings, by showing that mitochondrial respiration capacity was unaffected in AD brain and arguing that metabolic alterations in AD are unlikely to be driven by primary OxPhos deficiencies (96). Finally, exposing cell cultures, isolated mitochondria, and cells to amyloid-β oligomers — key players in AD — resulted in mitochondrial dysfunction (97), thus reversing the purported cause and effect.
Can impaired OxPhos and low ATP induce neurodegeneration?
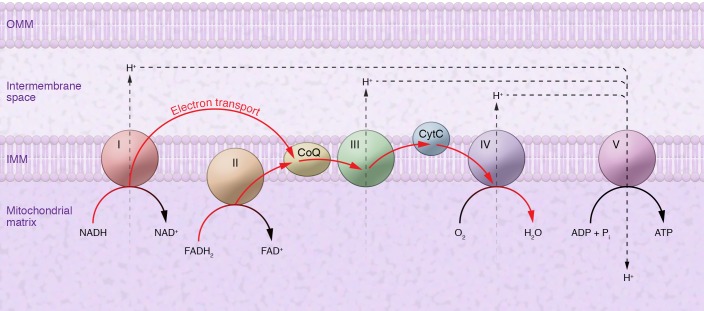
Before addressing the question of whether impaired OxPhos and low ATP can induce neurodegeneration, it is useful to summarize how mitochondrial energy is produced. From a broad view, oxidative energy metabolism comprises two elements: a respiratory chain that generates a proton gradient (derived from the NADH and FADH2 that are produced in the Krebs cycle) across the inner mitochondrial membrane (IMM), and an ATP synthase that uses this gradient to power the conversion of ADP to ATP. The respiratory chain complexes (CI, CIII, and CIV, but not CII) embedded in the IMM pump these protons “vertically” from the matrix to the intermembrane space (IMS), while transferring the NADH- and FADH2-derived electrons “horizontally” via a series of redox reactions: from CI and CII to coenzyme Q to CIII to cytochrome c to CIV and eventually to molecular oxygen (producing water) (Figure 1). The excess of H+ ions in the IMS relative to their paucity in the matrix sets up a proton gradient favoring the movement of H+ ions in the opposite direction, from the IMS to the matrix. These protons traverse the IMS through ATP synthase, which uses this “proton-motive force” to drive the conversion of ADP to ATP (98).
Figure 1. Simplified model of mitochondrial aerobic energy production (stoichiometries not implied).
The mitochondrial electron transport chain (ETC) is composed of enzymatic complexes (I–V) that transfer electrons from electron donors (NADH and FADH2) to electron acceptors embedded in the IMM via redox reactions, ultimately generating water. The ETC couples electron transport to the transfer of protons (H+) across the IMM, creating an electrochemical proton gradient that drives the synthesis of ATP by ATP synthase. OMM, outer mitochondrial membrane. CytC, cytochrome c; CoQ, coenzyme Q10.
Defects in energy synthesis can arise at many steps in this process. For example, problems in glycolysis or the Krebs cycle can reduce the production of NADH/FADH2; problems in proton pumping can reduce the strength of the gradient, reducing ATP synthesis efficiency; problems in electron flow can divert electrons from the respiratory chain to other molecules (e.g., proteins, lipids, DNA, and free oxygen [forming reactive oxygen species]); and alterations in the IMM’s permeability can allow protons to dissipate across this membrane, bypassing ATP synthase (a phenomenon called “uncoupling”), and so forth.
Moreover, mitochondria reside at the center of a complex network of metabolic pathways that can be modulated to counterbalance reductions in OxPhos to maintain cellular homeostasis. Thus, the correlation between an OxPhos defect and pathology is far from straightforward. Moreover, in any given cell, impaired mitochondria coexist with healthy mitochondria, which, above a certain threshold, will compensate for mitochondrial defects and permit a normal phenotype (99–101). This threshold, which is tissue-specific, depends on a number of factors (60). First, mitochondria, via changes in membrane lipid composition, can modulate the association of individual respiratory complexes into supercomplexes (102), thereby increasing the efficiency of ATP production, presumably by physically channeling redox substrates (e.g., coenzyme Q10 and cytochrome c), thereby mitigating the consequences of an OxPhos defect (103, 104).
Second, in most cell types mitochondria run at basal respiratory levels without using their total bioenergetic capacity (60). Only when a cell needs an additional surge of energy (e.g., because of increased activity or stress) do its mitochondria use their “reserve respiratory capacity” to increase substrate oxidation and/or ATP output (105). Thus, even in the presence of significant OxPhos defects, many cell types can avoid the consequences of reduced ATP by summoning this spare respiratory capacity. This plasticity may be the result of excess mt-mRNAs, mt-tRNAs, and/or a pool of respiratory complexes in “standby” (59) that act as a backup mechanism to compensate for OxPhos defects (60, 106).
Third, this reserve respiratory capacity can also be modulated by the different fuels feeding the Krebs cycle and OxPhos (60), in a tissue-dependent manner. For example, when faced with a general OxPhos defect, the brain can compensate so long as the diet is high in glucose (think grapes), because of its higher threshold for pyruvate utilization. However, a change to a diet high in fats and amino acids (think well-marbled steak) will not allow for such compensation, because of the brain’s low threshold for nonpyruvate substrates such as succinate (58).
If reserve capacity can compensate for a defect in mitochondrial respiration to maintain a steady level of ATP, to what extent can OxPhos activity be reduced before the ATP content is low enough to trigger cell death? While reports vary, some groups found that, depending on the tissue, reductions of 70% to 85% for CI and CIII, and approximately 60% for CIV, were necessary to produce a significant decrease in ATP production (107–110). Thus, reported reductions of 30% to 50% in CI or CIV activities in PD and AD, respectively, would not necessarily impact on ATP production significantly.
In support of this idea of reserve capacity, a T8993G mutation in mtDNA-encoded MTATP6, specifying the ATP6 subunit of ATP synthetase (complex V [CV]), results in a 50%–80% reduction in mitochondrial ATP production (111), but not in overall ATP content, as long as glucose is available (112). This mutation provokes a clinical picture that is not reminiscent of any common neurodegenerative diseases, but rather of a syndrome characterized by neuropathy, ataxia, and retinitis pigmentosa (known as NARP; ref. 113). On the other hand, at high mutation loads (>90%) this same mutation can provoke a maternally inherited form of Leigh’s syndrome (MILS) that can cause neuronal death in basal ganglia and midbrain (114, 115). Notably, the amount of ATP produced at the threshold for the NARP phenotype is only approximately 35% of the total ATP available (116, 117), implying that the remaining approximately 65% is still available for other non-OxPhos functions. Moreover, in these mitochondrial disorders, the loss of neurons observed within the affected brain areas follows a patchy, stochastic distribution (118, 119) rather than following a logic of differential susceptible subpopulations typical of neurodegenerative disorders. Indeed, in PD, nigral dopaminergic neurons die more than ventral tegmental area dopaminergic neurons (66); in ALS, motor neurons innervating fast fatiguing muscles die more than those innervating slow muscles (120); and in Huntington’s disease, striatal neurons expressing GABA and enkephalin die more than those expressing GABA and substance P (121).
Finally, mitochondria are often analyzed as an isolated subcellular fraction. For that reason, it is quite common to extrapolate specific respiratory defects in those organelles to an overall bioenergetic defect in the cell or tissue from which the mitochondrial fraction was derived. However, as noted above, mitochondria reside at the center of a complex metabolic network where multiple bidirectional mechanisms of adaptation are coregulated to maintain energy output (122). For example, the overall content of cellular ATP is the result of substrate-level phosphorylation of ADP via glycolysis in the cytoplasm and/or oxidative phosphorylation of ADP in the mitochondria. The latter is fueled mainly by the pyruvate produced during glycolysis, or to a lesser extent by fatty acid and/or amino acid oxidation (Figure 2). The rate and coupling of these pathways depend on cell type and are extremely adaptable to changes in energy demand and substrate availability (123). Under pathological conditions in which mitochondrial ATPase activity is reduced, most cells upregulate glycolysis and can completely bypass mitochondria to maintain sufficient levels of ATP (124). In fact, genetic or chemical inhibition of mitochondrial ATPase not only did not result in neurodegeneration, but actually prevented neuronal death by shifting ATP production from OxPhos to increased glycolysis (125, 126). In a second example, in ρ0 cells (cells lacking mtDNA and hence OxPhos activity), glycolytically derived ATP generated in the cytosol can travel back into the mitochondria (112) to maintain other functions, such as calcium buffering capacity and lipid synthesis.
