Abstract
In response to viral pathogens, the host upregulates antiviral genes that suppress translation of viral mRNAs. However, induction of such antiviral responses may not be exclusive to viruses, as the pathways lie at the intersection of broad inflammatory networks that can also be induced by bacterial pathogens. Using a model of Gram-negative sepsis, we show that propagation of kidney damage initiated by a bacterial origin ultimately involves antiviral responses that result in host translation shutdown. We determined that activation of the eukaryotic translation initiation factor 2-α kinase 2/eukaryotic translation initiation factor 2α (Eif2ak2/Eif2α) axis is the key mediator of translation initiation block in late-phase sepsis. Reversal of this axis mitigated kidney injury. Furthermore, temporal profiling of the kidney translatome revealed that multiple genes involved in formation of the initiation complex were translationally altered during bacterial sepsis. Collectively, our findings imply that translation shutdown is indifferent to the specific initiating pathogen and is an important determinant of tissue injury in sepsis.
Keywords: Inflammation, Nephrology
Keywords: Proteomics, Translation
Introduction
Bacterial sepsis remains the leading cause of death in intensive care units. It is a highly dynamic pathological state in which a wide array of pro- and antiinflammatory pathways are aberrantly activated (1). The resultant clinical outcome is dictated largely by the number of organs involved and severity of organ involvement. Severe renal failure often sets in gradually and significantly contributes to the mortality of late-phase sepsis (2, 3).
The cellular and molecular pathways involved in sepsis-induced organ failure are not well understood. For example, examination of postmortem kidney tissue from patients with severe sepsis revealed highly heterogeneous and rather nonspecific findings (4). Furthermore, under current clinical practice, renal biopsy is rarely performed in patients with sepsis-induced kidney injury, contributing to the stagnant progress in this field. To date, there are no therapeutic drugs approved for treating sepsis-induced organ failure (5).
Broad and unbiased omics, such as transcriptomics and proteomics, are increasingly used to uncover pathways that may be involved in the pathogenesis of sepsis. The power of these approaches can be further extended if they are used to examine omics changes at multiple time points. Indeed, temporal profiling is essential because the early and late phases of sepsis differ in pathogenesis. Furthermore, the quantitative relationship between specific mRNAs and their protein products is not linear and could be further distorted by the septic environment. Therefore, an ideal strategy could involve the investigation of mRNA and instantaneous protein translation, on the same detection scale and range (i.e., translatome; ref. 6), and at multiple time points.
In this study, we examined temporal changes in the translatome of the kidney in an animal model of bacterial sepsis. We identified translation shutdown as a key feature of late-phase sepsis. We further found that 5′ cap–dependent translation shutdown is mediated by viral response pathways — originating from a bacterial stimulus. The reversal of this shutdown ameliorates the degree of sepsis-induced kidney injury.
Results
Late-phase sepsis is characterized by global protein synthesis shutdown.
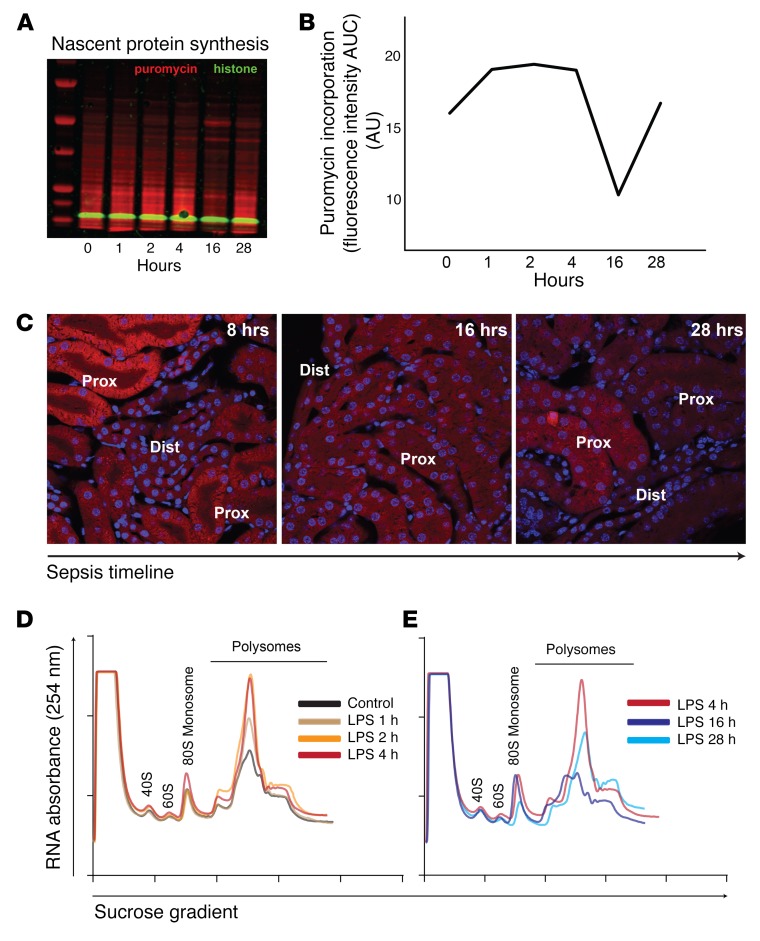
A moderate dose of LPS (5 mg/kg) results in robust induction of proinflammatory cytokines in tissues, including the kidney, without causing hemodynamic compromise (7, 8). Thus, this model is well suited for studying direct LPS effects on organs in vivo. Indeed, moderate endotoxemia causes severe acute kidney injury (AKI) due to direct interaction of LPS with its receptor, TLR4, expressed in the S1 proximal tubules (9, 10). The acute phase of sepsis-induced AKI (~4 hours) is characterized by tubular oxidative stress and tissue-derived proinflammatory cytokines and chemokines (8, 9). Coinciding with this inflammatory outburst, we found that the rate of nascent protein synthesis, determined by puromycin incorporation, increased as early as 1 hour after LPS challenge (Figure 1A). In contrast, late-phase sepsis (~16 hours) was characterized by global protein synthesis shutdown, which recovered spontaneously by 28 hours (Figure 1B). Imaging of nascent protein synthesis revealed that the observed changes in the whole-tissue lysate occurred primarily in proximal tubules (Figure 1C).
Figure 1. Sepsis causes a biphasic translation derangement.
