Abstract
Acetaldehyde dehydrogenase 2 (ALDH2) is a mitochondrial enzyme detoxifying acetaldehyde and endogenous lipid aldehydes; previous studies suggest a protective role of ALDH2 against cardiovascular disease (CVD). Around 40% of East Asians carrying the single nucleotide polymorphism (SNP) ALDH2 rs671 have an increased incidence of CVD. However, the role of ALDH2 in CVD beyond alcohol consumption remains poorly defined. Here we report that ALDH2/LDLR double knockout (DKO) mice have decreased atherosclerosis compared with LDLR-KO mice, whereas ALDH2/APOE-DKO mice have increased atherosclerosis, suggesting an unexpected interaction of ALDH2 with LDLR. Further studies demonstrate that in the absence of LDLR, AMPK phosphorylates ALDH2 at threonine 356 and enables its nuclear translocation. Nuclear ALDH2 interacts with HDAC3 and represses transcription of a lysosomal proton pump protein ATP6V0E2, critical for maintaining lysosomal function, autophagy, and degradation of oxidized low-density lipid protein. Interestingly, an interaction of cytosolic LDLR C-terminus with AMPK blocks ALDH2 phosphorylation and subsequent nuclear translocation, whereas ALDH2 rs671 mutant in human macrophages attenuates this interaction, which releases ALDH2 to the nucleus to suppress ATP6V0E2 expression, resulting in increased foam cells due to impaired lysosomal function. Our studies reveal a novel role of ALDH2 and LDLR in atherosclerosis and provide a molecular mechanism by which ALDH2 rs671 SNP increases CVD.
Keywords: Cell Biology, Metabolism
Keywords: Atherosclerosis, Cardiovascular disease, Macrophages
Introduction
Atherosclerosis is the major cause of cardiovascular disease (CVD), and macrophages play a central role in progression of atherosclerosis (1, 2). In the early stage of atherosclerosis, macrophages take up the oxidized low-density lipid protein (ox-LDL) through scavenger receptors and degrade it in lysosomes. The importance of lysosomal functions in macrophage foam cell formation has been increasingly recognized, and recent studies identified that autophagy and lysosome function were critically involved in cholesterol metabolism and that the overexpression of cholesteryl ester hydrolase in macrophages significantly reduces atherosclerosis (3, 4). Dysfunctional lysosomes may lead to the accumulation of lipids and foam cell formation, an early hallmark of atherosclerosis (5, 6). Furthermore, a recent study explored autophagy-lysosomal biogenesis as a potential therapeutic strategy for atherosclerosis (5, 7).
LDL receptor (LDLR) is an important factor for maintaining cholesterol homeostasis. LDLR is a cell-surface receptor that removes the cholesterol-rich LDL from plasma and maintains the cholesterol level in the blood circulation (8). The protein is encoded by the LDLR gene on chromosome 19, synthesized in ER, matured in Golgi, and finally transferred to the cytoplasm membrane. LDLR has a close relationship with the development of atherosclerosis. Human subjects with at least 1 LDLR abnormal allele manifest a high LDL-C level in their blood, termed familial hypercholesterolemia (FH), with markedly increased risk of coronary artery disease (CAD) due to the inefficient removal of LDL from the circulation (9, 10). Two mouse models, LDLR-deficient (LDLR-KO) (11) and apoprotein E–deficient (APOE-KO), have been extensively used for studying atherosclerosis since the 1990s (12). Moreover, proprotein convertase subtilisin/kexin type 9 (PCSK9) binds to LDLR and accelerates its degradation. PCSK9 has been explored as an attractive drug target for treating hypercholesterolemia and atherosclerosis (13, 14).
Acetaldehyde dehydrogenase 2 (ALDH2) is the mitochondrial isoform of aldehyde dehydrogenases, and it plays a key role in the metabolism of ethanol-derived acetaldehyde and other toxic lipid aldehydes generated from lipid peroxidation under oxidant stress (15). Some previous evidence suggests that ALDH2 protects against CVD through detoxification of endogenous lipid aldehydes, such as 4-hydroxy-2-nonenal (4-HNE) (16) and inhibiting inflammation in endothelial cells (ECs) (17). Moreover, 30%–50% of East Asians possess an ALDH2 rs671 mutant, which is responsible for an alcohol-flush reaction due to the poor metabolism of ethanol resulting from substantial loss of ALDH2 enzymatic activity in heterozygotes and homozygotes carrying the ALDH2 single nucleotide polymorphism (SNP) rs671 (18, 19). Epidemiological studies have shown that humans carrying the ALDH2 rs671 SNP are positively correlated with an increased risk of CVD, and the toxic effects of alcohol-derived acetaldehydes have been postulated to be primarily responsible for this cardiovascular liability (20). Paradoxically, however, humans with ALDH2 SNP tend to consume much less alcohol compared with noncarriers. The underlying mechanisms related to CVD beyond alcohol consumption remain largely unexplored.
In this study, we discovered what we believe is a novel proatherogenic effect of ALDH2, independent of its enzymatic activity. We made a serendipitous observation in that ALDH2/LDLR double knockout (DKO) mice had significantly decreased atherosclerotic plaques compared with LDLR-KO mice, which is opposite to ALDH2/APOE-DKO mice. Macrophages are the major cell types responsible for this phenotype. We further demonstrated that ALDH2 macrophages promote atherosclerosis by causing impaired lysosomal function and autophagy through interaction with AMPK, which leads to increased lipid deposition and foam cell formation due to impaired cholesterol hydrolysis in lysosome. LDLR with its cytosolic C-terminus blocks the interaction of ALDH2 and AMPK. Furthermore, even in the presence of LDLR, human ALDH2 rs671 mutant attenuates the interaction of LDLR and ALDH2 and increases the interaction of AMPK and ALDH2, which recapitulates the phenotype in LDLR-KO mice with increased atherosclerosis. Our study has identified what we believe is a novel molecular mechanism by which interaction of ALDH2 and LDLR regulates macrophage foam cell formation. This mechanism may apply to humans with ALDH2 SNP rs671 who have an increased incidence of CVD independent of alcohol consumption. Furthermore, our study warrants future investigation into whether AMPK activation by the widely used antidiabetic medication metformin potentially increases the risk of atherosclerosis in humans with ALDH2 rs671 SNP.
Results
ALDH2/LDLR-DKO mice attenuate atherosclerotic plaque formation compared with LDLR-KO mice, whereas ALDH2/APOE-DKO mice show increased plaque areas compared with APOE-KO mice.