Figure 2. Mitochondrial metabolic network.
Many metabolic pathways converge to maintain energy output. While most cells use glucose/pyruvate for ATP synthesis, oxidation of fatty acids and amino acids can also be used in response to changes in the cellular environment and availability of substrates. These mechanisms ultimately converge onto the Krebs cycle to produce NADH and FADH2, which in turn feed the mitochondrial ETC. Possible mechanisms of OxPhos deficiency in neurodegenerative diseases involving the AKT/PKB pathway and pyruvate metabolism are shown. G6P, glucose 6-phosphate.
Overall, we can conclude that, clinically speaking, neurodegenerative disorders have little resemblance to primary mitochondrial disorders. Moreover, of all known mtDNA alterations in authentic mitochondrial disease, few have been proven to result in dementia, even in long-lived patients, and none have been unambiguously linked to the development of neurodegeneration.
Possible mechanism of OxPhos deficiency in neurodegenerative disease
Despite the controversies, we cannot exclude the fact that alterations in OxPhos occur in neurodegeneration. In the course of neurodegeneration, deficits in bioenergetics do often arise and could reduce the capacity of compromised neurons to withstand the actual disease process. However, we have espoused here the view that the OxPhos abnormalities documented in neurodegenerative disorders could be nonspecific features of dying cells. But even if this is the case, the question remains of how these mitochondrial alterations arise in the first place. Although several potential mechanisms may be proposed, including defects in mitochondrial autophagy, especially in the context of PD mutations in PINK1 and PARKIN, or in mitochondrial dynamics that have been reviewed elsewhere (127, 128), herein we would like to offer two other possible scenarios.
Alterations in pyruvate metabolism: the case of PD.
Neuronal resilience to defects in glycolysis or OxPhos within a certain range is the consequence of the balanced cooperation between mitochondrial OxPhos and glycolysis (and to a lesser extent the pentose phosphate pathway) to maintain ATP levels. In the brain, metabolic flexibility depends not only on the interplay between glycolytic and OxPhos pathways and the nature of the fuel, but also on the communication between astrocytes and neurons (129). Briefly, astrocytic metabolism relies mostly on glycolysis to produce lactate to send to the neuron (129). In the neuron, lactate is converted into pyruvate (129), which then enters the mitochondria via the mitochondrial pyruvate carriers (130) to be converted into acetyl-CoA by pyruvate dehydrogenase (PDH) (131), feeding the Krebs cycle to power OxPhos.
Maintaining proper cell bioenergetics in these pathways requires coordination between glycolysis, lactate uptake, pyruvate entry into mitochondria, and OxPhos (131). One of the main orchestrators behind this coordinated effort is AKT, which, when activated, translocates to mitochondria, increasing the binding of the glycolytic enzyme hexokinase II (HKII) to the mitochondrial surface (132). The localization of HKII on mitochondrial membranes promotes both glycolysis and OxPhos (133), while activated AKT induces the expression of hexokinase and the glucose transporters (134, 135). Disruption of these pathways can alter glucose metabolism and pyruvate entry into mitochondria, reducing OxPhos (131, 136). Consequently, it is possible that during neurodegeneration, mitochondrial OxPhos deficit can be reinterpreted as a consequence of prior and sustained reductions in the production and transport of pyruvate to the mitochondria (137).
Interestingly, aberrant glucose and pyruvate metabolism has been described in AD and PD (138), and both disorders have been linked to type 2 diabetes (T2D) (139). Specifically, 60% of PD patients suffer various degrees of insulin resistance (140). Conversely, T2D patients have a 35% increased risk of developing PD (140–142) and a 65% increased risk of developing AD (143).
Early studies comparing metabolism in PD patients versus controls showed significantly lower preclinical glucose utilization (144) and reduced pyruvate oxidation in PD patient fibroblasts (145). In addition, significant elevations in pyruvate and decreases in succinate, malate, citrate, and acetate in PD patient blood suggest the presence of defects in PDH complex activity and in genes encoding PDH-interacting proteins (146). Similar reductions in glucose metabolism were also detected in animal models of PD (147–150) and in cells incubated with the neurotoxins MPTP or 6-hydroxydopamine (6-OHDA) (151). While the connections between glucose and PD had often been discussed as expected consequences of reduced CI activity, a hypothetical and early alteration in glucose metabolism would result in similar reductions in mitochondrial respiration (136).
Relevant to this possibility, dopaminergic neurons have been shown to be quite sensitive to hypoglycemic conditions, resulting in reduced tonic firing and membrane hyperpolarization (152). Moreover, in vitro glucose deprivation in neuronal cultures reduces dopaminergic uptake and promotes cytosolic aggregation of α-synuclein and dopaminergic neuron death (153). These data are also in support of reports showing that α-synuclein aggregation occurs prior to significant reductions in mitochondrial activity (154).
While the mechanism behind these alterations in glucose metabolism is unknown, the literature discusses several possibilities (155). For instance, AKT activation, a key regulator of glucose metabolism (134), has been shown to be significantly reduced in the substantia nigra of PD patients (156) and in cultured cells exposed to 6-OHDA (157). Furthermore, mutations in genes related to familial PD such as DJ-1 or PINK1 have also been linked to diminished AKT signaling (158). Remarkably, even some AKT (159) and GSK3B polymorphisms (160) are associated with a higher PD risk. Finally, another familial PD gene, PARKIN, is implicated in the regulation of the glycolysis-OxPhos connection (161) by binding and ubiquitylating pyruvate kinase M2 (PKM2) (162), decreasing its activity and glucose metabolic rate (131).
Alterations in ER-mitochondria contact sites: the case of AD.
In the last decade, much evidence has been marshalled demonstrating that mitochondria are indeed “social organelles” by means of their membrane-to-membrane interaction with other organelles. Among these, ER domains that contact mitochondria, or mitochondria-associated ER membranes (MAMs; Figure 3), have been defined as functional platforms that contain a subset of enzymatic activities involved in regulating calcium, lipid homeostasis, mitochondrial dynamics, and a number of other functions previously ascribed to isolated mitochondria (163, 164). MAM domains form lipid raft–like structures that are regulated by their lipid milieu and by the proteins embedded within them. Thus, alterations in the conformation or subcellular localization of MAM-resident proteins or in MAM-associated lipid pathways can induce alterations in MAM-regulated functions and the crosstalk between ER and mitochondria. Of special relevance to our discussion is that many proteins related to neurodegeneration, especially in AD and PD, have been shown to translocate to MAM and participate in regulating these ER domains and their communication with mitochondria.
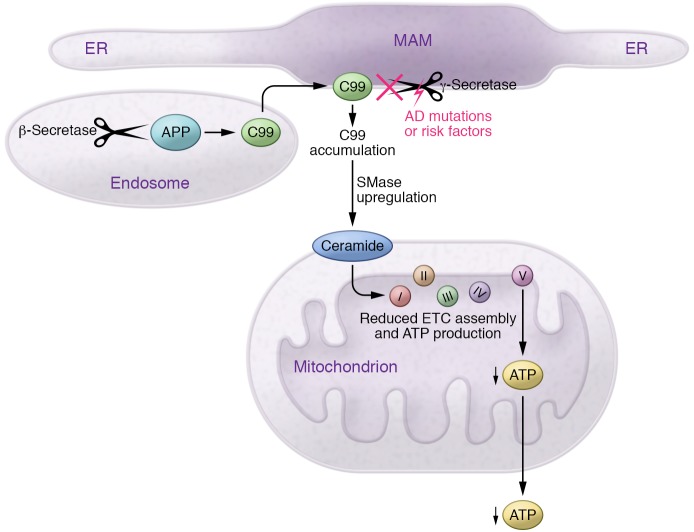
Figure 3. Mitochondria-associated membrane and AD.