(A and B) Time-course analysis of global protein synthesis in a murine model of sepsis (LPS, 5 mg/kg tail vein, i.v.), as determined by incorporation of puromycin to growing polypeptide chains in vivo. Kidneys were harvested 30 minutes after puromycin administration at various time points in the sepsis timeline. Representative Western blot and its quantification are shown. (C) Imaging of nascent protein synthesis in kidney tissues using O-propargyl-puromycin (OPP), an alkyne analog of puromycin, in vivo. Alexa Fluor 555–azide was conjugated to OPP by copper(I)-catalyzed azide-alkyne cycloaddition (Click chemistry). Prox, proximal tubules; dist, distal tubules/collecting ducts. Original magnification, ×60. (D and E) Polysomal profiling of kidney extracts from mice treated with LPS for indicated durations. Cycloheximide was injected at the time of sacrifice to prevent the release of ribosomes from the cognate mRNA. Ribosomal subunit 40S, 60S, mono-ribosome (80S), and polyribosomes were separated using sucrose density gradient. (D) Increased mono-ribosome and poly-ribosome signals indicating increased protein synthesis are observed in the early phases of sepsis (LPS, 1, 2, and 4 hours). (E) Increased mono-ribosome, but decreased poly-ribosome, fractions indicating initiation block and decreased protein synthesis are observed 16 hours after LPS administration. Representative UV absorbance traces from each time point are overlaid, and for comparison, an identical trace (LPS, 4 hours) is shown in both D and E. Polysome-to-monosome ratios: 3.0, 3.5, 4.7, 3.1, 1.9, and 3.7 at 0, 1, 2, 4, 16, and 28 hours, respectively.
We further characterized protein synthesis by performing polysome profiling. We found that, in early phase sepsis, there were increased monosome and polysome fractions, indicative of increased global translation (Figure 1D). In contrast, the profile in late-phase sepsis showed sustained monosome fraction, but decreased polysome fraction (Figure 1E). This is strongly indicative of translation initiation block (11). Collectively, these findings suggest that the rate of translation varies in the sepsis timeline and that global translation shutdown is a hallmark of late-phase sepsis.
Nascent proteomics reveals protein synthesis shutdown in major metabolic pathways.
To further examine global translation shutdown in late-phase sepsis, we next performed nascent proteomics in which only newly made proteins are labeled, enriched, and analyzed by mass spectrometry (Figure 2A and Supplemental Figure 1A). This approach offers 2 major advantages over general proteomics. First, with the enrichment strategy, excessive signals from abundant proteins and background noise are eliminated. Second, nascent proteomics provides a snapshot of proteins that are currently being synthesized, rather than a cumulative protein profile that is confounded by protein half-lives and turnover rates. As shown in Figure 2B and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI123284DS1), nascent proteomics recapitulated the profound protein synthesis shutdown 16 hours after LPS administration. We found that the majority of proteins involved in vital metabolic pathways, such as the TCA cycle and peroxisome metabolism, were downregulated at this time point (Figure 3, A and B). We noted that the ATP/AMP ratio declined after the 16-hour time point, suggesting that the loss of energy was the result rather than the cause of global translation shutdown (Figure 3, C and D). Interestingly, a small set of genes was found to be resistant to global translation shutdown. For example, the levels of Acod1/Irg1, an enzyme involved in the synthesis of antimicrobial metabolite itaconate, and Nos2, inducible nitric oxide synthase, continued to increase over time (Figure 3, A and B). Pathway enrichment analysis suggests that many of these resistant genes are involved in cellular processes, such as mRNA processing and chaperoning of protein folding (Supplemental Figure 1, B–D).
Figure 2. Translation shutdown as determined by nascent proteomics.
(A) Overview of nascent proteomics protocol. (B) Approximately 6,000 proteins were identified using label-free quantitative mass spectrometry (Supplemental Tables 1 and 2). Principal component analysis biplot illustrating relationships between experimental groups (black) and individual nascent proteins (red) in the 2D space (PC1, PC2). n = 3 for each time point. Median protein values are used for data display.
Figure 3. Multiple metabolic pathways are affected by translation shutdown during late-phase sepsis.
(A and B) Nascent proteins detected are mapped to the KEGG TCA cycle and peroxisome pathways. Arrowheads point to Acod1/Irg1 and Nos2. (C and D) Representative HPLC chromatograms of kidney tissue nucleotides and quantitation of nucleotide ratios are shown. n = 3 per condition. P values determined by Student’s t test (2 sided, nonpaired). Error bars show SD.
In summary, late-phase sepsis is characterized by translation initiation block causing translation shutdown that affects critical metabolic pathways.
Comprehensive characterization of the translatome in the sepsis timeline.
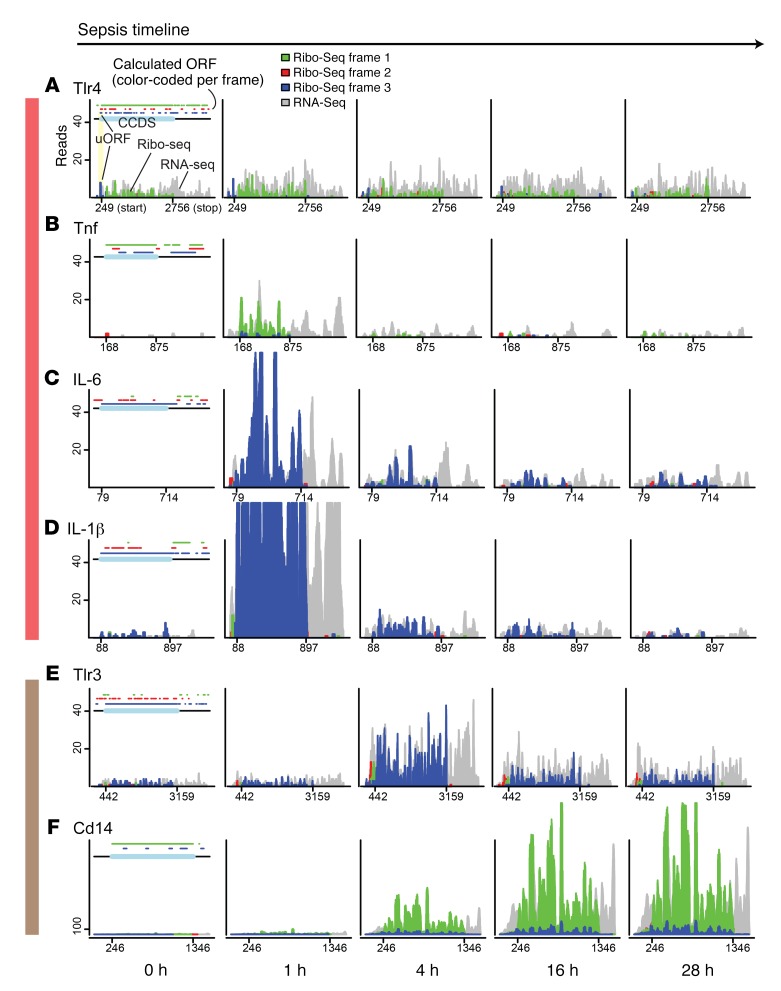
To obtain a more mechanistic insight into the temporal evolution of translation dysregulation in vivo, we analyzed kidneys from septic animals using the Ribo-Seq approach (Supplemental Figure 2A). Ribo-Seq provides a global snapshot of actively translated mRNAs at single nucleotide resolution (ribosome footprint) (6). The position of the ribosome footprint directly correlates with the codon that is being translated at the time of tissue harvest. Our Ribo-Seq display strategy and quality control measurements are shown in Figure 4A and Supplemental Figure 2, B–F. For example, we show in Figure 4A that the calculated codon frame of the TLR4 consensus coding sequence (CCDS) is frame 1. The actual translating ribosomes are indeed synchronized to frame 1, and the ribosomal coverage ends precisely at the stop codon, demonstrating the high fidelity of ribosome movement (triplet periodicity) and high-quality tissue Ribo-Seq data. Figure 4A also shows that LPS did not alter TLR4 translation kinetics or translation efficiency (Ribo-Seq to RNA-Seq coverage ratio) in the course of sepsis. Additionally, we noted the presence of an upstream open reading frame (uORF) (frame 3, blue) that is spanned exactly by corresponding ribosomal reads in the same blue periodicity. This indicates that this uORF is potentially functional. Although discovery of encoding uORF is not the focus of this study, we identified a number of potentially functional uORFs across the genome in this sepsis model, as shown in the subsequent figures.