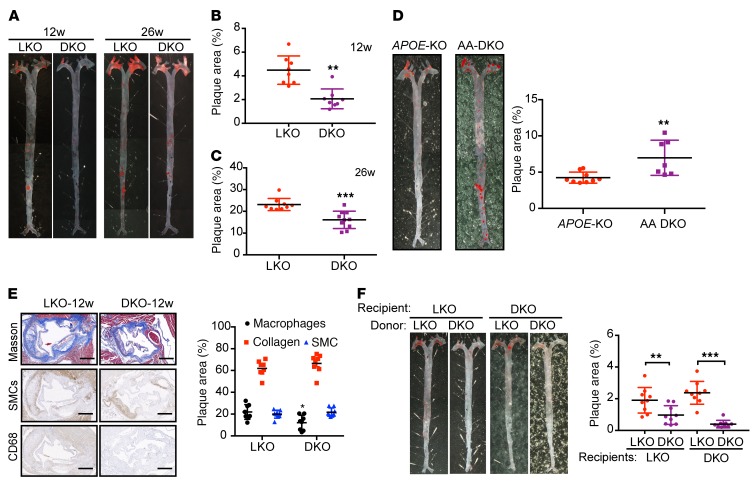
To investigate the role of ALDH2 in atherosclerosis, we crossed ALDH2-KO mice onto LDLR-KO background mice to generate ALDH2/LDLR-DKO mice and then fed them a Western diet (WD) for 12 or 26 weeks. Surprisingly, when compared with LDLR-KO mice, male ALDH2/LDLR-DKO mice had significantly decreased en face areas of atherosclerotic lesions in aorta at 2 different time points, even though body weight (BW), ratio of heart weight to body weight, level of total glycerides (TG), and total cholesterol (TC) were not significantly changed (Figure 1A; 4.13% ± 0.52% vs. 2.06% ± 0.30%, P < 0.01, n = 8 at 12 weeks, Figure 1B; 23.12% ± 0.93% vs. 15.31% ± 1.13%, P < 0.001, n = 9–10 at 26 weeks, Figure 1C) (Supplemental Figure 1, A–D, n = 9–10; supplemental material available online with this article; https://doi.org/10.1172/JCI122064DS1). To validate the phenotype, we replicated the 26-week time point and found a similar trend (ALDH2/LDLR-DKO vs. LDLR-KO: 20.40% ± 1.41% vs. 15.16% ± 0.78%, P < 0.01, n = 15) (Supplemental Figure 1E). And consistent with en face results, H&E staining for aortic root lesions showed that LDLR-KO mice had more aortic root lesions than ALDH2/LDLR-KO mice (male, 12 weeks and 26 weeks, Supplemental Figure 1F). The same trends were observed for female mice (Supplemental Figure 1G; 3.81% ± 0.37% vs. 2.06% ± 0.26%, P < 0.01, n = 9 at 12 weeks, Supplemental Figure 1H; 14.59% ± 1.01% vs. 9.923% ± 0.93%, P < 0.01, n = 8, at 26 weeks, Supplemental Figure 1I). These observations were contrary to those in ALDH2/APOE-DKO mice, in which ALDH2/APOE-KO had a significantly increased atherosclerotic area compared with APOE-KO (4.24% ± 0.25% vs. 6.98% ± 0.92%, P < 0.01, n = 7–9 at 12 weeks, Figure 1D), consistent with a previous study in which ALDH2 knockdown by ALDH2-RNAi lentivirus promoted atherosclerosis in APOE-KO mice with increased plaque area, more macrophage infiltration, less collagen, and smooth muscle cells (Supplemental Figure 2A) (17).
Figure 1. ALDH2-KO decreases areas of atherosclerotic plaque in LDLR-KO background mice, but increases areas of atherosclerotic plaque in APOE-KO background mice.
(A) Representative en face Sudan IV staining. (B, C) Quantification of plaque areas of aortas from male mice fed WD for 12 weeks (B, n = 8) and 26 weeks (C, n = 9–10, conducted twice). (D) Representative en face Sudan IV staining (n = 7–9) and quantification of Sudan IV–positive areas of aortas from male APOE-KO and APOE/ALDH2-DKO (AA-DKO) mice fed WD for 12 weeks. (E) Representative IHC and quantification of macrophages, collagen, and SMCs after 12 weeks of WD feeding (n = 8). Scale bar: 400 μm. (F) Representative en face Sudan IV staining and quantification of Sudan IV–positive areas of aortas from male LDLR-KO mice (left, n = 9) and LDLR/ALDH2-DKO mice (right, n = 9) transplanted with LDLR-KO and ALDH2/LDLR-DKO bone marrow. Statistical comparisons were made using a 2-tailed Student’s t test. All data are mean ± SD. **P < 0.01, ***P < 0.001. Original magnification for A, D, and F: ×6.3.
We next examined the cell types responsible for the decreased plaque formation in ALDH2/LDLR-DKO mice by immunohistochemical (IHC) staining of collagen (using Masson staining), α-smooth muscle cells (SMCs), and macrophages (CD68) in mouse aorta. In ALDH2/LDLR-DKO mice, we found that macrophages were significantly decreased after 12 weeks (P < 0.05, Figure 1E) and 26 weeks (P < 0.05, Supplemental Figure 2B) of WD feeding, whereas the SMCs were slightly decreased only at 26 weeks. To further examine the roles of macrophages in decreasing atherosclerosis with ALDH2-KO in LDLR-KO background, we transplanted the bone marrow of ALDH2/LDLR-DKO and LDLR-KO mice to LDLR-KO and ALDH2/LDLR-DKO mice, respectively, and fed the mice a WD for 12 weeks after 8 weeks of recovery. Analysis of en face atherosclerotic lesions showed that transplanting ALDH2/LDLR-DKO bone marrow to LDLR-KO mice decreased the formation of atherosclerotic plaque compared with transplanting bone marrow from LDLR-KO to LDLR-KO mice (1.9% ± 0.27% vs. 0.97% ± 0.20%, P < 0.01, n = 9, Figure 1F). Consistently, transplanting bone marrow from ALDH2/LDLR-DKO to ALDH2/LDLR-DKO mice resulted in decreased atherosclerotic plaques compared with transplanting bone marrow from LDLR-KO to ALDH2/LDLR-DKO mice (2.317% ± 0.43% vs. 0.49% ± 0.13%, P < 0.01, n = 9, Figure 1F). All these data show that macrophages are primarily responsible for the decreased atherosclerosis in ALDH2/LDLR-DKO mice compared with LDLR-KO mice.
Taken together, ALDH2-KO on a LDLR-KO background significantly decreased the atherosclerotic plaque formation, contrary to the phenotypes observed in ALDH2-KO on APOE-KO background, suggesting an unexpected mechanism involving an interaction of ALDH2 with LDLR in macrophages.
ALDH2 regulates macrophage foam cell formation in an LDLR-dependent manner through modulation of lysosomal function and cholesteryl ester (CE) hydrolysis.