MAM is a specialized, lipid raft–like subdomain of the ER that communicates with mitochondria. APP is processed first by β-secretase in endosomes to produce C99. C99 then translocates to the ER (by an unknown mechanism), where it is cleaved rapidly by MAM-localized γ-secretase. In AD, however, this process is impaired, leading to the abnormal accumulation of C99 in the MAM, which correlates with increased conversion of sphingomyelin to ceramide via an upregulation of sphingomyelinase activity. In turn, the elevated ceramide compromises ETC assembly and ATP production.
For example, we and others have shown that α-synuclein can translocate to MAM regions (165, 166), supporting the reported affinity of α-synuclein for lipid raft–like domains and its previously reported association with mitochondrial membranes. In addition, mutations in α-synuclein alter the activities localized in these domains, such as lipid metabolism (165) and calcium regulation (167).
In AD (Figure 3), presenilins, amyloid precursor protein, and γ-secretase activity were found to be enriched at MAM domains (168–170), which could explain previous reports suggesting a mitochondrial localization of these AD-related proteins (171, 172). Notably, as in PD, activities localized in MAM (including lipid homeostasis and calcium regulation) are altered in both familial and sporadic AD (173–175). These MAM-associated disturbances can impact mitochondrial biology by several mechanisms.
First, ER-mitochondria connections regulate calcium homeostasis, which is well known to be altered in AD (167, 176, 177). Among its many functions, mitochondrial calcium buffering capacity regulates the intracellular concentration of calcium, not only by buffering local changes in the cytosol, but also via highly regulated contacts with the plasma membrane and/or the ER (178, 179). Therefore, alterations in the communication between mitochondria and these organelles can significantly impact the entry of calcium into mitochondria, affecting overall calcium signaling in the cell (180, 181). Notably, calcium concentration is a regulator of rate-limiting enzymes in the Krebs cycle, and increases in calcium in the mitochondrial matrix activate the Krebs cycle and OxPhos (182). Thus, mitochondrial calcium buffering capacity not only balances intracellular calcium concentration, but also enables the coregulation of mitochondrial bioenergetics and ATP requirements in the cell. However, over a certain threshold, calcium increases in mitochondria can collapse membrane potential and induce the opening of the mitochondrial transition pore, triggering mitochondrial dysfunction and cell death (183). Taken together, these data imply that alterations in ER-mitochondria contact sites could initiate OxPhos defects through changes in calcium regulation, and likely play a significant role in AD pathogenesis.
Second, as mentioned above, MAMs are involved in the regulation of lipid homeostasis, including sphingolipid metabolism. Recent data indicate that brain tissues from AD models and AD patient–derived cells display significant increases in sphingomyelinase-mediated sphingomyelin hydrolysis within MAM domains (175). This increased sphingomyelinase activity reduced sphingomyelin content and elevated ceramide (the product of sphingomyelin hydrolysis) at ER-mitochondria contact sites and on mitochondrial membranes (175). Pharmacological reduction of ceramide levels in AD models and in AD patient cells rescued mitochondrial respiratory deficits (175), underscoring the detrimental effect of ceramides on OxPhos regulation (184, 185). These results illustrate how bioenergetic defects are plausible consequences of alterations in the regulation of sphingolipid turnover at MAM domains (185).
Several possibilities have been suggested to explain the mechanism behind sphingolipid-mediated disturbances in AD. Elevations in ceramide could alter mitochondrial membrane properties, hampering the conformation or assembly of respiratory complexes in the IMM (173) and negatively impacting respiratory rate (186). In addition, upon exceeding a threshold, ceramide can form channels in the outer mitochondrial membrane large enough to allow soluble proteins to translocate into the cytosol (187). These ceramide channels bind to, and are regulated by, pro- and antiapoptotic proteins, making this lipid an important player in the induction of apoptosis (187). We also note that increases in the local concentration of ceramide at the MAM “glue” mitochondria to tubulin via the outer membrane protein porin (also called the voltage-dependent anion channel [VDAC]) (155), which regulates mitochondrial transport, subcellular distribution, and division (188).
Third, one of the best-known activities localized to MAM regions is phospholipid synthesis and regulation (163). Alterations in MAM can affect the phospholipid milieu of mitochondrial membranes, including the signature mitochondrial lipid, cardiolipin (189, 190). Cardiolipin interacts with and regulates a number of mitochondrial proteins, including OxPhos complexes (191). In fact, this lipid is required for the proper assembly and functioning of all membrane-bound respiratory complexes and ATP synthase (192). Cardiolipin is also involved in the formation and stabilization of mitochondrial supercomplexes (193). Unsurprisingly, alterations in the concentration and/or transacylation of cardiolipin are associated with mitochondrial OxPhos defects (194) and with specific mitochondrial disorders, such as Barth syndrome (195, 196). Cardiolipin could also influence bioenergetics indirectly, including regulation of the orientation of the ADP/ATP carrier in the IMM (197) via modulation of mitochondrial creatine kinase activity (198), or even via regulation of BAX oligomeric pores on mitochondrial outer membrane (199).
Concluding remarks
In the past three decades, tremendous progress has been achieved in our understanding of mitochondrial biology in health and disease. Deciphering the mitochondrial genome provided insights into the catalog of genetic mutations linked to primary mitochondrial diseases and to the observation that the same genetic mutation may produce a similar defect, i.e., OxPhos deficiency, and yet the clinical presentation may be varied. This realization prompted investigators to embark on studies based on the following: if mitochondrial mutations (using OxPhos deficiency as a proxy) can give rise to different neurological syndromes that had no known etiology, then other neurological disorders could actually be unsuspected mitochondrial diseases. Almost serendipitously, PD and AD emerged as perfect candidates for the concept of a mitochondrial etiology, followed by other neurodegenerative disorders. In many instances, while studies found no consistent genetic basis, they documented alterations of OxPhos activity in patients as well as in animals and cellular models of human neurodegenerations.
We have argued in this Review that many of these findings, while appealing, are generally correlative and fail to demonstrate causality. We have also contended that the interpretation of OxPhos defects in neurodegeneration too often stems from studies of isolated mitochondria. These facts did not deter many investigators from regarding neurodegenerative disorders as atypical forms of mitochondrial diseases. Admittedly, even if such a view may not be accurate, and it turns out that OxPhos alterations are mere consequences of initial insults that trigger the disease, a deficit in bioenergetics can still contribute to the pathogenesis and be a valuable therapeutic target. Thus, it will be important to pursue investigations aimed at elucidating how bioenergetics can be improved. Investigations should also be encouraged to elucidate the origin of an OxPhos defect in neurodegeneration, even if it is a secondary alteration, because the most effective therapy for neurodegenerations such as AD or PD is likely going to be mechanism-specific. As such, we have proposed herein two “out-of-the-box” mechanisms in an attempt to broaden the conversation about how OxPhos arises in, and contributes to, neurodegeneration. Indeed, the lack of answers after more than 30 years and dozens of published papers should make us wonder whether the clues mentioned in the introduction are misleading us and whether it is time to rethink the search.
Acknowledgments
The authors are supported by the Henry J. and Erna D. Leir Foundation, the Art2Cure Initiative, and the Michael J. Fox Foundation (to CGL), the NIH (grants AG045335 and AG056387 to EAG, HD032062 to EAS, and NS107442-01, EB 009041, and NS099862 to SP), the US Department of Defense (W81WXWH-08-1-0465 to SP and W911F-15-1-069 to EAS), and the Marriott Mitochondrial Disorders Collaborative Research Network (to EAS).
Version 1. 01/02/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2019;129(1):34–45.
https://doi.org/10.1172/JCI120848.
Contributor Information
Estela Area-Gomez, Email: eag2118@columbia.edu.
Cristina Guardia-Laguarta, Email: cg2645@cumc.columbia.edu.
Eric A. Schon, Email: eas3@columbia.edu.
Serge Przedborski, Email: sp30@columbia.edu.