Figure 4. Translatome time course reveals propagation of TLR4-mediated signaling.
(A–F) Ribo-Seq analysis of select genes under indicated conditions. Details for data visualization strategy are described in Supplemental Figure 2. RNA-Seq reads (gray) and Ribo-Seq reads (red, blue, green) were mapped to the mouse transcriptome. The x axes denote mRNA positions (position 1 is the start of 5′ UTR and x-max position is the end of 3′ UTR). The y axes denote read coverage. Green, red, and blue colors in the histogram correspond to ribosome frames 1, 2, and 3, respectively. On the top of each control figure (0 hour), calculated ORF positions (defined by ATG start codon, TAG/TAA/TGA stop codons) and their associated frame colors as determined by modulo operation are shown. Annotated CCDS is highlighted in light blue on the top, and its start and stop positions are shown on the x axis. When multiple isoforms are present, a protein coding transcript with best curated isoform is shown (e.g., gold transcript in Ensembl). All read lengths are displayed (median, 29 nt). The color-coded vertical bars (red, brown) correspond to pathways highlighted in Figure 5J.
Analysis of the translatome landscape in sepsis revealed major time-dependent changes. We show in Figure 4, B–D, that MyD88-dependent NF-κB signaling (the conventional TLR4 proinflammatory pathway) was immediately activated in vivo. NF-κB–induced cytokines, such as IL-6 and IL-1β, were significantly increased, both transcriptionally and translationally, 1 hour after LPS injection (Figure 4). Remarkably, the expression of NF-κB–induced genes was rapidly downregulated and returned to near baseline levels by 4 hours. This rapid downregulation can be explained by the concomitant (rather than sequential) expression of NF-κB inhibitory molecules at the 1-hour time point (e.g., Nfkbia, Tnfaip3, Tnip3; Supplemental Figure 3) as well as upregulation of the mRNA decay machinery (e.g., adenylate-uridylate–rich [AU-rich] element binding protein Zfp36 and endonuclease Zc3h12a; Supplemental Figure 3).
The subsequent phase of this model of bacterial sepsis was characterized by upregulation of antiviral signaling pathways. For example, activation of the MyD88-independent TIR domain–containing adapter-inducing IFN-β (TRIF) pathway leads to the induction of IFN and IFN-induced antiviral genes (12). Indeed, we found that a wide array of antiviral genes were simultaneously upregulated at 4 hours and that many of these genes exhibited sustained translation up to 28 hours after LPS (Figure 5, A–D, and Supplemental Figure 4). These antiviral genes included Zc3hav1, Rsad2, and Oas1a, and they played crucial roles in intracellular antiviral responses. Expressions of Tlr3 and Cd14, important molecules that trigger TRIF signaling, were also increased (Figure 4, E and F). Additionally, the widely used kidney injury marker Havcr1/Kim1 — which is a hepatitis A virus receptor — also started to increase at 4 hours (Supplemental Figure 4).
Figure 5. Propagation of TLR4-mediated signaling elicits antiviral response pathways.
(A–I) Ribo-Seq analysis of select genes involved in viral response and Eif2ak2 (PKR) activation. Ribo-Seq reads (red, green, blue) are superimposed as a front layer over RNA-Seq reads (gray). (J) Schematic of TLR4-mediated signaling pathways is shown. The background colors correspond to the colors of vertical bars in Figure 4 (red, brown) and Figure 5 (beige, blue).
In Supplemental Figures 5–12, we show comprehensive high-resolution renal translatome analysis in the sepsis timeline. The data can be further mined by the scientific community as a platform to uncover novel, sepsis timeline–specific therapeutic targets that may be uniquely identifiable by Ribo-Seq. For example, we show in Figure 5 that 2 genes in the viral pathway, Mx1 and Irf7, exhibited a complex translatome profile in which high uORF ribosome signal and low translation efficiency became prominent during the course of sepsis. This translatome profile is consistent with cap-independent translation mechanisms, such as those using internal ribosome entry site (IRES) (13). We also found that in vivo treatment with lactimidomycin/puromycin, a combination of compounds that is used for identification of initiation start sites (14), revealed differential ribosomal kinetics. For instance, the antiviral genes Rsad2 and Oas1a have similar translatome features (no uORF, similar 5′ UTR and coding sequence (CDS) lengths, similar temporal responses to LPS; Figure 5B and Supplemental Figure 4A). Remarkably, however, lactimidomycin/puromycin treatment resulted in 2 distinct ribosomal coverage patterns (Supplemental Figure 13, A and B). The initiation site of Rsad2 was highlighted as expected. In contrast, Oas1a ribosomes remained unaffected by the treatment, suggesting that their ribosomal kinetics are markedly different.
Propagation of TLR4-mediated signaling converges to antiviral response pathways that result in global protein synthesis shutdown.
Although TLR4 is a receptor for LPS in the outer membrane of Gram-negative bacteria, our data indicated that pathways involved in viral infection are the salient feature of endotoxin-induced kidney injury during the subacute to late phases (4 to 16 hours) of sepsis. One important mechanism by which the host represses viral replication is translation shutdown mediated by eukaryotic translation initiation factor 2-α kinase 2 (Eif2ak2) (PKR) (15). Eif2ak2 activation is triggered by dsRNA binding to Eif2ak2, leading to dimerization and autophosphorylation. Activated Eif2ak2 in turn inhibits 5′ cap–dependent translation via phosphorylation of eukaryotic translation initiation factor 2α (eIF2α). Eif2α is the main regulatory subunit of the GTP/Met-tRNA ternary complex, a part of the larger translation initiation complex. Using a cell-culture system, we verified that Eif2ak2 activation causes Eif2a phosphorylation and that Eif2ak2 inhibition prevents Eif2α phosphorylation (Supplemental Figure 13C).