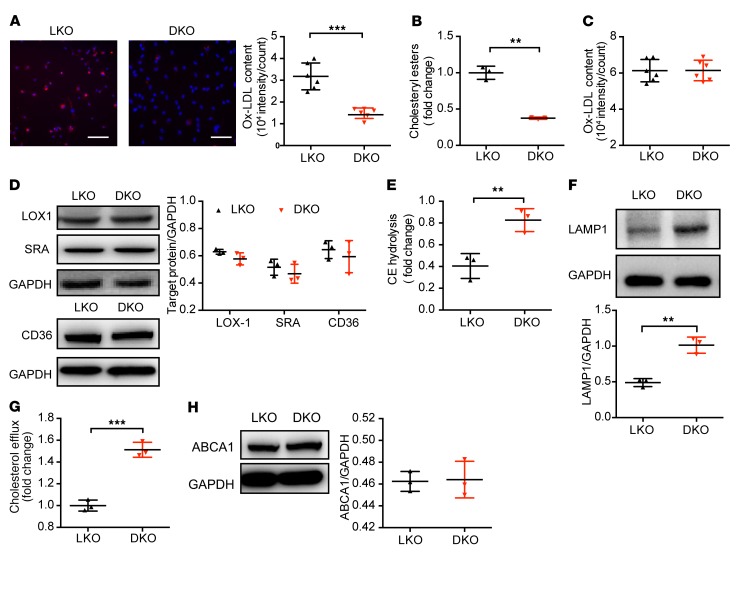
It is well established that macrophage foam cells are formed from excessive accumulation of ox-LDL. We examined the macrophage phagocytotic capability with different genetic backgrounds and found that when compared with ALDH2/LDLR-DKO, LDLR-KO led to increased levels of ox-LDL and CE, a hallmark of macrophage foam cells (Figure 2, A and B), whereas ALDH2-KO had no effect on ox-LDL and CE in the presence of LDLR (Supplemental Figure 3, A and B), consistent with the hypothesis that ALDH2 affects foam cell formation by interaction with LDLR. These in vitro observations were consistent with the decreased plaque formation in ALDH2/LDLR-DKO compared with LDLR-KO mice.
Figure 2. ALDH2/LDLR-DKO macrophages exhibit decreased foam cells compared with LDLR-KO macrophages through restoring the lysosomal function and increased CE hydrolysis, without affecting ox-LDL binding and ABCA1 expression.
(A) ALDH2-KO decreases foam cell formation in LDLR-KO (LKO) BMDMs. Quantification is also shown (n = 6). The ox-LDL signals are shown in red and Hoechst signals are shown in blue. Scale bar: 100 μm. (B) CE decreased in ALDH2/LDLR-DKO (DKO) BMDMs compared with LKO macrophages (n = 3). (C) Binding to ox-LDL in LKO and ALDH2/LDLR-DKO BMDMs (n = 6). (D) Expression of LOX1, SRA, and CD36 in LKO and ALDH2/LDLR-DKO BMDMs (n = 3). (E) CE hydrolysis significantly increased in ALDH2/LDLR-DKO BMDMs compared with LKO macrophages (n = 3). (F) Expression of lysosome function marker LAMP1 in LDLR-KO and ALDH2/LDLR-DKO macrophages (n = 3). (G) Cholesterol efflux increased by ALDH2-KO in LDLR-KO BMDMs (n = 3). (H) ABCA1 expression in LKO and DKO BMDMs (n = 3). Statistical comparisons were made using 2-tailed Student’s t test. All data are mean ± SD. **P < 0.01, ***P < 0.001.
Macrophages play an important role in cholesterol homeostasis through balancing ox-LDL uptake, degradation (hydrolysis in lysosome), and efflux. Dysregulation of any of these processes may eventually lead to foam cell formation. We investigated whether the ox-LDL binding was regulated by ALDH2 and found that the binding of ox-LDL did not change significantly in LDLR-KO and ALDH2/LDLR-DKO macrophages or WT and ALDH2-KO macrophages (Figure 2C and Supplemental Figure 3C), consistent with the similar expression levels of the 3 major scavenger receptors responsible for ox-LDL uptake (LOX1, SRA, and CD36) (Figure 2D and Supplemental Figure 3D). These results suggest that the interaction of ALDH2 with LDLR does not affect binding of ox-LDL in macrophages. Then, we measured the hydrolysis of CE and the expression of lysosome function markers (LAMP1) in macrophages from LDLR-KO and ALDH2/LDLR-DKO mice after treatment with ox-LDL (Figure 2, E and F). Interestingly, CE hydrolysis and LAMP1 expression were significantly decreased in LDLR-KO macrophages compared with those from ALDH2/LDLR-DKO macrophages (Figure 2, E and F), but were not changed in WT and ALDH2-KO macrophages (Supplemental Figure 3, E and F), suggesting that in the absence of LDLR, ALDH2 impairs lysosomal function and CE hydrolysis but this effect is lost when LDLR is present. We measured cholesterol efflux in different genetic macrophages and found that cholesterol efflux was also increased in ox-LDL–loaded ALDH2/LDLR-DKO macrophages compared with LDLR-KO macrophages (Figure 2G), but it was not significantly changed in WT and ALDH2-KO macrophages (Supplemental Figure 3G). Interestingly, however, the expressions of the transporter responsible for cholesterol efflux, ABCA1, and enzyme for cholesteryl ester formation, ACAT1, were not significantly changed (Figure 2H, Supplemental Figure 3H, and Supplemental Figure 4), suggesting that the increased efflux was most likely due to the increased lysosomal hydrolysis of CE instead of subsequent reesterification of cholesterol and efflux.
Collectively, these data demonstrate that the interaction of ALDH2 and LDLR affects foam cell formation primarily through modulating macrophage lysosomal function (see below) without changing the binding, reesterification of cholesterol, or expressions of transporter ABCA1 for cholesterol efflux.
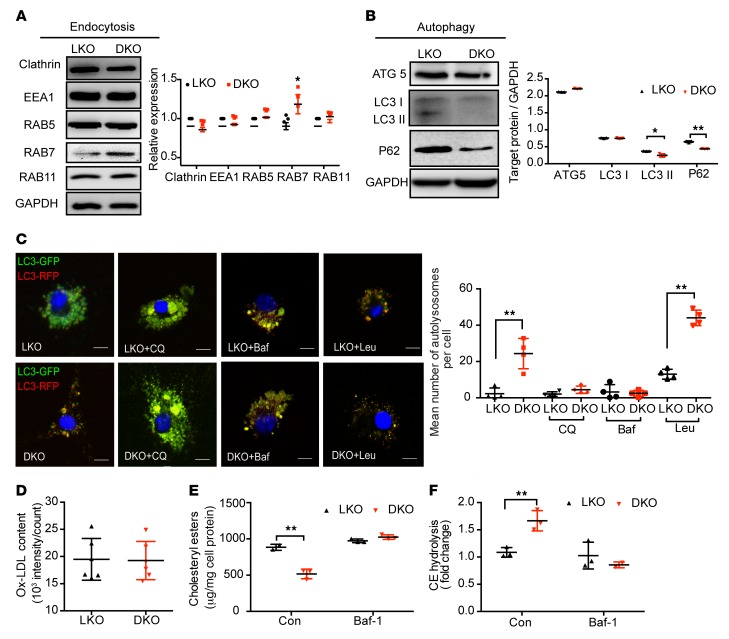
Decreased macrophage foam cell formation in ALDH2/LDLR-DKO compared with LDLR-KO mice is due to the increased lysosomal function and autophagy.