References
- 1.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70(6):1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20(7):745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cashman CR, Höke A. Mechanisms of distal axonal degeneration in peripheral neuropathies. Neurosci Lett. 2015;596:33–50. doi: 10.1016/j.neulet.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13(12):878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thyagarajan D, et al. A novel mitochondrial 12SrRNA point mutation in parkinsonism, deafness, and neuropathy. Ann Neurol. 2000;48(5):730–736. doi: 10.1002/1531-8249(200011)48:5<730::AID-ANA6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Kovacs GG. Tauopathies. Handb Clin Neurol. 2017;145:355–368. doi: 10.1016/B978-0-12-802395-2.00025-0. [DOI] [PubMed] [Google Scholar]
- 7.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Ikebe S, et al. Increase of deleted mitochondrial DNA in the striatum in Parkinson’s disease and senescence. Biochem Biophys Res Commun. 1990;170(3):1044–1048. doi: 10.1016/0006-291X(90)90497-B. [DOI] [PubMed] [Google Scholar]
- 9.Ozawa T, Tanaka M, Ikebe S, Ohno K, Kondo T, Mizuno Y. Quantitative determination of deleted mitochondrial DNA relative to normal DNA in parkinsonian striatum by a kinetic PCR analysis. Biochem Biophys Res Commun. 1990;172(2):483–489. doi: 10.1016/0006-291X(90)90698-M. [DOI] [PubMed] [Google Scholar]
- 10.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 11.Lin MT, et al. Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease. Ann Neurol. 2012;71(6):850–854. doi: 10.1002/ana.23568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker WD, Parks JK. Mitochondrial ND5 mutations in idiopathic Parkinson’s disease. Biochem Biophys Res Commun. 2005;326(3):667–669. doi: 10.1016/j.bbrc.2004.11.093. [DOI] [PubMed] [Google Scholar]
- 13.Luoma P, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364(9437):875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 14.Schapira AH, Holt IJ, Sweeney M, Harding AE, Jenner P, Marsden CD. Mitochondrial DNA analysis in Parkinson’s disease. Mov Disord. 1990;5(4):294–297. doi: 10.1002/mds.870050406. [DOI] [PubMed] [Google Scholar]
- 15.Lestienne P, Nelson J, Riederer P, Jellinger K, Reichmann H. Normal mitochondrial genome in brain from patients with Parkinson’s disease and complex I defect. J Neurochem. 1990;55(5):1810–1812. doi: 10.1111/j.1471-4159.1990.tb04973.x. [DOI] [PubMed] [Google Scholar]
- 16.Shoffner JM, Watts RL, Juncos JL, Torroni A, Wallace DC. Mitochondrial oxidative phosphorylation defects in Parkinson’s disease. Ann Neurol. 1991;30(3):332–339. doi: 10.1002/ana.410300304. [DOI] [PubMed] [Google Scholar]
- 17.Mann VM, Cooper JM, Schapira AH. Quantitation of a mitochondrial DNA deletion in Parkinson’s disease. FEBS Lett. 1992;299(3):218–222. doi: 10.1016/0014-5793(92)80118-Z. [DOI] [PubMed] [Google Scholar]
- 18.Ozawa T. Mechanism of somatic mitochondrial DNA mutations associated with age and diseases. Biochim Biophys Acta. 1995;1271(1):177–189. doi: 10.1016/0925-4439(95)00026-z. [DOI] [PubMed] [Google Scholar]
- 19.Kirches E. Do mtDNA mutations participate in the pathogenesis of sporadic parkinson’s disease? Curr Genomics. 2009;10(8):585–593. doi: 10.2174/138920209789503879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lücking CB, Kösel S, Mehraein P, Graeber MB. Absence of the mitochondrial A7237T mutation in Parkinson’s disease. Biochem Biophys Res Commun. 1995;211(2):700–704. doi: 10.1006/bbrc.1995.1868. [DOI] [PubMed] [Google Scholar]
- 21.Tan EK, Khajavi M, Thornby JI, Nagamitsu S, Jankovic J, Ashizawa T. Variability and validity of polymorphism association studies in Parkinson’s disease. Neurology. 2000;55(4):533–538. doi: 10.1212/WNL.55.4.533. [DOI] [PubMed] [Google Scholar]
- 22.Brown MD, et al. Mitochondrial DNA sequence analysis of four Alzheimer’s and Parkinson’s disease patients. Am J Med Genet. 1996;61(3):283–289. doi: 10.1002/(SICI)1096-8628(19960122)61:3<283::AID-AJMG15>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 23.Simon DK, Mayeux R, Marder K, Kowall NW, Beal MF, Johns DR. Mitochondrial DNA mutations in complex I and tRNA genes in Parkinson’s disease. Neurology. 2000;54(3):703–709. doi: 10.1212/WNL.54.3.703. [DOI] [PubMed] [Google Scholar]
- 24.van der Walt JM, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72(4):804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onyango I, Khan S, Miller B, Swerdlow R, Trimmer P, Bennett P. Mitochondrial genomic contribution to mitochondrial dysfunction in Alzheimer’s disease. J Alzheimers Dis. 2006;9(2):183–193. doi: 10.3233/JAD-2006-9210. [DOI] [PubMed] [Google Scholar]
- 26.Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101(29):10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11(2):133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 28.Lin FH, et al. Detection of point mutations in codon 331 of mitochondrial NADH dehydrogenase subunit 2 in Alzheimer’s brains. Biochem Biophys Res Commun. 1992;182(1):238–246. doi: 10.1016/S0006-291X(05)80136-6. [DOI] [PubMed] [Google Scholar]
- 29.Payami H, Hoffbuhr K. Lack of evidence for maternal effect in familial Alzheimer’s disease. Genet Epidemiol. 1993;10(6):461–464. doi: 10.1002/gepi.1370100622. [DOI] [PubMed] [Google Scholar]
- 30.Kösel S, Egensperger R, Mehraein P, Graeber MB. No association of mutations at nucleotide 5460 of mitochondrial NADH dehydrogenase with Alzheimer’s disease. Biochem Biophys Res Commun. 1994;203(2):745–749. doi: 10.1006/bbrc.1994.2245. [DOI] [PubMed] [Google Scholar]
- 31.Wragg MA, Talbot CJ, Morris JC, Lendon CL, Goate AM. No association found between Alzheimer’s disease and a mitochondrial tRNA glutamine gene variant. Neurosci Lett. 1995;201(2):107–110. doi: 10.1016/0304-3940(95)12146-3. [DOI] [PubMed] [Google Scholar]
- 32.Hutchin TP, Heath PR, Pearson RC, Sinclair AJ. Mitochondrial DNA mutations in Alzheimer’s disease. Biochem Biophys Res Commun. 1997;241(2):221–225. doi: 10.1006/bbrc.1997.7793. [DOI] [PubMed] [Google Scholar]
- 33.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92(15):6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis RE, et al. Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1997;94(9):4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Petruzzella V, Chen X, Schon EA. Is a point mutation in the mitochondrial ND2 gene associated with Alzheimer’s disease. Biochem Biophys Res Commun. 1992;186(1):491–497. doi: 10.1016/S0006-291X(05)80834-4. [DOI] [PubMed] [Google Scholar]
- 36.Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244(4902):346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- 37.Corral-Debrinski M, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23(2):471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 38.Hamblet NS, Castora FJ. Elevated levels of the Kearns-Sayre syndrome mitochondrial DNA deletion in temporal cortex of Alzheimer’s patients. Mutat Res. 1997;379(2):253–262. doi: 10.1016/S0027-5107(97)00158-9. [DOI] [PubMed] [Google Scholar]
- 39.Wei YH. Mitochondrial DNA alterations as ageing-associated molecular events. Mutat Res. 1992;275(3–6):145–155. doi: 10.1016/0921-8734(92)90019-l. [DOI] [PubMed] [Google Scholar]
- 40.Blanchard BJ, Park T, Fripp WJ, Lerman LS, Ingram VM. A mitochondrial DNA deletion in normally aging and in Alzheimer brain tissue. Neuroreport. 1993;4(6):799–802. doi: 10.1097/00001756-199306000-00051. [DOI] [PubMed] [Google Scholar]