Accordingly, we hypothesized that the global translation shutdown observed in the LPS model could be due to activation of this antiviral response pathway. Indeed, we found that in vivo expression levels of Eif2ak2 increased at 4 hours after LPS injection and remained elevated, coinciding with the time at which antiviral response genes were upregulated (Figure 5, E and F, and Figure 6, A and B). We also found that phosphorylation of Eif2α started to increase at 4 hours (Figure 6A). These changes suggest that Eif2ak2-mediated inactivation of the Eif2 complex contributed to the deleterious global translation shutdown. Importantly, none of the other Eif2α kinases, namely Eif2ak1 (HRI), Eif2ak3 (PERK), and Eif2ak4 (GCN2), showed changes in their translatome profiles, suggesting that Eif2α phosphorylation was specifically induced by Eif2ak2, the antiviral response gene (Supplemental Figure 13, D–G). In addition, the translation of Eif2ak2 phosphatases Ppp1r15a (Gadd34) and Ppp1r15b (CreP) remained unchanged (despite the increased mRNA expression in the case of Gadd34), which in effect could also contribute to the sustained phosphorylation of Eif2α (Figure 5, G and H). Collectively, these changes support a role for the Eif2ak2/Eif2α axis in mediating global translation shutdown. Phosphorylated Eif2α (p-Eif2α), in turn, was implicated in the activation of the stress responsive Atf4 pathway, known to be involved in either recovery or abortive gene activation (Figure 5, I and J, and Supplemental Figure 14, A–E). The lack of translational activation of proapoptotic Ddit3 (CHOP) might indicate a preponderance of recovery activities in the late phase of the sublethal LPS model (Supplemental Figure 14D).
Figure 6. Translation shutdown mediated by the Eif2ak2/Eif2α axis contributes to sepsis-induced kidney injury.
(A) Mice were treated with 5 mg/kg LPS i.v. for indicated durations, and kidneys were analyzed by Western blot for Eif2ak2, Eif2α, and Ser51 p-Eif2α (arrow). (B) Select protein levels as determined by nascent proteomics are shown. (C and D) In vivo effects of ISRIB (5 mg/kg i.p.) on protein synthesis (puromycin incorporation in the kidney; ISRIB for 16 hours) and tissue expression of Eif2ak2, Eif2α, and p-Eif2α (kidneys harvested 16 hours after 5 mg/kg LPS i.v. with or without ISRIB i.p. for 16 hours). LPS increased Eif2ak2 and p-Eif2α. This increase was not affected by ISRIB. (E) Serum creatinine levels 24 hours after LPS with or without ISRIB treatment administered at indicated time points. *P < 0.05 vs. LPS without ISRIB treatment by Student’s t test (2 sided, nonpaired). Error bars show SD.
Finally, to demonstrate the causal role of translation shutdown in inducing AKI, we used the integrated stress response inhibitor (ISRIB), a compound that stabilizes the guanine nucleotide exchange factor Eif2b in an activated state, thereby rescuing translation despite the presence of p-Eif2α (16, 17). When given early in the course of sepsis in vivo, ISRIB reversed the effect of p-Eif2α on translation and led to significant renoprotection without affecting the levels of Eif2ak2, Eif2α, or p-Eif2α (Figure 6, C–E, and Supplemental Figure 15A). Ribo-Seq analysis revealed that ISRIB increased translation of multiple genes involved in general metabolism as well as genes that are indispensable for effective transcription and translation, such as Med13l (a component of the RNA polymerase II mediator complex), Taf4b (a subunit of RNA polymerase II preinitiation complex), and Eef1g (eukaryotic translation elongation factor 1G; Figure 7, A–C, and Supplemental Figure 15, B and C). In contrast, Nrf1 translation was reduced in animals that received ISRIB (Figure 7A). Nrf1 is a direct transcription factor of Eif2α. The reduced Nrf1 expression is likely through a negative feedback mechanism in response to increased overall translation in the ISRIB-treated animals. The overall cytokine/chemokine profiles were similar between animals treated with and without ISRIB (Supplemental Figure 15, D and E). Additionally, we show in Supplemental Figure 15, F and G, that the inhibition of Eif2ak2 by 2-aminopurine or C16 (thereby limiting Eif2α phosphorylation; refs. 18, 19) afforded renoprotective effects comparable to those observed with ISRIB treatment. Finally, we show in Supplemental Figure 15, H and I, that ISRIB prevented translation shutdown and conferred renoprotection in a model of polymicrobial sepsis using cecal ligation and puncture. Note that, when given at later time points, ISRIB and 2-aminopurine were ineffective. We show in the next section that this lack of therapeutic effects at later time points is due in part to the emergence of Eif2α-independent mechanisms that inhibit translation. This underscores the importance of timing therapy in the dynamic timeline of sepsis. Furthermore, while timely and transient reversal of translation shutdown is protective, chronic inhibition of Eif2α phosphorylation may have deleterious effects. This is because, as noted above, Eif2α phosphorylation is also involved in the adaptive stress-responsive Atf4 pathway.
Figure 7. ISRIB reverses the effect of p-Eif2α on translation.
(A) Ribo-Seq analysis of kidney extracts from mice treated with LPS for 16 hours with or without ISRIB. Smear plot in which top 10 differentially translated genes are highlighted in red (edgeR exactTest with BH adjusted P values < 0.05, n = 3 per condition). The x axis denote mean log counts per million (logCPM), and the y axis denotes log fold change (logFC). ISRIB was administered 1 hour after LPS. (B and C) Comparative translatome count values mapped to the TCA cycle and peroxisome pathways. (D) S1 proximal tubules from kidneys of control or septic mice were laser microdissected. Original magnification, ×60. Glom, glomerulus. (E) S2/S3 proximal tubules from indicated conditions were manually microdissected. Original magnification, ×5. (F and G) Transcriptomics analysis was performed on S1 and S2/S3 segments (n = 4 for each condition). Select gene expression levels are shown. P values were determined by Student’s t test (2 sided, nonpaired). The boxplot middle line shows the median, the box edges show the 25th and 75th percentiles, and the whiskers show the ×1.5 interquartile range.
In summary, we showed that Eif2ak2/Eif2α-mediated translation shutdown is an important contributor to sepsis-induced kidney injury. To extend the findings, we also performed laser and manual microdissection of S1 and S2/S3 proximal tubular segments, respectively, and obtained transcriptomics analysis (Figure 7, D–G). We found that the bulk kidney Eif2ak2 signal specifically originated from S2/S3 proximal tubules. S2/S3 are indeed the main cell types that exhibit LPS-induced injury in vivo, as we previously reported (9, 10, 20). Nevertheless, the role of Eif2ak2 in other cell types cannot be ruled out.
Multiple mechanisms contribute to translation shutdown in late-phase sepsis.