Emerging evidence demonstrates that endocytosis and autophagy are important for ox-LDL metabolism in macrophages. We examined how these 2 functions were involved in the decreased foam cell formation in ALDH2/LDLR-DKO macrophages due to the interaction of LDLR and ALDH2. We found that expression of RAB7 was significantly elevated (Figure 3A) whereas the protein levels of LC3 II and P62 were significantly decreased in ALDH2/LDLR-DKO macrophages compared with those from LDLR-KO mice (Figure 3B), suggesting that ALDH2/LDLR-DKO rescued the impaired autophagy by increasing lysosomal function which leads to an increased endocytosis (increased expression of RAB7, Figure 3A and Supplemental Figure 5A) and autophagic flux (decreased expression of P62, Figure 3B). Consistently, fluorescent confocal microscopy indicated that the number of autolysosomes (RFP, indicator of an acidic environment, which is critical for maintaining normal lysosome function) was increased in ALDH2/LDLR-DKO macrophages and that this increase was completely diminished by treatments with autophagic inhibitors chloroquine (CQ) or bafilomycin A1 (Baf-A1) to neutralize lysosomal pH (Figure 3C). Interestingly, leupeptin A (Leu), an inhibitor of autophagy without affecting lysosomal pH, did not affect the increase of autolysosome numbers in ALDH2/LDLR-DKO macrophages compared with LDLR-KO macrophages, suggesting that ALDH2 regulates autophagy through modulation of lysosomal function. Moreover, Baf-A1 also diminished the increased uptake of ox-LDL in LDLR-KO macrophages to the same level as those in ALDH2/LDLR-DKO macrophages (Figure 3D). These observations agreed with the fact that compared with ALDH2/LDLR-DKO macrophages, accumulation of CE (caused by the decreased CE hydrolysis in LDLR-KO macrophages and Baf-A1 treatment) completely eliminated these differences (Figure 3, E and F). Interestingly, autophagic flux is inhibited in ALDH2-KO macrophages compared with WT macrophages without a significant change of endocytosis, ox-LDL uptake, CE content, and hydrolysis (Supplemental Figure 5, C–F), consistent with the hypothesis that ALDH2 interacts with LDLR and regulates autophagy. Taken together, these results demonstrate that impaired lysosomal function is responsible for foam cell formation in LDLR-KO macrophages, and that this effect is eliminated in ALDH2/LDLR-DKO mice, suggesting that ALDH2 interacts with LDLR to cause lysosomal dysfunction and foam cell formation in macrophages through some unknown mechanism.
Figure 3. LDLR/ALDH2-DKO leads to increased macrophage endocytosis, autophagy, and CE hydrolysis in lysosome compared with LDLR-KO.
(A and B) Endocytosis (A) and autophagy (B) are increased in ALDH2/LDLR-DKO macrophages compared with LDLR-KO (LKO; n = 3). (C) The number of autolysosomes is increased in ALDH2/LDLR-DKO macrophages compared with LKO (n = 4). Scale bars: 5 μm. (D–F) Inhibition of autophagy by Baf-A1 treatment diminishes increased foam cell formation (D, n = 6) and cholesteryl ester accumulation (E, n = 3) due to impaired CE hydrolysis (F, n = 3) in LKO macrophages compared with those from DKO. Statistical comparisons were made using 2-tailed Student’s t test. All data are mean ± SD. *P < 0.05, **P < 0.01.
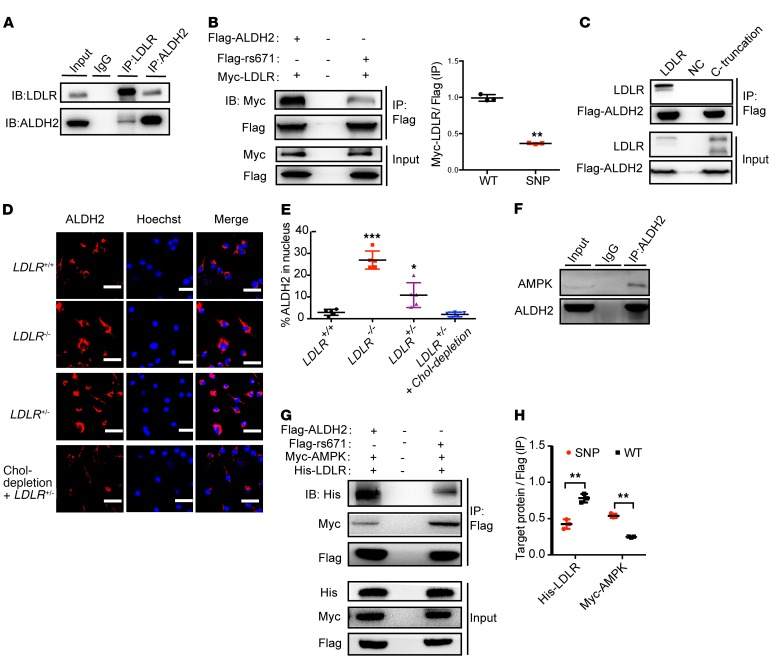
LDLR modulates nuclear translocation of ALDH2 through physical binding to ALDH2, and ALDH2 rs671 mutant decreases this interaction.
To explore the underlying mechanism by which ALDH2 regulates macrophage foam cell formation through interaction with LDLR, we first examined ALDH2 enzymatic activity in macrophages from mice with different genetic backgrounds. We found that knocking out ALDH2 alone, or an absence of LDLR (ALDH2/LDLR-DKO), almost completely destroyed the ALDH2 activity (Supplemental Figure 5G), whereas ALDH2 activity was not changed in WT and LDLR-KO macrophages. However, foam cell formation was significantly attenuated in ALDH2/LDLR-DKO macrophages compared with LDLR-KO macrophages, suggesting that LDLR and ALDH2 interactions on foam cell formation are independent of ALDH2 enzymatic function. These observations prompted us to hypothesize that ALDH2 may exert a nonenzymatic function, presumably by translocation to the nucleus, to regulate genes important for lysosomal function and autophagy, and that LDLR blocks this nuclear translocation by physically binding to ALDH2. To support this hypothesis, we first performed coimmunoprecipitation (Co-IP) experiments with Myc-tagged LDLR and Flag-tagged ALDH2 in 293T cells. As shown in Supplemental Figure 6A, LDLR and ALDH2 appeared to physically bind together. Similar results were observed in primary macrophage cells (Figure 4A). Importantly, ALDH2 rs671 SNP mutant attenuates the binding with LDLR (Figure 4B). To investigate how LDLR interacts with ALDH2, we truncated the C-terminus of LDLR and found that this truncation caused complete loss of binding to ALDH2 (Figure 4C), whereas a mutant (N812A) at the C-terminus of LDLR significantly attenuated the binding of ALDH2 to LDLR (Supplemental Figure 6B), suggesting that the C-terminus in the cytosolic region of LDLR is the critical region of binding to ALDH2. Next, we carried out immunofluorescence experiments and found out that ALDH2 translocated to the nucleus in the absence of LDLR, whereas LDLR expression in WT inhibited this translocation (Supplemental Figure 6C). Similarly, a gene dose effect was observed for LDLR to inhibit the nuclear translocation of ALDH2: heterozygous LDLR+/– has an intermediate effect compared with LDLR-KO. In addition, upregulation of LDLR by cholesterol depletion has a similar effect on inhibition of ALDH2 nuclear translocation as on that in WT macrophages (Figure 4, D and E). Interestingly, ALDH2 rs671 mutant promoted ALDH2 translocation even in the presence of LDLR (Supplemental Figure 6, D and E). Together, these data support the hypothesis that LDLR regulates the nuclear translocation of ALDH2 through physical binding to ALDH2, whereas ALDH2 rs671 mutant decreases this interaction.