- 41.Chandrasekaran K, Giordano T, Brady DR, Stoll J, Martin LJ, Rapoport SI. Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Brain Res Mol Brain Res. 1994;24(1–4):336–340. doi: 10.1016/0169-328x(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 42.Fukuyama R, Hatanpää K, Rapoport SI, Chandrasekaran K. Gene expression of ND4, a subunit of complex I of oxidative phosphorylation in mitochondria, is decreased in temporal cortex of brains of Alzheimer’s disease patients. Brain Res. 1996;713(1–2):290–293. doi: 10.1016/0006-8993(95)01517-5. [DOI] [PubMed] [Google Scholar]
- 43.Aksenov MY, et al. The expression of several mitochondrial and nuclear genes encoding the subunits of electron transport chain enzyme complexes, cytochrome c oxidase, and NADH dehydrogenase, in different brain regions in Alzheimer’s disease. Neurochem Res. 1999;24(6):767–774. doi: 10.1023/A:1020783614031. [DOI] [PubMed] [Google Scholar]
- 44.Simonian NA, Hyman BT. Functional alterations in Alzheimer’s disease: selective loss of mitochondrial-encoded cytochrome oxidase mRNA in the hippocampal formation. J Neuropathol Exp Neurol. 1994;53(5):508–512. doi: 10.1097/00005072-199409000-00010. [DOI] [PubMed] [Google Scholar]
- 45.Hatanpää K, Chandrasekaran K, Brady DR, Rapoport SI. No association between Alzheimer plaques and decreased levels of cytochrome oxidase subunit mRNA, a marker of neuronal energy metabolism. Brain Res Mol Brain Res. 1998;59(1):13–21. doi: 10.1016/S0169-328X(98)00117-X. [DOI] [PubMed] [Google Scholar]
- 46.Simonian NA, Hyman BT. Functional alterations in Alzheimer’s disease: selective loss of mitochondrial-encoded cytochrome oxidase mRNA in the hippocampal formation. J Neuropathol Exp Neurol. 1994;53(5):508–512. doi: 10.1097/00005072-199409000-00010. [DOI] [PubMed] [Google Scholar]
- 47.Phillips NR, Simpkins JW, Roby RK. Mitochondrial DNA deletions in Alzheimer’s brains: a review. Alzheimers Dement. 2014;10(3):393–400. doi: 10.1016/j.jalz.2013.04.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang SW, Zhang D, Chung HD, Zassenhaus HP. The frequency of point mutations in mitochondrial DNA is elevated in the Alzheimer’s brain. Biochem Biophys Res Commun. 2000;273(1):203–208. doi: 10.1006/bbrc.2000.2885. [DOI] [PubMed] [Google Scholar]
- 49.Blanch M, Mosquera JL, Ansoleaga B, Ferrer I, Barrachina M. Altered mitochondrial DNA methylation pattern in alzheimer disease-related pathology and in Parkinson disease. Am J Pathol. 2016;186(2):385–397. doi: 10.1016/j.ajpath.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 50.Liang WS, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirano M, et al. Apparent mtDNA heteroplasmy in Alzheimer’s disease patients and in normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc Natl Acad Sci U S A. 1997;94(26):14894–14899. doi: 10.1073/pnas.94.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoekstra JG, Hipp MJ, Montine TJ, Kennedy SR. Mitochondrial DNA mutations increase in early stage Alzheimer disease and are inconsistent with oxidative damage. Ann Neurol. 2016;80(2):301–306. doi: 10.1002/ana.24709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canugovi C, Shamanna RA, Croteau DL, Bohr VA. Base excision DNA repair levels in mitochondrial lysates of Alzheimer’s disease. Neurobiol Aging. 2014;35(6):1293–1300. doi: 10.1016/j.neurobiolaging.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou Y, Song H, Croteau DL, Akbari M, Bohr VA. Genome instability in Alzheimer disease. Mech Ageing Dev. 2017;161(pt A):83–94. doi: 10.1016/j.mad.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lezza AM, et al. Mitochondrial DNA 4977 bp deletion and OH8dG levels correlate in the brain of aged subjects but not Alzheimer’s disease patients. FASEB J. 1999;13(9):1083–1088. doi: 10.1096/fasebj.13.9.1083. [DOI] [PubMed] [Google Scholar]
- 56.Williams SL, Mash DC, Züchner S, Moraes CT. Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet. 2013;9(12):e1003990. doi: 10.1371/journal.pgen.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossignol R, Malgat M, Mazat JP, Letellier T. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J Biol Chem. 1999;274(47):33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 59.Grivennikova VG, Serebryanaya DV, Isakova EP, Belozerskaya TA, Vinogradov AD. The transition between active and de-activated forms of NADH:ubiquinone oxidoreductase (Complex I) in the mitochondrial membrane of Neurospora crassa. Biochem J. 2003;369(pt 3):619–626. doi: 10.1042/BJ20021165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370(pt 3):751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lunnon K, et al. Mitochondrial genes are altered in blood early in Alzheimer’s disease. Neurobiol Aging. 2017;53:36–47. doi: 10.1016/j.neurobiolaging.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 62.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348(26):2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 63.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219(4587):979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 64.Ramsay RR, Dadgar J, Trevor A, Singer TP. Energy-driven uptake of N-methyl-4-phenylpyridine by brain mitochondria mediates the neurotoxicity of MPTP. Life Sci. 1986;39(7):581–588. doi: 10.1016/0024-3205(86)90037-8. [DOI] [PubMed] [Google Scholar]
- 65.Ramsay RR, Salach JI, Dadgar J, Singer TP. Inhibition of mitochondrial NADH dehydrogenase by pyridine derivatives and its possible relation to experimental and idiopathic parkinsonism. Biochem Biophys Res Commun. 1986;135(1):269–275. doi: 10.1016/0006-291X(86)90972-1. [DOI] [PubMed] [Google Scholar]
- 66.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39(6):889–909. doi: 10.1016/S0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 67.Greenamyre JT, Sherer TB, Betarbet R, Panov AV. Complex I and Parkinson’s disease. IUBMB Life. 2001;52(3–5):135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 68.Sherer TB, et al. An in vitro model of Parkinson’s disease: linking mitochondrial impairment to altered α-synuclein metabolism and oxidative damage. J Neurosci. 2002;22(16):7006–7015. doi: 10.1523/JNEUROSCI.22-16-07006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lannuzel A, et al. The mitochondrial complex I inhibitor annonacin is toxic to mesencephalic dopaminergic neurons by impairment of energy metabolism. Neuroscience. 2003;121(2):287–296. doi: 10.1016/S0306-4522(03)00441-X. [DOI] [PubMed] [Google Scholar]
- 70.Bindoff LA, Birch-Machin M, Cartlidge NE, Parker WD, Jr , Turnbull DM. Mitochondrial function in Parkinson’s disease. Lancet. 1989;2(8653):49. doi: 10.1016/s0140-6736(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 71.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1(8649):1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 72.Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54(3):823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 73.Parker WD, Jr , Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol. 1989;26(6):719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 74.Haas RH, et al. Low platelet mitochondrial complex I and complex II/III activity in early untreated Parkinson’s disease. Ann Neurol. 1995;37(6):714–722. doi: 10.1002/ana.410370604. [DOI] [PubMed] [Google Scholar]
- 75.Mizuno Y, et al. Deficiencies in complex I subunits of the respiratory chain in Parkinson’s disease. Biochem Biophys Res Commun. 1989;163(3):1450–1455. doi: 10.1016/0006-291X(89)91141-8. [DOI] [PubMed] [Google Scholar]
- 76.Orth M, Schapira AH. Mitochondrial involvement in Parkinson’s disease. Neurochem Int. 2002;40(6):533–541. doi: 10.1016/S0197-0186(01)00124-3. [DOI] [PubMed] [Google Scholar]
- 77.Przedborski S, et al. Chronic levodopa administration alters cerebral mitochondrial respiratory chain activity. Ann Neurol. 1993;34(5):715–723. doi: 10.1002/ana.410340515. [DOI] [PubMed] [Google Scholar]