The formation of the 5′ cap initiation complex is a highly regulated process that requires assembly of multiple subunits in a specific order. Therefore, sustained phosphorylation of Eif2α could sufficiently explain the global translation shutdown in sepsis. However, the narrow therapeutic window of ISRIB and 2-aminopurine suggested that dysregulation of other components of the initiation complex might also contribute to depressed translation. Indeed, we found that translation of cap-binding protein Eif4e was decreased even though the mRNA level was unaffected (Figure 8A). We also found that Eif4ebp3, a repressor of Eif4e assembly to the cap complex, was increased (Figure 8B). In addition, the RNA helicase Eif4a2 was translationally downregulated (Figure 8C and Supplemental Figure 12E). Taken together, the changes in these components can also repress the early steps of cap-dependent translation initiation (eIF4F complex formation; Figure 8G and Supplemental Figure 16).
Figure 8. Multiple mechanisms contribute to translation shutdown in late-phase sepsis.
(A–F) Ribo-Seq analysis of select genes involved in translation (in mice). (A) For clarity, the layer order for Eif4e was reversed, with RNA-Seq reads (gray) shown as front layer (right 2 panels). (G) Schematic of the 5′ cap initiation complex.
Outside of the core cap machinery, late-phase sepsis was also associated with changes in noncanonical initiation factors. Malignant T cell amplified sequence 1 (Mcts1) was recently reported to be an important regulator of translation reinitiation downstream of uORFs (21). Thus, the decreased expression of Mcts1 observed in sepsis (Figure 8D) could have broad repressive effects on multiple genes that harbor ribosome-engaged uORFs (e.g., Mx1, Irf7, Eif2ak2, Ppp1r15a, Ppp1r15b, Atf4 in Figure 5; Zc3h12a in Supplemental Figure 3; Plvap, Kdr in Supplemental Figure 9; Nphs1 in Supplemental Figure 10; Sesn2, Ddit3 in Supplemental Figure 14; Eif4g2 in Supplemental Figure 16). We also noted that Eif6 expression level was increased and remained elevated at 28 hours (Figure 8E). Eif6 is a multifaceted molecule that prevents the association of 60S and 40S ribosomal subunits to form the 80S initiation complex. Interestingly, such an antiassociation factor is reported to promote translation under certain experimental conditions (22, 23). Whether the increased level of Eif6 in our sepsis model reflects compensatory or recovery response to translation shutdown remains to be determined. We also show in Figure 8F that the pre-mRNA cap–binding molecule Ncbp2 levels decreased over the course of sepsis. This could negatively affect the kinetics of nascent mRNAs and their pioneer round of translation in the nucleus (24).
Finally, the burst of protein synthesis during the early phase of sepsis could cause ER stress and contribute to the global translation shutdown. Indeed, we show in Supplemental Figure 16F, that Xbp1, a characteristic marker for ER stress, was increased at 4 hours and remained elevated up to 28 hours. More specifically, activation of Xbp1 requires unconventional splicing, which induces a frameshift and generates a read-through protein product (25). Ribo-Seq clearly illustrated the genesis of the spliced Xbp1 isoform (Supplemental Figure 16, G and H).
In aggregate, high-resolution translatome analysis provided a framework for analyzing the previously underappreciated translation shutdown that characterizes late-phase sepsis and highlighted potential therapeutic targets.
EIF2AK2 expression is increased in kidneys from patients with sepsis-induced AKI.
Finally, we asked whether human sepsis–related AKI has a signature comparable to the one found in the murine model. We obtained archived kidney biopsy samples showing a primary pathologic diagnosis of acute tubular necrosis (ATN). From these biopsy samples, we identified 5 cases of bacterial sepsis and 10 various non–sepsis-related cases (Figure 9K and Supplemental Table 3). The histopathology of sepsis-induced kidney injury was indistinguishable from that of other causes of ATN. We performed RNA-Seq and found that the overall transcriptomic signatures of biopsies from sepsis versus non–sepsis-related AKI were also indistinguishable (Figure 9A). However, we found that expression of EIF2AK2 (but not other EIF2AK genes) was increased in patients with sepsis, especially so in the patient with prolonged and recurrent sepsis (Figure 9G). Thus, EIF2AK2 could potentially be a useful sepsis-specific kidney injury marker and help further stratify the sepsis timeline. Multiple other genes in the human renal biopsies from sepsis-related AKI also exhibited time-specific changes that paralleled those observed in the mouse translatome (Figure 9, B–I; Supplemental Figures 5 and 6 for mice). Note that, although the sepsis timeline could be divided into early and late phases in both humans and mice, the actual time scale is likely different between the 2 species. Furthermore, defining early and late sepsis is difficult when comparing single–time-point data from different patients. More robust conclusions can be made when data are obtained from one patient at various points in the sepsis timeline.
Figure 9. Eif2ak2 expression is increased in the kidneys of septic patients.
(A–I) RNA-Seq was performed on archived human kidney biopsy samples with final pathologic diagnosis of ATN. These samples were divided into 2 groups based on clinical information: sepsis-related AKI and non–sepsis-related AKI (referred to as others; Supplemental Table 3). Sepsis* refers to 1 patient in the sepsis-related AKI group who had prolonged and repeated episodes of sepsis. Serum creatinine from these patients is shown in J. P values were determined by Student’s t test (2 sided, nonpaired). The boxplot middle line shows the median, the box edges show the 25th and 75th percentiles, and the whiskers show ×1.5 interquartile range. (K) Overview of human kidney biopsy analysis.
Discussion
The development of kidney injury in sepsis is a clinical tipping point, after which the survival rate severely declines. The mechanisms of the downward spiral leading to complete organ failure and death are poorly understood. The current paradigm views inflammation as the centerpiece of sepsis pathology (1, 5, 26–28). However, this paradigm falls short of connecting inflammation to the phenomenon of terminal “total organ shutdown.” Using high-resolution translatomics and nascent proteomics, we dissected a chain of events that was initiated by a defined single-entry point (i.e., the interaction of LPS and its receptor TLR4). This interaction resulted in extensive dynamic changes in the translatome landscape. Upregulation of the conventional MyD88-dependent NF-κB pathway was characteristic of the early phase of sepsis (1 hour). This was followed by activation of the MyD88-independent TRIF pathway, which led to the production of various antiviral proteins (4 hours). This “viral response phase” was prolonged, possibly through the sustained upregulation of amplifiers of TRIF signaling, such as CD14, as well as crosstalk between endogenous nucleotide fragments and cytoplasmic or endosomal viral sensors such as RIG-1 and TLR3. Activation of Eif2ak2 and other antiviral genes resulted in cap-dependent translation shutdown in late-phase sepsis (16 hours). This role of Eif2ak2 in sepsis extends its other well-described functions, such as those related to inflammasome biology (29). We propose that such global translation shutdown is an important cause of organ failure in sepsis.
Our analysis of the translatome using Ribo-Seq data revealed various changes in the expression of uORFs in specific genes during sepsis. The functional properties of uORFs are largely unknown and require detailed molecular investigation for each gene (30). This is because the regulation is believed to be specific to each cognate 5′ UTR mRNA structure (31–33). Functions ascribed to uORFs include slowing down the translation of CDS, creating a nidus to facilitate the recruitment of the initiation complex, and in some cases, acting as a source of short peptides that regulate translation. Elucidating specific functions of uORFs could contribute to novel therapeutic approaches to controlling inflammation and translation.