Figure 4. LDLR inhibits but ALDH2 rs671 mutant increases nuclear translocation of ALDH2 through interaction with AMPK.
(A) LDLR directly interacts with ALDH2 in BMDMs (n = 3). (B) ALDH2 rs671 mutant pulls down much less LDLR compared with WT ALDH2 (n = 3). (C) ALDH2 does not bind to LDLR when LDLR C-terminal is truncated (n = 3). (D and E) LDLR gene-dose–dependent inhibition of ALDH2 translocation. (D) LDLR upregulation decreased ALDH2 translocation by cholesterol depletion. Scale bars: 100 μm. Quantification is shown in E (n = 5). (F) ALDH2 directly binds to AMPK in LDLR-KO BMDMs. (G and H) ALDH2 rs671 mutant pulls down more AMPK compared with WT ALDH2 by cotransfection of Flag-tagged ALDH2, Myc-tagged AMPK, and His-tagged LDLR (G) and quantification (H, n = 3). Statistical comparisons were made using 2-tailed Student’s t test (B and H) or ANOVA (E). All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
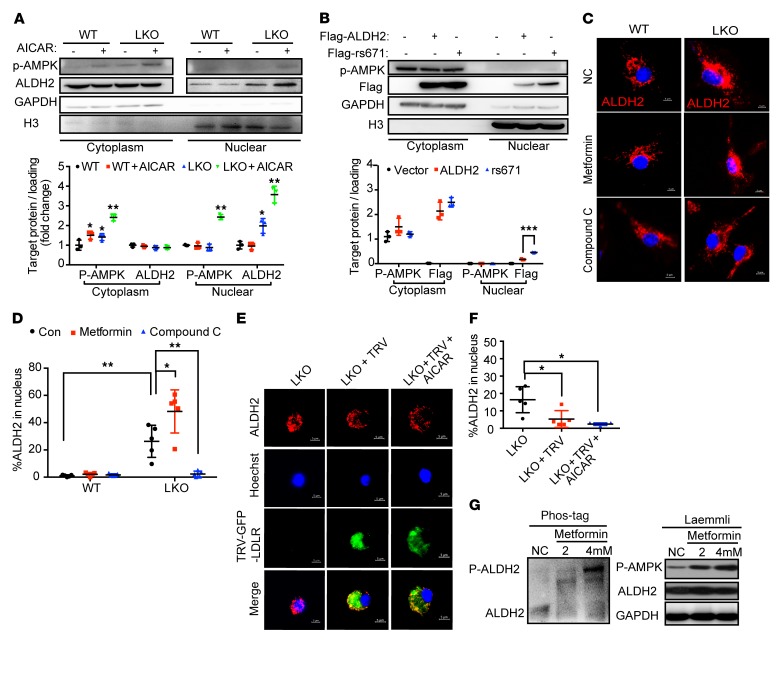
AMPK promotes ALDH2 translocation by phosphorylating ALDH2 in macrophages in the absence of LDLR or ALDH2 rs671 mutant.
Previous studies suggested that ALDH2 was a substrate for AMPK (21), and we hypothesized that ALDH2 phosphorylated by AMPK played an important role in regulating ALDH2 nuclear translocation. To determine whether ALDH2 was phosphorylated by AMPK in LDLR-KO macrophages, we performed high-resolution mass spectrometry–based phosphoproteomics to identify the potential phosphorylation sites of ALDH2 in LDLR-KO macrophages. In LDLR-KO macrophages, we identified 2 phosphorylation sites: threonine 356 (T356, Supplemental Figure 7A) and tyrosine 148 (Y148, Supplemental Figure 7B). Furthermore, in LDLR-KO macrophages treated with AMPK agonist AICAR (5-aminoimidazole-4-carboxamide1-β-D-ribofuranoside), we confirmed the phosphorylation at T356 (Supplemental Figure 7A), demonstrating that AMPK phosphorylates ALDH2 at T356 in LDLR-KO macrophages. To provide further evidence that AMPK could phosphorylate ALDH2, we performed Co-IP experiments and demonstrated that ALDH2 indeed interacted with AMPK in 293T cells (Supplemental Figure 8A) and LDLR-KO bone marrow–derived macrophages (BMDMs) (Figure 4F). Moreover, ALDH2 rs671 mutant increased the binding of ALDH2 and AMPK but decreased the binding of ALDH2 and LDLR (Figure 4, G and H; Supplemental Figure 8B). Interestingly, we also observed a gene dose effect of LDLR on inhibition of ALDH2 phosphorylation in LDLR–/– and LDLR+/– macrophages: increased expression of LDLR from –/– to +/– led to decreased ALDH2 phosphorylation, and upregulation of LDLR by cholesterol depletion further inhibited ALDH2 phosphorylation (Supplemental Figure 8C). Next, to investigate whether AMPK enabled translocation of ALDH2 to the nucleus, we performed nuclear fractionation experiments and found that more ALDH2 translocated into the nucleus in LDLR-KO BMDMs with AICAR treatment (Figure 5A). Moreover, ALDH2 rs671 mutant promoted ALDH2 translocation, which is consistent with the increased binding with AMPK (Figure 5B). To further confirm that AMPK promoted ALDH2 translocation, we performed a time course study with AMPK activation and demonstrated that ALDH2 initially resided in mitochondria, and that AMPK activation by AICAR enabled the translocation from mitochondria to nucleus as indicated by the increased green fluorescence at 60 and 90 minutes (Supplemental Figure 8D). Furthermore, a widely used antidiabetic agent and known AMPK activator, metformin, also increased ALDH2 translocation, whereas inhibition of AMPK by Compound C (CC) blocked the nuclear translocation of ALDH2 in LDLR-KO macrophages (Figure 5, C and D). Moreover, we transfected retrovirus with LDLR and GFP (GFP-TRV-LDLR) in LDLR-KO BMDMs and observed that LDLR blocked the translocation of ALDH2 to the nucleus in LDLR-KO BMDMs (Figure 5E). Furthermore, the nuclear translocation of ALDH2 can be blocked by expression of LDLR in LDLR-KO macrophages even in the presence of AICAR (Figure 5, E and F). Next, we found that metformin dose-dependently increased the phosphorylation of ALDH2 in LDLR-KO macrophages (Figure 5G) but not in WT macrophages (Supplemental Figure 8E). Taken together, these results unambiguously demonstrated that AMPK phosphorylated ALDH2 and enabled its nuclear translocation in the absence of LDLR or ALDH2 rs671 mutant.
Figure 5. AMPK phosphorylates ALDH2 and promotes ALDH2 translocation in the absence of LDLR or ALDH2 rs671 mutant.