- 78.Przedborski S, Jackson-Lewis V, Fahn S. Antiparkinsonian therapies and brain mitochondrial complex I activity. Mov Disord. 1995;10(3):312–317. doi: 10.1002/mds.870100314. [DOI] [PubMed] [Google Scholar]
- 79.Saraiva AA, Borges MM, Madeira MD, Tavares MA, Paula-Barbosa MM. Mitochondrial abnormalities in cortical dendrites from patients with Alzheimer’s disease. J Submicrosc Cytol. 1985;17(3):459–464. [PubMed] [Google Scholar]
- 80.Sumpter PQ, Mann DM, Davies CA, Yates PO, Snowden JS, Neary D. An ultrastructural analysis of the effects of accumulation of neurofibrillary tangle in pyramidal neurons of the cerebral cortex in Alzheimer’s disease. Neuropathol Appl Neurobiol. 1986;12(3):305–319. doi: 10.1111/j.1365-2990.1986.tb00142.x. [DOI] [PubMed] [Google Scholar]
- 81.Peterson C, Goldman JE. Alterations in calcium content and biochemical processes in cultured skin fibroblasts from aged and Alzheimer donors. Proc Natl Acad Sci U S A. 1986;83(8):2758–2762. doi: 10.1073/pnas.83.8.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swerdlow RH, Koppel S, Weidling I, Hayley C, Ji Y, Wilkins HM. Mitochondria, cybrids, aging, and Alzheimer’s disease. Prog Mol Biol Transl Sci. 2017;146:259–302. doi: 10.1016/bs.pmbts.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arduíno DM, Esteves AR, Swerdlow RH, Cardoso SM. A cybrid cell model for the assessment of the link between mitochondrial deficits and sporadic Parkinson’s disease. Methods Mol Biol. 2015;1265:415–424. doi: 10.1007/978-1-4939-2288-8_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheehan JP, et al. Calcium homeostasis and reactive oxygen species production in cells transformed by mitochondria from individuals with sporadic Alzheimer’s disease. J Neurosci. 1997;17(12):4612–4622. doi: 10.1523/JNEUROSCI.17-12-04612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swerdlow RH, et al. Origin and functional consequences of the complex I defect in Parkinson’s disease. Ann Neurol. 1996;40(4):663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- 86.Swerdlow RH, et al. Cybrids in Alzheimer’s disease: a cellular model of the disease? Neurology. 1997;49(4):918–925. doi: 10.1212/WNL.49.4.918. [DOI] [PubMed] [Google Scholar]
- 87.Cadonic C, Sabbir MG, Albensi BC. Mechanisms of mitochondrial dysfunction in Alzheimer’s disease. Mol Neurobiol. 2016;53(9):6078–6090. doi: 10.1007/s12035-015-9515-5. [DOI] [PubMed] [Google Scholar]
- 88.Parker WD, Jr , Filley CM, Parks JK. Cytochrome oxidase deficiency in Alzheimer’s disease. Neurology. 1990;40(8):1302–1303. doi: 10.1212/WNL.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 89.Parker WD, Jr , et al. Reduced platelet cytochrome c oxidase activity in Alzheimer’s disease. Neurology. 1994;44(6):1086–1090. doi: 10.1212/WNL.44.6.1086. [DOI] [PubMed] [Google Scholar]
- 90.Parker WD, Jr , Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer’s disease brain. Neurology. 1994;44(6):1090–1096. doi: 10.1212/WNL.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 91.Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer’s disease. J Neurochem. 1994;63(6):2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 92.Maurer I, Zierz S, Möller HJ. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol Aging. 2000;21(3):455–462. doi: 10.1016/S0197-4580(00)00112-3. [DOI] [PubMed] [Google Scholar]
- 93.Cooper JM, Wischik C, Schapira AH. Mitochondrial function in Alzheimer’s disease. Lancet. 1993;341(8850):969–970. doi: 10.1016/0140-6736(93)91269-r. [DOI] [PubMed] [Google Scholar]
- 94.Molina JA, et al. Respiratory chain enzyme activities in isolated mitochondria of lymphocytes from patients with Alzheimer’s disease. Neurology. 1997;48(3):636–638. doi: 10.1212/WNL.48.3.636. [DOI] [PubMed] [Google Scholar]
- 95.Kish SJ, et al. Decreased brain protein levels of cytochrome oxidase subunits in Alzheimer’s disease and in hereditary spinocerebellar ataxia disorders: a nonspecific change? J Neurochem. 1999;72(2):700–707. doi: 10.1046/j.1471-4159.1999.0720700.x. [DOI] [PubMed] [Google Scholar]
- 96.Sims NR, Finegan JM, Blass JP, Bowen DM, Neary D. Mitochondrial function in brain tissue in primary degenerative dementia. Brain Res. 1987;436(1):30–38. doi: 10.1016/0006-8993(87)91553-8. [DOI] [PubMed] [Google Scholar]
- 97.Kaminsky YG, Tikhonova LA, Kosenko EA. Critical analysis of Alzheimer’s amyloid-beta toxicity to mitochondria. Front Biosci (Landmark Ed) 2015;20:173–197. doi: 10.2741/4304. [DOI] [PubMed] [Google Scholar]
- 98.Mazat JP, Ransac S, Heiske M, Devin A, Rigoulet M. Mitochondrial energetic metabolism-some general principles. IUBMB Life. 2013;65(3):171–179. doi: 10.1002/iub.1138. [DOI] [PubMed] [Google Scholar]
- 99.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 100.Celotto AM, Chiu WK, Van Voorhies W, Palladino MJ. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS One. 2011;6(10):e25823. doi: 10.1371/journal.pone.0025823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci U S A. 2014;111(29):10654–10659. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Acin-Perez R, Enriquez JA. The function of the respiratory supercomplexes: the plasticity model. Biochim Biophys Acta. 2014;1837(4):444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 103.D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum Mol Genet. 2006;15(13):2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- 104.Lopez-Fabuel I, Resch-Beusher M, Carabias-Carrasco M, Almeida A, Bolanos JP. Mitochondrial complex I activity is conditioned by supercomplex I-III2-IV assembly in brain cells: relevance for Parkinson’s disease. Neurochem Res. 2017;42(6):1676–1682. doi: 10.1007/s11064-017-2191-2. [DOI] [PubMed] [Google Scholar]
- 105.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191(1-3):288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pathak RU, Davey GP. Complex I and energy thresholds in the brain. Biochim Biophys Acta. 2008;1777(7–8):777–782. doi: 10.1016/j.bbabio.2008.05.443. [DOI] [PubMed] [Google Scholar]
- 107.Boulet L, Karpati G, Shoubridge EA. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF) Am J Hum Genet. 1992;51(6):1187–1200. [PMC free article] [PubMed] [Google Scholar]
- 108.Letellier T, Heinrich R, Malgat M, Mazat JP. The kinetic basis of threshold effects observed in mitochondrial diseases: a systemic approach. Biochem J. 1994;302(pt 1):171–174. doi: 10.1042/bj3020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davey GP, Clark JB. Threshold effects and control of oxidative phosphorylation in nonsynaptic rat brain mitochondria. J Neurochem. 1996;66(4):1617–1624. doi: 10.1046/j.1471-4159.1996.66041617.x. [DOI] [PubMed] [Google Scholar]
- 110.Barrientos A, Moraes CT. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J Biol Chem. 1999;274(23):16188–16197. doi: 10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- 111.Manfredi G, Yang L, Gajewski CD, Mattiazzi M. Measurements of ATP in mammalian cells. Methods. 2002;26(4):317–326. doi: 10.1016/S1046-2023(02)00037-3. [DOI] [PubMed] [Google Scholar]
- 112.Gajewski CD, Yang L, Schon EA, Manfredi G. New insights into the bioenergetics of mitochondrial disorders using intracellular ATP reporters. Mol Biol Cell. 2003;14(9):3628–3635. doi: 10.1091/mbc.e02-12-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46(3):428–433. [PMC free article] [PubMed] [Google Scholar]
- 114.Leigh D. Subacute necrotizing encephalomyelopathy in an infant. J Neurol Neurosurg Psychiatry. 1951;14(3):216–221. doi: 10.1136/jnnp.14.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Thorburn DR, Rahman J, Rahman S. Mitochondrial DNA-associated Leigh syndrome and NARP. In: Adam MP, et al., eds. GeneReviews. Seattle, Washington, USA: University of Washington; 1993. https://www.ncbi.nlm.nih.gov/books/NBK1173/ Updated September 28, 2017. Accessed November 1, 2018.