In humans, translatome analysis of renal biopsy samples is yet to be established. Indeed, special protocols need to be implemented that will allow rapid and efficient handling of the biopsy in order not to lose information about highly time-sensitive events, such as ribosomal mRNA dynamics. Nevertheless, our transcriptomics analysis of available human biopsy samples correlated well with the mouse translatome data, especially for genes that are regulated transcriptionally. As expected, the scale of the sepsis timeline differs between rodents and humans. However, the relative changes over time were comparable. Cell-type–specific responses could not be determined due to the use of bulk tissue.
In conclusion, combined examination of the translatome, transcriptome, and nascent proteome provides a new platform that allows the identification of temporal aberration of inflammation and translation in sepsis. Our data support a model in which upregulation of antiviral response pathways links the early propagation of inflammation to systemic translation shutdown in the later phases of sepsis.
Methods
Animals and cells.
Male mouse strain C57BL/6J was obtained from the Jackson Laboratory. Mice were 8–12 weeks of age and weighed 20–30 g. Animals were subjected to a single-dose, 5 mg/kg LPS tail vein i.v. injection (E. coli serotype 0111:B4, MilliporeSigma). Untreated mice received an equivalent volume of sterile, normal saline vehicle. Cecal ligation and puncture were performed under isoflurane anesthesia; 75% of the mouse cecum was ligated and punctured twice with a 27-gauge needle. Animals were resuscitated with 500 μl of normal saline s.c. No antibiotics were used. ISRIB (5 mg/kg, diluted in DMSO) was administered i.p. at the time of abdominal closure. The following reagents were also used in vivo under indicated experiments: puromycin (40 mg/kg i.p.; MilliporeSigma), O-propargyl-puromycin (an alkyne analog of puromycin, 40 mg/kg i.p.; Jena Bioscience), lactimidomycin (1 mg/kg i.p., stock in DMSO, diluted in saline; MilliporeSigma), ISRIB (5 mg/kg i.p., diluted in DMSO, MilliporeSigma), 2-aminopurine (10 mg/kg i.p., stock in PBS/glacial acetic acid 200:1, diluted in saline; MilliporeSigma), and C16 (0.16 mg/kg i.p., stock in DMSO; MilliporeSigma). RAW 264.7 murine macrophage cells were cultured using high-glucose DMEM with 10%FBS and treated with LPS (500 ng/ml), 2-aminopurine (1 mM), polyinosinic:polycytidylic acid (poly[I:C]) (50 μg/ml), or their vehicles.
Click chemistry and immunofluorescence microscopy.
Mice were injected with O-propargyl puromycin (40 mg/kg) i.p. 30 minutes before tissue harvest. Kidneys were fixed with 4% paraformaldehyde, sectioned (10 μm), and deparaffinized. Tissues were incubated for 30 minutes with 100 mM Tris Base, 1 mM CuSO4, 100 mM ascorbic acid, 100 μM Alexa Fluor 555 azide (Molecular Probes), and 0.2% Triton X-100 at room temperature (34). Tissues were then washed in PBS with 0.1% Triton X-100 for 30 minutes 3 times, counterstained with DAPI for 10 minutes, washed in PBS 3 times, and imaged with an Olympus FV1000-MPE confocal/multiphoton microscope.
Western blotting.
For in vivo puromycin incorporation into nascent proteins, mice were injected with puromcyin (40 mg/kg) i.p. and tissues were harvested 30 minutes later. Tissues were extracted with RIPA buffer with an additional 5 mM EDTA, 5 mM EGTA, Halt protease inhibitors (Pierce), phosStop inhibitor (Roche), and benzonase nuclease (EMD Millipore). Total protein levels were determined using a modified Lowry assay (Bio-Rad). Equal amounts of kidney proteins (20 μg), and in some cases, equal volumes of extracts per kidney, were mixed with NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) with 100 mM of DTT and separated by electrophoreses on NuPage 4%–12% Bis-Tris gels and transferred to PVDF membranes. Antibodies used included the following: anti-puromycin antibody (1 μg/ml; Developmental Studies Hybridoma Bank), PKR–Alexa Fluor 680 (B-10, Santa Cruz Biotechnology Inc., catalog sc-6282), p-eIF2α (Ser51, Invitrogen, catalog PIPA537800), eIF2α (Cell Signaling Technology, catalog 9722), ATF4 (Creb2) Alexa Fluor 680 (B-3, Santa Cruz Biotechnology Inc., catalog sc-390063), and histone H3 (Cell Signaling Technology, catalog 9715).
Polyribosomal profiling.
For polyribosome profiling of kidney tissues, cardiac perfusion was performed with 6 ml of cycloheximide (100 μg/ml in PBS). Harvested kidneys were immediately placed in a lysis buffer consisting of 1% Triton X-100, 0.1% deoxycholate, 20 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, EDTA-free Protease Inhibitor Cocktail Tablet (Roche), 50 μg/ml cycloheximide, and RNAsin (1:500 dilution). Tissues were homogenized at 4°C using a Precellys tissue homogenizer. Tissue homogenates were incubated on ice for 10 minutes, then centrifuged at 9,600 g for 10 minutes. The supernatant was added to the top of a sucrose gradient generated by BioComp Gradient Master (10% sucrose on top of 50% sucrose in 20 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, and 50 mg/ml cycloheximide) and centrifuged at 283,800 g for 2 hours at 4°C. The gradients were harvested from the top in a Biocomp harvester (Biocomp Instruments), and the RNA content of eluted ribosomal fractions was continuously monitored with UV absorbance at 254 nm.
Nascent proteomics.
Mice were injected with LPS i.v. at indicated time points, and O-propargyl puromycin (40 mg/kg) was injected i.p. 30 minutes before sacrifice. One kidney from each animal was homogenized in cold RIPA lysis buffer with protease inhibitor cocktail and benzonase, incubated on ice for 10 minutes, and centrifuged at 9,600 g for 10 minutes. Supernatants were mixed with 8M urea and processed for copper-based Click chemistry reaction using a Click-&-Go Protein Enrichment Kit (Click Chemistry Tools, 1151). Samples were incubated in the copper catalyst solution with azide-agarose resin for 16 hours, followed by rigorous washing, including SDS ×5, 8M urea ×10, and 20% acetonitrile ×10. Enriched proteins were digested directly from the resin, and two-thirds of the recovered volume of nascent proteins obtained from each kidney were submitted for mass spectrometry analysis. Label-free quantitative analysis was performed using the Orbitrap Fusion Lumos Tribrid mass spectrometer at the Indiana University Proteomics Core.
HPLC.