(A) AMPK activation promotes ALDH2 nuclear translocation in LDLR-KO BMDMs by cellular fractionation (LKO; n = 3). (B) ALDH2 rs671 mutant increases the translocation of ALDH2 in 293T cells (n = 3). (C) AMPK activation leads to ALDH2 nuclear translocation, whereas inhibition of AMPK blocks nuclear translocation of ALDH2. Scale bars: 5 μm. (D) Quantification of ALDH2 in the nucleus (n = 5). (E) LDLR blocks the translocation of ALDH2 even AMPK activation. Scale bars: 5 μm. Quantification shown in F (n = 5). (G) AMPK activation leads to a dose-dependent increase of ALDH2 phosphorylation in the absence of LDLR in LKO macrophages by SuperSep Phos-tag SDS-PAGE. Statistical comparisons were made using ANOVA. All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
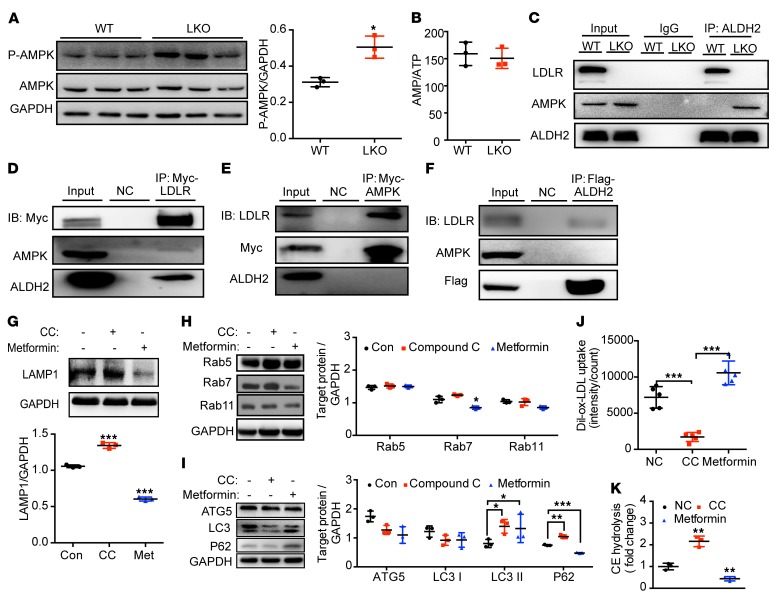
LDLR inhibits the binding of ALDH2 with AMPK, and loss of LDLR leads to the increased activation of AMPK in macrophages.
We next investigated how ALDH2, LDLR, and AMPK interact. We found that LDLR-KO increased the phosphorylation of AMPK (Figure 6A). Intriguingly, however, we did not observe significant changes for the mRNA levels of the upstream genes of AMPK: Lkb1, Camkk2, Mo25, Strad, and Tak1 (Supplemental Figure 9A). Furthermore, there is no difference for the levels of AMP, ADP, ATP, and AMP/ATP, with or without ox-LDL in LDLR-KO and WT macrophages (Figure 6B and Supplemental Figure 9, B–D). These results suggest that AMPK activation by LDLR-KO is not likely through conventional pathways. Instead, LDLR might physically interfere with the phosphorylation of AMPK through interaction with AMPK and inhibition of the binding of ALDH2 to AMPK. To support our hypothesis, we performed Co-IP experiments in WT and LDLR-KO BMDMs and found that LDLR blocked the interaction of ALDH2 and AMPK (Figure 6C). In 293T cells, we found that when ALDH2 or AMPK was overexpressed, both proteins bound to LDLR, whereas LDLR only interacted with ALDH2 and AMPK when LDLR was overexpressed (Figure 6, D–F). These results demonstrate that ALDH2 and AMPK preferentially interact with LDLR when LDLR presents, suggesting that physical interaction of LDLR and AMPK presumably blocks the phosphorylation sites of AMPK. Consistently, AMPK is activated (increased levels of P-AMPK) in the absence of LDLR without affecting the upstream genes of AMPK (Figure 6A).
Figure 6. AMPK regulates lysosomal function, endocytosis, autophagy, and foam cell formation in an LDLR-dependent manner.
(A) LDLR-KO increases the AMPK activation in BMDMs (n = 3). (B) Ratio of AMP/ATP in macrophages from LDLR-KO and WT treated with ox-LDL (n = 3). (C) LDLR inhibits the binding of ALDH2 and AMPK in macrophages. (D–F) AMPK and ALDH2 prefer to bind to LDLR. LDLR pulls down AMPK and ALDH2 (D). AMPK pulls down LDLR but not ALDH2 (E). ALDH2 pulls down LDLR but not AMPK (F). (G) AMPK activation by metformin decreases LAMP1 expression, whereas AMPK inhibition by compound C leads to increased LAMP1 expression in LDLR-KO BMDMs (LKO; n = 3). (H and I) AMPK activation by metformin decreases endocytosis (H) and autophagy (I), whereas AMPK inhibition by compound C leads to increased endocytosis (H) and autophagy (I) in LKO BMDMs (n = 3). (J) AMPK activation leads to increased foam cell formation, whereas AMPK inhibition results in decreased foam cell formation (n = 5). (K) AMPK activation decreases CE hydrolysis, whereas AMPK inhibition increases CE hydrolysis (n = 3). Statistical comparisons were made using 2-tailed Student’s t test (A and B) or ANOVA (G, H–K). All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
AMPK plays an important role in regulating endocytosis and autophagy for LDLR-regulated ox-LDL metabolism in macrophages.
To confirm the role of AMPK in regulating foam cell formation through modulating lysosomal function and autophagy, we treated LDLR-KO and WT macrophages with CC and metformin, respectively. As expected, LAMP1 expression was increased by AMPK inhibition with CC treatment but decreased by AMPK activation with metformin (Figure 6G, Supplemental Figure 10A) in ox-LDL–loaded LDLR-KO macrophages. Furthermore, to examine how AMPK regulates endocytosis and autophagy, we showed that in LDLR-KO BMDMs, AMPK activation by metformin decreased endocytosis (decreased RAB7 expression) and autophagy (increased P62 and LC3 II expression), whereas AMPK inhibition by CC led to increased endocytosis (increased RAB7 expression) and autophagy (decreased P62 and LC3 II expression) (Figure 6, H and I). Consistently, in WT BMDMs, AMPK activation by metformin increased autophagy (decreased P62), whereas AMPK inhibition by CC led to decreased autophagy (increased P62) without affecting endocytosis (Supplemental Figure 10, B and C). Notably, AMPK activation or inhibition only affects the protein levels of P62 without changing its transcription (Supplemental Figure 11). Next, we found that AMPK activation by metformin promoted foam cell formation by decreasing CE hydrolysis, whereas AMPK inhibition by CC treatment significantly attenuated foam cell formation by increasing CE hydrolysis in ox-LDL–loaded LDLR-KO macrophages (Figure 6, J and K). Together, these results demonstrate that AMPK mediates ox-LDL metabolism, autophagy, and foam cell formation through interactions with ALDH2 and LDLR.