- 116.Manfredi G, et al. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J Biol Chem. 1999;274(14):9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 117.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 118.Rojo A, et al. NARP-MILS syndrome caused by 8993 T>G mitochondrial DNA mutation: a clinical, genetic and neuropathological study. Acta Neuropathol. 2006;111(6):610–616. doi: 10.1007/s00401-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 119.Schon EA, Santra S, Pallotti F, Girvin ME. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin Cell Dev Biol. 2001;12(6):441–448. doi: 10.1006/scdb.2001.0281. [DOI] [PubMed] [Google Scholar]
- 120.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 121.Deng YP, Albin RL, Penney JB, Young AB, Anderson KD, Reiner A. Differential loss of striatal projection systems in Huntington’s disease: a quantitative immunohistochemical study. J Chem Neuroanat. 2004;27(3):143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 122.Reder C. Metabolic control theory: a structural approach. J Theor Biol. 1988;135(2):175–201. doi: 10.1016/S0022-5193(88)80073-0. [DOI] [PubMed] [Google Scholar]
- 123.Goodpaster BH, Sparks LM. Metabolic flexibility in health and disease. Cell Metab. 2017;25(5):1027–1036. doi: 10.1016/j.cmet.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee M, Yoon JH. Metabolic interplay between glycolysis and mitochondrial oxidation: the reverse Warburg effect and its therapeutic implication. World J Biol Chem. 2015;6(3):148–161. doi: 10.4331/wjbc.v6.i3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Formentini L, et al. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014;33(7):762–778. doi: 10.1002/embj.201386392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goldberg J, et al. The mitochondrial ATP synthase is a shared drug target for aging and dementia. Aging Cell. 2018;17(2):e12715. doi: 10.1111/acel.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23(2):64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol. 2015;11(1):11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 129.Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14(6):724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 130.McCommis KS, Finck BN. Mitochondrial pyruvate transport: a historical perspective and future research directions. Biochem J. 2015;466(3):443–454. doi: 10.1042/BJ20141171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71(14):2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015;22(2):364. doi: 10.1038/cdd.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem. 2013;288(33):23798–23806. doi: 10.1074/jbc.M113.482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13(10):444–451. doi: 10.1016/S1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- 135.Beg M, Abdullah N, Thowfeik FS, Altorki NK, McGraw TE. Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1-mediated glucose uptake. Elife. 2017;6:e26896. doi: 10.7554/eLife.26896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Isaev NK, Stel’mashuk EV, Zorov DB. Cellular mechanisms of brain hypoglycemia. Biochemistry Mosc. 2007;72(5):471–478. doi: 10.1134/S0006297907050021. [DOI] [PubMed] [Google Scholar]
- 137.Bender T, Martinou JC. The mitochondrial pyruvate carrier in health and disease: to carry or not to carry? Biochim Biophys Acta. 2016;1863(10):2436–2442. doi: 10.1016/j.bbamcr.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 138.Banerjee K, Munshi S, Frank DE, Gibson GE. Abnormal glucose metabolism in Alzheimer’s disease: relation to autophagy/mitophagy and therapeutic approaches. Neurochem Res. 2015;40(12):2557–2569. doi: 10.1007/s11064-015-1631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Santiago JA, Potashkin JA. Shared dysregulated pathways lead to Parkinson’s disease and diabetes. Trends Mol Med. 2013;19(3):176–186. doi: 10.1016/j.molmed.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 140.Hu G, Jousilahti P, Bidel S, Antikainen R, Tuomilehto J. Type 2 diabetes and the risk of Parkinson’s disease. Diabetes Care. 2007;30(4):842–847. doi: 10.2337/dc06-2011. [DOI] [PubMed] [Google Scholar]
- 141.Palacios N, et al. Obesity, diabetes, and risk of Parkinson’s disease. Mov Disord. 2011;26(12):2253–2259. doi: 10.1002/mds.23855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang X, et al. Serum cholesterol and the progression of Parkinson’s disease: results from DATATOP. PLoS One. 2011;6(8):e22854. doi: 10.1371/journal.pone.0022854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arvanitakis Z, Wilson RS, Bienias JL, Evans DA, Bennett DA. Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Arch Neurol. 2004;61(5):661–666. doi: 10.1001/archneur.61.5.661. [DOI] [PubMed] [Google Scholar]
- 144.Martin WR, Hayden MR. Cerebral glucose and dopa metabolism in movement disorders. Can J Neurol Sci. 1987;14(3 suppl):448–451. doi: 10.1017/s0317167100037896. [DOI] [PubMed] [Google Scholar]
- 145.Mytilineou C, Werner P, Molinari S, Di Rocco A, Cohen G, Yahr MD. Impaired oxidative decarboxylation of pyruvate in fibroblasts from patients with Parkinson’s disease. J Neural Transm Park Dis Dement Sect. 1994;8(3):223–228. doi: 10.1007/BF02260943. [DOI] [PubMed] [Google Scholar]
- 146.Ahmed SS, Santosh W, Kumar S, Christlet HT. Metabolic profiling of Parkinson’s disease: evidence of biomarker from gene expression analysis and rapid neural network detection. J Biomed Sci. 2009;16:63. doi: 10.1186/1423-0127-16-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schwartzman RJ, Alexander GM. Changes in the local cerebral metabolic rate for glucose in the MPTP primate model of Parkinson’s disease. Adv Neurol. 1987;45:171–173. [PubMed] [Google Scholar]
- 148.Schwartzman RJ, Alexander GM. Changes in the local cerebral metabolic rate for glucose in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) primate model of Parkinson’s disease. Brain Res. 1985;358(1–2):137–143. doi: 10.1016/0006-8993(85)90957-6. [DOI] [PubMed] [Google Scholar]
- 149.Lucignani G, Namba H, Nehlig A, Porrino LJ, Kennedy C, Sokoloff L. Effects of insulin on local cerebral glucose utilization in the rat. J Cereb Blood Flow Metab. 1987;7(3):309–314. doi: 10.1038/jcbfm.1987.68. [DOI] [PubMed] [Google Scholar]
- 150.Palombo E, Porrino LJ, Bankiewicz KS, Crane AM, Sokoloff L, Kopin IJ. Local cerebral glucose utilization in monkeys with hemiparkinsonism induced by intracarotid infusion of the neurotoxin MPTP. J Neurosci. 1990;10(3):860–869. doi: 10.1523/JNEUROSCI.10-03-00860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mazzio E, Soliman KF. Pyruvic acid cytoprotection against 1-methyl-4-phenylpyridinium, 6-hydroxydopamine and hydrogen peroxide toxicities in vitro. Neurosci Lett. 2003;337(2):77–80. doi: 10.1016/S0304-3940(02)01327-7. [DOI] [PubMed] [Google Scholar]
- 152.Guatteo E, et al. Dopamine-containing neurons are silenced by energy deprivation: a defensive response or beginning of cell death? Neurotoxicology. 2005;26(5):857–868. doi: 10.1016/j.neuro.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 153.Bellucci A, Collo G, Sarnico I, Battistin L, Missale C, Spano P. Alpha-synuclein aggregation and cell death triggered by energy deprivation and dopamine overload are counteracted by D2/D3 receptor activation. J Neurochem. 2008;106(2):560–577. doi: 10.1111/j.1471-4159.2008.05406.x. [DOI] [PubMed] [Google Scholar]