To extract nucleotides, one kidney from each anesthetized mouse was removed and immediately immersed in liquid nitrogen. Kidneys were pulverized under liquid nitrogen with mortar and pestle and suspended in 30% ice-cold acetonitrile. Supernatants were gassed with nitrogen gas to evaporate acetonitrile, and the nucleotide levels were measured by HPLC (Agilent 1100 series) with a UV detector at 254 nm. The HPLC buffer consisted of 2 mM Paired-Ion Chromatography Reagent A (Waters), 10 mM ammonium phosphate, and 20% acetonitrile, and the buffer was run isocratically at 2 ml/min. The column used was a Nova-Pack C18 4-μm, 8 × 100 mm Radial-Pak Cartridge (Waters).
Ribo-Seq analysis.
Kidneys were snap-frozen and pulverized under liquid nitrogen. In some experiments, 1 mg/kg lactimidomycin and 40 mg/kg puromycin were administered i.p., 30 minutes and 15 minutes before sacrifice, respectively. Cryo-lysis was done using a Precellys tissue homogenizer in a polysome buffer consisting of Illumina TruSeq Polysome Buffer, Riboguard (Lucigen), Triton X-100, DTT, DNAse I, cycloheximide, and NP40. Tissue lysates were centrifuged at 20,000 g for 10 minutes. Equal volumes of supernatant aliquots were made for Ribo-Seq and RNA-Seq workflows (35). RNA concentration was determined using Qubit assays. Polysome digestion was performed in polysome buffer without detergent (20 mM Tris, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml cycloheximide, 50 U Lucigen RNase-I) for 45 minutes at room temperature, and the reaction was stopped with 10 μl SUPERase (Thermo Fisher Scientific). Ribosome-protected fragments were isolated using 1M sucrose cushion plus 20 U/ml SUPERase, and centrifuged at 541,700 g at 4°C in a TLA100.4 rotor using thick wall polycarbonate tubes (Beckman) for 1 hour. RNA fragments were purified from TRIzol resuspended ribosomal pellets using the Direct-zol kit and treated with T4 polynucleotide kinase to heal the 2′ to 3′ cyclic phosphate. We then proceeded with adaptor ligation and standard library construction steps. Ribosomal RNAs were depleted using Ribo-Zero Gold (Illumina). Sequencing was performed at Cofactor Genomics using Illumina NextSeq 500 (approximately 70 million reads per library, single-end 75 bp reads). Mapping was done using bowtie2 against the mm10 transcriptome as well as the mm10 genome.
Manual and laser microdissection and microarray analysis.
Mouse kidneys were sliced, and tubular microdissection was performed in ice-cold dissection solution (135 mM NaCl, 1 mM Na2HPO4, 1.2 mM Na2SO4, 1.2 mM MgSO4, 5 mM KCl, 2 mM CaCl2, 5.5 mM glucose, 5 mM HEPES, 0.1% BSA; pH 7.4) using a Leica MZ 12.5 stereomicroscope equipped with a Leica MC 170 HD digital microscope camera and ThermaZone cooling device, as described previously (10). RNA was extracted using the PicoPure RNA Kit (Life Technologies). For S1 proximal tubule laser microdissection, kidney sections (10 μm thick) were placed on polyphenylene sulfate membrane slides (Leica). Immunofluorescence staining was performed as described previously, and dissection was performed on a Leica LMD6000 microscope (10). S1 segments were recognized as they extended from Bowman’s space. Three consecutive sections (from the same kidney tissue) were pooled to increase the amount of RNA. RNA extraction was done using the PicoPure Kit (Applied Biosystems). RNA was labeled using the standard protocol for the Affymetrix WT Pico kit. Individually labeled samples were hybridized to the Mouse Gene 2.0 ST GeneChips for 17 hours, then washed, stained, and scanned with the standard protocol, using Affymetrix GeneChip Command Console software.
PCR.
RNA extraction from snap-frozen kidneys was performed using TRIzol, and the extracted RNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Life Technologies). TaqMan gene expression assays used included kidney injury molecule-1 (Mm00506686_m1) and glyceraldehyde-3-phosphate dehydrogenase (Mm99999915_g1). Real-time quantitative PCR amplifications were performed for 40 cycles using ViiA 7 Real-Time PCR systems. The ΔΔCt method was used to analyze the relative changes in gene expression. For Xbp1 PCR, the following primers were used: Xbp1F: AGTTAAGAACACGCTTGGGAAT; Xbp1R: AAGATGTTCTGGGGAGGTGAC; S-Xbp1F: CTGAGTCCGAATCAGGTGCAG; and S-Xbp1R: GTCCATGGGAAGATGTTCTGG
Metabolomic profiling.
Metabolomic analysis was performed at Metabolon Inc. Snap-frozen kidney tissues were processed following the Metabolon standard extraction method (60% methanol). The extracts were analyzed by gas chromatography–mass spectrometry and ultra-high performance liquid chromatography–tandem mass spectrometry platforms.
Human kidney biopsy and RNA-Seq.
Archived kidney biopsy samples were retrieved from the Indiana Biobank. The database was queried for primary pathology diagnoses of ATN, and 15 cases were selected after excluding solid organ transplant and liver-failure cases. Of those 15 cases, 5 were found to have documented sepsis. Bulk tissues were subjected to RNA-Seq at the Indiana University Center for Medical Genomics Core. The Illumina TruSeq RNA Access Library Prep Kit was used for library construction. Sequences were performed with 2 × 75 bp paired-end configuration on HiSeq4000 using a HiSeq 3000/4000 PE SBS Kit (Illumina), and the sequenced data were mapped to the hg38 human genome using the STAR RNA-Seq aligner.
Cytokine/chemokine multiplex.
Analysis of kidney homogenate cytokines/chemokines was performed using Milliplex MAP Mouse Cytokine/Chemokine Magnetic Bead Panel–Premixed 32 Plex (MCYTMAG-70K-PX32; Millipore). Proteins were extracted with RIPA buffer, and total protein concentrations were adjusted to 0.4 μg/μl per well according to the manufacturer’s instructions. Analysis was performed by the Multiplex Analysis Core at Indiana University.
Data analysis.
Data were analyzed for statistical significance and visualization with R software 3.4.3. The following packages were used: ggbiplot (Figure 2B); levelplot (Figure 6B, Supplemental Table 1, Supplemental Figure 5G); Pathview (Figure 3, A and B, and Figure 7, A and B); hclust (euclidean, ward.D) and cutree (Supplemental Figure 1B, Supplemental Figure 7, and Supplemental Figure 8); clusterProfiler (Supplemental Figure 1, C and D); loess (Supplemental Figure 15D); annotationDbi, GenomicRanges, GenomicAlignments, GenomicFeatures, rbamtools, Biostrings, riboSeqR (36), biomaRt and TxDb.Mmusculus.UCSC.mm10.knownGene for Figures 4, 5, and 8 and Supplemental Figures 3–6 and 9–14; Riboprofiling (37) (Supplemental Figure 2, C–E); and bedtools and riboWaltz (Supplemental Figure 2F). Cell-type selective markers in Supplemental Figures 9–11 were based on ref. 38. RNA-Seq data and part of Ribo-Seq data were analyzed using edgeR (Figure 7A, Figure 9, A–I, and Supplemental Figure 15B). P values were adjusted with the FDR method as indicated. To calculate translation efficiency (Supplemental Figure 12), trimming was done from start to start+15 nt position and from end–15 nt position to end position in order to eliminate cumulative signals from initiation and termination events (Supplemental Figure 2E). Reads mapped to the trimmed CDS regions were counted for Ribo-Seq and RNA-Seq. Genes with relatively low expression were filtered out (<100 reads), and 1 protein coding transcript per gene is shown (predominantly Ensembl 201 transcripts) using ggplots.