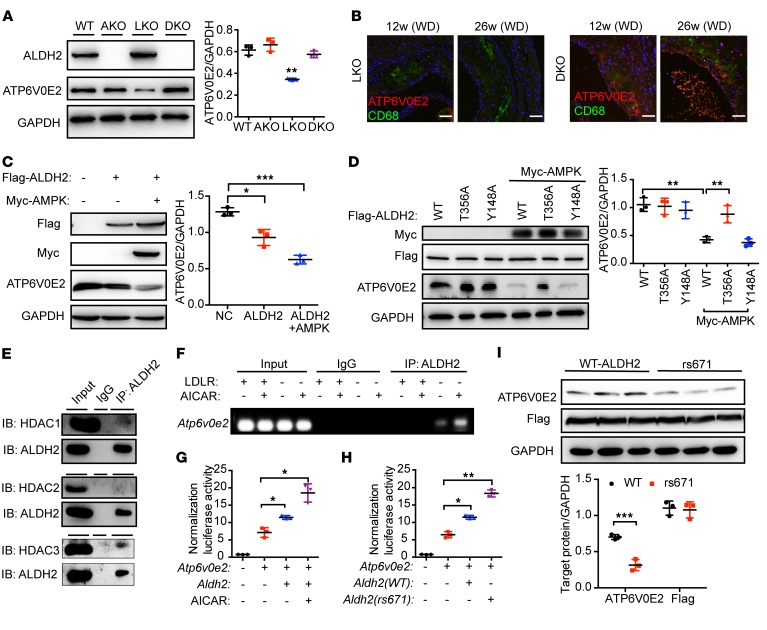
Nuclear translocated ALDH2 regulates lysosomal function through downregulation of the transcription of Atp6v0e2 in LDLR-KO or ALDH2 rs671 mutant.
To investigate the functional consequences after ALDH2 enters the nucleus, we performed transcriptomic RNA-Seq analysis to map the genes that were regulated by ALDH2 using ALDH2/LDLR-DKO and LDLR-KO macrophages (Supplemental Figures 12–15). We found that a lysosomal function–related gene, Atp6v0e2, coding H+-ATPase, was ranked as the third most upregulated gene (Supplemental Table 1). Due to the importance of lysosomal function and cholesterol efflux in macrophage metabolism of ox-LDL, ATP6V0E2 mRNA levels were further validated by quantitative PCR (Supplemental Figure 16). Expression of ATP6V0E2 was significantly regulated by LDLR. There was no significant difference between the RNA levels of Atp6v0e2 in WT and ALDH2-KO, whereas Atp6v0e2 was significantly downregulated in LDLR-KO, consistent with the increased uptake of ox-LDL in LDLR-KO and decreased expression of lysosomal biogenesis protein LAMP1 (Figure 2, A, B, and F). Interestingly, even though the mRNA level of Atp6v0e2 in ALDH2/LDLR-DKO was higher than mRNA levels in WT, ALDH2-KO, and LDLR-KO (about 20-fold for LDLR-KO), the ATP6V0E2 protein level in LDLR-KO macrophages was half of the protein levels in WT, ALDH2-KO, and ALDH2/LDLR-DKO macrophages (Figure 7A). Moreover, we examined the protein expression of ATP6V0E2 in the macrophages of aorta from LDLR-KO and ALDH2/LDLR-DKO mice fed a WD for 12 and 26 weeks, respectively, by immunofluorescence (green, CD68, marker for macrophages; red, ATP6V0E2). As shown in Figure 7B, ATP6V0E2 expression was substantially increased in aorta from ALDH2/LDLR-DKO mice compared with that from LDLR-KO mice, consistent with the hypothesis that expression of ATP6V0E2 is important for regulating foam cell formation. Moreover, when both AMPK and ALDH2 were overexpressed in 293T cells, ATP6V0E2 expression was significantly decreased (Figure 7C). Overexpressed AMPK and a mutant ALDH2 T356A site did not change ATP6V0E2 expression, whereas WT and mutant ALDH2 Y148A decreased ATP6V0E2 expression (Figure 7D). Similar results were obtained in the treatment with metformin (Supplemental Figure 17). These results demonstrated that T356, rather than Y148, in ALDH2 was responsible for regulating ATP6V0E2 expression by AMPK. Furthermore, LDLR dose-dependently decreased ALDH2 phosphorylation and increased ATP6V0E2 expression in 293T cells (Supplemental Figure 18A). Moreover, our previous data demonstrated that the LDLR C-terminus was involved in the interaction of LDLR and ALDH2 (Figure 4C and Supplemental Figure 6B). To investigate if this interaction regulates ATP6V0E2 expression, we inhibited the endocytotic pathway by knocking down the expression of clathrin heavy chain (CHC) and observed that inhibiting LDLR from entering the cytoplasm decreased ATP6V0E2 expression through downregulation of RAB5 and RAB7 (Supplemental Figure 18B). Next, to find how ALDH2 regulates ATP6V0E2, we tested the interactions of ALDH2 and the HDAC family by Co-IP in macrophages. We found that ALDH2 had stronger interaction with HDAC3 than with HDAC1 and HDAC2 in LDLR-KO BMDMs (Figure 7E). We next performed chromatin immunoprecipitation (ChIP) assay to investigate whether ALDH2 bound to HDAC3 and regulated the transcription of ATP6V0E2. We designed the primers on the promoter region stretched over a transcription start site (TSS) of ATP6V0E2, and found that ALDH2 regulated the transcription of ATP6V0E2 in LDLR-KO BMDMs and that AMPK activation by AICAR enhanced this effect (Figure 7F). Next, we constructed an ATP6V0E2 promoter-luciferase plasmid and studied luciferase activity to test whether ALDH2 could regulate the transcriptional activity of ATP6V0E2. As shown in Figure 7G, ALDH2 indeed increased the luciferase activity of ATP6V0E2, which was further enhanced by treating with AICAR in 293T cells. To further examine the role of HDAC3 in ALDH2 regulation of ATP6V0E2, we performed a ChIP experiment to measure the binding of HDAC3 to the ATP6V0E2 promoter in LDLR-KO and ALDH2/LDLR-DKO BMDMs treated with metformin. In Supplemental Figure 19A, consistent with previous data, HDAC3 only bound to the ATP6V0E2 promoter in the presence of ALDH2. We also used an HDAC3-specific inhibitor RGF966 to treat the macrophages from LDLR-KO and WT mice. Inhibition of HDAC3 led to increased levels of H3K27ac in both WT and LDLR-KO macrophages, whereas the decreased expression of ATP6V0E2 in LDLR-KO macrophages was rescued to the same levels as WT (Supplemental Figure 19B). Furthermore, ALDH2 rs671 mutant also increased the regulation of ATP6V0E2 transcriptional activity (Figure 7H) and resulted in decreased protein expression of ATP6V0E2 (Figure 7I).
Figure 7. Nuclear translocated ALDH2 regulates the transcription of ATP6V0E2, a critical protein for lysosomal function, endocytosis, and autophagy, and ALDH2 rs671 polymorphism decreases ATP6V0E2 expression.