- 154.Zaltieri M, Longhena F, Pizzi M, Missale C, Spano P, Bellucci A. Mitochondrial dysfunction and α-synuclein synaptic pathology in Parkinson’s disease: who’s on first? Parkinsons Dis. 2015;2015:108029. doi: 10.1155/2015/108029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kong JN, et al. Novel function of ceramide for regulation of mitochondrial ATP release in astrocytes. J Lipid Res. 2018;59(3):488–506. doi: 10.1194/jlr.M081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Timmons S, Coakley MF, Moloney AM, O’Neill C. Akt signal transduction dysfunction in Parkinson’s disease. Neurosci Lett. 2009;467(1):30–35. doi: 10.1016/j.neulet.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 157.Malagelada C, Jin ZH, Greene LA. RTP801 is induced in Parkinson’s disease and mediates neuron death by inhibiting Akt phosphorylation/activation. J Neurosci. 2008;28(53):14363–14371. doi: 10.1523/JNEUROSCI.3928-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Greene LA, Levy O, Malagelada C. Akt as a victim, villain and potential hero in Parkinson’s disease pathophysiology and treatment. Cell Mol Neurobiol. 2011;31(7):969–978. doi: 10.1007/s10571-011-9671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiromerisiou G, Hadjigeorgiou GM, Papadimitriou A, Katsarogiannis E, Gourbali V, Singleton AB. Association between AKT1 gene and Parkinson’s disease: a protective haplotype. Neurosci Lett. 2008;436(2):232–234. doi: 10.1016/j.neulet.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kwok JB, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson’s disease. Ann Neurol. 2005;58(6):829–839. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- 161.Zhang C, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108(39):16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu K, et al. Parkin regulates the activity of Pyruvate kinase M2. J Biol Chem. 2016;291(19):10307–10317. doi: 10.1074/jbc.M115.703066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta. 2014;1841(4):595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 164.Herrera-Cruz MS, Simmen T. Over six decades of discovery and characterization of the architecture at mitochondria-associated membranes (MAMs) Adv Exp Med Biol. 2017;997:13–31. doi: 10.1007/978-981-10-4567-7_2. [DOI] [PubMed] [Google Scholar]
- 165.Guardia-Laguarta C, et al. α-Synuclein is localized to mitochondria-associated ER membranes. J Neurosci. 2014;34(1):249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Paillusson S, et al. α-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca(2+) homeostasis and mitochondrial ATP production. Acta Neuropathol. 2017;134(1):129–149. doi: 10.1007/s00401-017-1704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Calì T, Ottolini D, Brini M. Mitochondrial Ca(2+) as a key regulator of mitochondrial activities. Adv Exp Med Biol. 2012;942:53–73. doi: 10.1007/978-94-007-2869-1_3. [DOI] [PubMed] [Google Scholar]
- 168.Area-Gomez E, et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol. 2009;175(5):1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Newman M, Ebrahimie E, Lardelli M. Using the zebrafish model for Alzheimer’s disease research. Front Genet. 2014;5:189. doi: 10.3389/fgene.2014.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schreiner B, Hedskog L, Wiehager B, Ankarcrona M. Amyloid-β peptides are generated in mitochondria-associated endoplasmic reticulum membranes. J Alzheimers Dis. 2015;43(2):369–374. doi: 10.3233/JAD-132543. [DOI] [PubMed] [Google Scholar]
- 171.Hansson CA, et al. Nicastrin, presenilin, APH-1, and PEN-2 form active gamma-secretase complexes in mitochondria. J Biol Chem. 2004;279(49):51654–51660. doi: 10.1074/jbc.M404500200. [DOI] [PubMed] [Google Scholar]
- 172.Pavlov PF, et al. Mitochondrial γ-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB J. 2011;25(1):78–88. doi: 10.1096/fj.10-157230. [DOI] [PubMed] [Google Scholar]
- 173.Area-Gomez E, et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012;31(21):4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hedskog L, et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc Natl Acad Sci U S A. 2013;110(19):7916–7921. doi: 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pera M, et al. Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J. 2017;36(22):3356–3371. doi: 10.15252/embj.201796797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Supnet C, Bezprozvanny I. The dysregulation of intracellular calcium in Alzheimer disease. Cell Calcium. 2010;47(2):183–189. doi: 10.1016/j.ceca.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gibson GE, Thakkar A. Interactions of mitochondria/metabolism and calcium regulation in Alzheimer’s disease: a calcinist point of view. Neurochem Res. 2017;42(6):1636–1648. doi: 10.1007/s11064-017-2182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Burgoyne T, Patel S, Eden ER. Calcium signaling at ER membrane contact sites. Biochim Biophys Acta. 2015;1853(9):2012–2017. doi: 10.1016/j.bbamcr.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 179.Szymański J, et al. Interaction of mitochondria with the endoplasmic reticulum and plasma membrane in calcium homeostasis, lipid trafficking and mitochondrial structure. Int J Mol Sci. 2017;18(7):E1576. doi: 10.3390/ijms18071576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Marchi S, et al. Endoplasmic reticulum-mitochondria communication through Ca(2+) signaling: the importance of mitochondria-associated membranes (MAMs) Adv Exp Med Biol. 2017;997:49–67. doi: 10.1007/978-981-10-4567-7_4. [DOI] [PubMed] [Google Scholar]
- 181.Marchi S, et al. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium. 2018;69:62–72. doi: 10.1016/j.ceca.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 182.Tarasov AI, Griffiths EJ, Rutter GA. Regulation of ATP production by mitochondrial Ca(2+) Cell Calcium. 2012;52(1):28–35. doi: 10.1016/j.ceca.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Keinan N, Pahima H, Ben-Hail D, Shoshan-Barmatz V. The role of calcium in VDAC1 oligomerization and mitochondria-mediated apoptosis. Biochim Biophys Acta. 2013;1833(7):1745–1754. doi: 10.1016/j.bbamcr.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 184.Yu J, et al. JNK3 signaling pathway activates ceramide synthase leading to mitochondrial dysfunction. J Biol Chem. 2007;282(35):25940–25949. doi: 10.1074/jbc.M701812200. [DOI] [PubMed] [Google Scholar]
- 185.Kogot-Levin A, Saada A. Ceramide and the mitochondrial respiratory chain. Biochimie. 2014;100:88–94. doi: 10.1016/j.biochi.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 186.Lapuente-Brun E, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 187.Colombini M. Ceramide channels and mitochondrial outer membrane permeability. J Bioenerg Biomembr. 2017;49(1):57–64. doi: 10.1007/s10863-016-9646-z. [DOI] [PubMed] [Google Scholar]
- 188.Ausman J, et al. Ceramide-induced BOK promotes mitochondrial fission in preeclampsia. Cell Death Dis. 2018;9(3):298. doi: 10.1038/s41419-018-0360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Flis VV, Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol. 2013;5(6):a013235. doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.van Vliet AR, Verfaillie T, Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochim Biophys Acta. 2014;1843(10):2253–2262. doi: 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 191.Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39(3):257–288. doi: 10.1016/S0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 192.Acehan D, Malhotra A, Xu Y, Ren M, Stokes DL, Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J. 2011;100(9):2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pfeiffer K, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278(52):52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 194.Gohil VM, Hayes P, Matsuyama S, Schägger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J Biol Chem. 2004;279(41):42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
- 195.Xu Y, et al. Loss of protein association causes cardiolipin degradation in Barth syndrome. Nat Chem Biol. 2016;12(8):641–647. doi: 10.1038/nchembio.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Corcelli A, Schlame M. Cardiolipin as key lipid of mitochondria in health and disease. 2nd Edition, Florence, Italy, September 30-October 1, 2015. Chem Phys Lipids. 2016;198:10–12. doi: 10.1016/j.chemphyslip.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 197.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182(5):937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schlattner U, et al. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim Biophys Acta. 2009;1788(10):2032–2047. doi: 10.1016/j.bbamem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 199.Lai YC, Li CC, Sung TC, Chang CW, Lan YJ, Chiang YW. The role of cardiolipin in promoting the membrane pore-forming activity of BAX oligomers. Biochim Biophys Acta Biomembr. 2019;1861(1):268–280. doi: 10.1016/j.bbamem.2018.06.014. [DOI] [PubMed] [Google Scholar]