Data availability.
The Ribo-Seq and transcriptomics data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE120877, GSE120879, GSE122274). The proteomics data are provided in Supplemental Table 2.
Study approval.
All animal protocols were approved by the Indiana University Institutional Animal Care Committee and conform to the NIH (Guide for the Care and Use of Laboratory Animals, National Academies Press, 2011). The study in humans was approved by the Indiana University Institutional Review Board (protocol no. 1601431846). As only archived human biopsies were used in this study, the Institutional Review Board determined that informed consent was not required.
Author contributions
TH, BM, and PCD designed and coordinated the study. TH, BM, FS, AZ, and ZP performed experiments. TH and JM performed data analyses. TH, BM, and PCD interpreted data. MTE provided clinical data and human kidney biopsy samples. TH and PCD wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Raghu Mirmira (Center for Diabetes and Metabolic Diseases, Indiana University School of Medicine) for polysomal profiling analysis. RNA sequencing and microarray analyses were performed by staff at the Center for Medical Genomics at the Indiana University School of Medicine. Proteomics analysis was performed by staff at the Proteomics Core at the Indiana University School of Medicine. We also thank staff at the Multiplex Analysis Core at the Indiana University Melvin and Bren Simon Cancer Center for providing support in sample analysis. Measurement of serum creatinine concentration was performed at the University of Texas Southwestern O’Brien Kidney Research Core Center, using capillary electrophoresis. This work was supported by NIH grant K08-DK113223 and a Paul Teschan research grant (Dialysis Clinics Inc.) to TH, R01-DK080063 and a Veterans Affairs Merit grant (1I01BX002901) to PCD, and K08-DK107864 to MTE. This work was also supported by Indiana University O’Brien Center grant P30-DK079312.
Version 1. 12/03/2018
Electronic publication
Version 2. 01/02/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(1):296–309.https://doi.org/10.1172/JCI123284.
See the related Commentary at Targeting protein translation to prevent septic kidney injury.
Contributor Information
Takashi Hato, Email: thato@iu.edu.
Bernhard Maier, Email: bfmaier@iupui.edu.
Farooq Syed, Email: fsyed@iupui.edu.
Jered Myslinski, Email: jmyslins@iu.edu.
Amy Zollman, Email: alzollma@iu.edu.
Zoya Plotkin, Email: zplotkin@iupui.edu.
References
- 1.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(21):2063. doi: 10.1056/NEJMc1312359. [DOI] [PubMed] [Google Scholar]
- 2.Uchino S, et al. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA. 2005;294(7):813–818. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- 3.Lai TS, et al. Risk of developing severe sepsis after acute kidney injury: a population-based cohort study. Crit Care. 2013;17(5):R231. doi: 10.1186/cc13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takasu O, et al. Mechanisms of cardiac and renal dysfunction in patients dying of sepsis. Am J Respir Crit Care Med. 2013;187(5):509–517. doi: 10.1164/rccm.201211-1983OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, Kurosawa S, Remick DG. The pathogenesis of sepsis. Annu Rev Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16(11):651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano D, et al. Reduction of tubular flow rate as a mechanism of oliguria in the early phase of endotoxemia revealed by intravital imaging. J Am Soc Nephrol. 2015;26(12):3035–3044. doi: 10.1681/ASN.2014060577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hato T, et al. The macrophage mediates the renoprotective effects of endotoxin preconditioning. J Am Soc Nephrol. 2015;26(6):1347–1362. doi: 10.1681/ASN.2014060561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalakeche R, et al. Endotoxin uptake by S1 proximal tubular segment causes oxidative stress in the downstream S2 segment. J Am Soc Nephrol. 2011;22(8):1505–1516. doi: 10.1681/ASN.2011020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hato T, et al. Endotoxin preconditioning reprograms S1 tubules and macrophages to protect the kidney. J Am Soc Nephrol. 2018;29(1):104–117. doi: 10.1681/ASN.2017060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyer EE, Moore MJ. Redefining the translational status of 80S monosomes. Cell. 2016;164(4):757–769. doi: 10.1016/j.cell.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 2015;12(2):147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García MA, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70(4):1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai JC, et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science. 2018;359(6383):eaaq0939. doi: 10.1126/science.aaq0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015;4:05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingrand S, Barrier L, Lafay-Chebassier C, Fauconneau B, Page G, Hugon J. The oxindole/imidazole derivative C16 reduces in vivo brain PKR activation. FEBS Lett. 2007;581(23):4473–4478. doi: 10.1016/j.febslet.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Hato T, et al. Two-photon intravital fluorescence lifetime imaging of the kidney reveals cell-type specific metabolic signatures. J Am Soc Nephrol. 2017;28(8):2420–2430. doi: 10.1681/ASN.2016101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleich S, et al. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512(7513):208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandin V, et al. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455(7213):684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brina D, et al. eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat Commun. 2015;6:8261. doi: 10.1038/ncomms9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142(3):368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 26.Deutschman CS, Tracey KJ. Sepsis: current dogma and new perspectives. Immunity. 2014;40(4):463–475. doi: 10.1016/j.immuni.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 27.van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017;17(7):407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 28.Zarjou A, Agarwal A. Sepsis and acute kidney injury. J Am Soc Nephrol. 2011;22(6):999–1006. doi: 10.1681/ASN.2010050484. [DOI] [PubMed] [Google Scholar]
- 29.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olexiouk V, Crappé J, Verbruggen S, Verhegen K, Martens L, Menschaert G. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016;44(D1):D324–D329. doi: 10.1093/nar/gkv1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weingarten-Gabbay S, et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351(6270):aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 33.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012;109(2):413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGlincy NJ, Ingolia NT. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung BY, et al. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-Seq data analysis. RNA. 2015;21(10):1731–1745. doi: 10.1261/rna.052548.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popa A, et al. RiboProfiling: a Bioconductor package for standard Ribo-Seq pipeline processing. F1000Res. 2016;5:1309. doi: 10.12688/f1000research.8964.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. doi: 10.1101/348615. doi: 10.1101/348615. Clark J, Chen L, Chou C, Jung H, Knepper MA. Cell-type selective markers represented in whole-kidney RNA-Seq data. BioRχiv. Published June 15, 2018. Accessed November 27, 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ribo-Seq and transcriptomics data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE120877, GSE120879, GSE122274). The proteomics data are provided in Supplemental Table 2.