(A) ALDH2/LDLR-DKO leads to significant upregulation of ATP6V0E2 expression compared with LDLR-KO (LKO; n = 3). (B) ATP6V0E2 (red) colocalizes with macrophages (CD68, green) and ATP6V0E2 expression is significantly increased in the aorta of DKO mice. Scale bars: 100 μm. (C) Overexpressed AMPK and ALDH2 decrease ATP6V0E2 expression in 293T cells (n = 3). (D) ALDH2 T356A not Y148A mutant rescues decreased ATP6V0E2 expression, which is caused by overexpressed ALDH2 and AMPK (n = 3). (E) Nuclear translocated ALDH2 binds to HDAC3. (F and G) Nuclear translocated ALDH2 regulates transcription of ATP6V0E2. In the absence of LDLR, nuclear translocated ALDH2 binds to ATP6V0E2 promoter (F) and regulates transcriptional activity (G), which is enhanced by AMPK activation (n = 3). (H) ALDH2 rs671 enhanced transcriptional activity of ATP6V0E2. (I) The rs671 mutant decreased ATP6V0E2 protein expression (n = 3). Statistical comparisons were made using 2-tailed Student’s t test (I) or ANOVA (A, C, D, G, and H). All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Taken together, our data demonstrate that in the macrophages of LDLR-KO or ALDH2 rs671 mutant, AMPK phosphorylates ALDH2 at T356, which enables its nuclear translocation. Once in the nucleus, ALDH2 binds to HDAC3 and suppresses the transcription and protein expression of ATP6V0E2.
Human macrophages from subjects carrying ALDH2 rs671 SNP have increased foam cell formation caused by the downregulation of ATP6V0E2 and autophagy due to increased nuclear translocation of ALDH2.
To investigate the clinical relevance of ALDH2-regulated macrophage formation, we recruited human subjects carrying ALDH2 rs671 SNP (heterozygous ALDH2*2/2*1, n = 10) and control (ALDH2*1, n = 16) to isolate peripheral blood monocytes (PBMCs) (Supplemental Table 4). After differentiation into macrophages, we treated the cells with ox-LDL and observed significantly decreased ATP6V0E2 mRNA levels in macrophages from ALDH2*2/2*1 compared with those from ALDH2*1 (Figure 8A). Furthermore, ATP6V0E2 protein levels were significantly decreased, whereas the P62 levels were increased, in ALDH2*2/2*1 compared with ALDH2*1 (Figure 8B), suggesting an impaired autophagy due to the downregulation of ATP6V0E2. Consistently, the cholesteryl ester hydrolysis was significantly attenuated in ALDH2*2/2*1 macrophages (Figure 8C). Furthermore, confocal studies clearly indicated that more ALDH2 in ALDH2*2/2*1 macrophages translocated into nucleus than in ALDH2*1 macrophages. This effect was augmented upon AMPK activation by metformin treatment (Figure 8D). These in vitro studies provide strong evidence that human ALDH2 SNP rs671 potentially promotes atherosclerosis through modulation of macrophage foam cell formation, which represents a novel molecular mechanism by which ALDH2 rs671 polymorphism increases the risk of CVD. Interestingly, our study suggests that metformin may increase the risk of atherosclerosis in humans with ALDH2 rs671 SNP, which warrants future investigation.
Figure 8. Human macrophages from subjects carrying ALDH2 rs671 SNP have increased foam cell formation caused by the downregulation of ATP6V0E2, and autophagy due to the increased nuclear translocation of ALDH2.
(A) Macrophages from ALDH2*2/2*1 (n = 10) have lower ATP6V0E2 mRNA levels than those from ALDH2*1 (n = 16). (B) ALDH2 rs671 mutant increased autophagy in human macrophages treated with ox-LDL (n = 3). (C) ALDH2 rs671 mutant decreases CE hydrolysis in human macrophages (ALDH2*1, n = 16; ALDH2*2/2*1, n = 10). (D) ALDH2 rs671 SNP increases ALDH2 translocation in human macrophages treated with metformin (n = 5). Scale bars: 5 μm. Statistical comparisons were made using 2-tailed Student’s t test. All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
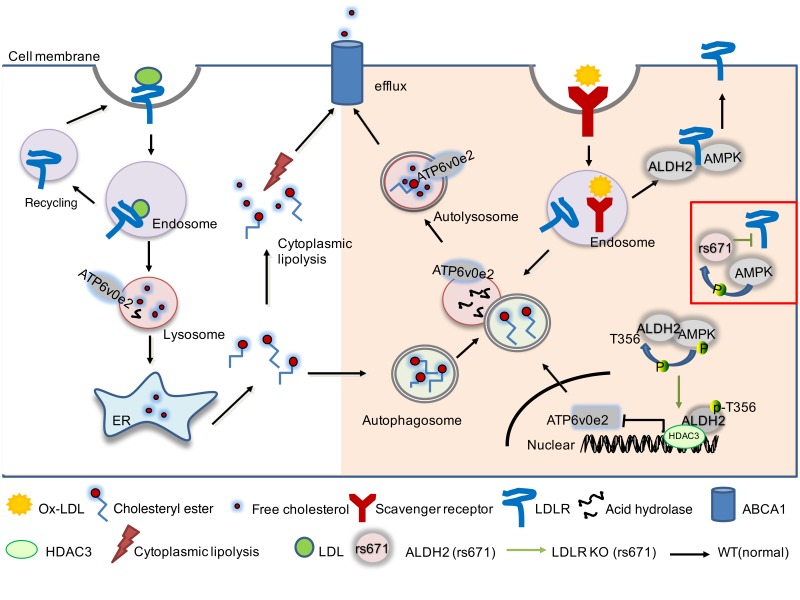
CVD remains the leading cause of death worldwide, and atherosclerosis is the major factor underlying CVD (22). A body of epidemiological evidence has linked the ALDH2 SNP rs671 (E487K) to an increased risk of CVD in around 40% of East Asians carrying this SNP, but the underlying molecular mechanisms remain poorly defined (23). ALDH2 primarily resides in mitochondria and exerts a detoxification function of ethanol-derived acetaldehyde and other endogenous lipid aldehydes, such as 4-HNE. Previous studies supported a protective role of ALDH2 in CVD based on the detoxification function and the toxic effects of acetaldehyde have been postulated to be responsible for this cardiovascular effect. Paradoxically, the carriers of ALDH2 SNP rs671 tend to consume less alcohol compared with the noncarriers; thus, the underlying mechanism for the association of ALDH2 SNP and CVD independent of alcohol remains to be investigated. Our study has identified what we believe is a novel molecular mechanism by which ALDH2 SNP rs671 increased atherosclerosis through AMPK-phosphorylated ALDH2, translocating into the nucleus and repressing gene expression of ATP6V0E2, which is responsible for maintaining normal lysosomal function, autophagy, and ox-LDL degradation (Figure 9).
Figure 9. ALDH2 regulates macrophage foam cell formation through interacting with LDLR and AMPK.
The translocation of AMPK phosphorylated ALDH2 to the nucleus to regulate expression of ATP6V0E2, a critical protein for lysosomal function, endocytosis, and autophagy.
It is well established that dysregulation of cholesterol metabolism leads to atherosclerosis. In macrophages, after binding to LDLR, LDL is transferred to lysosome through endosome, whereas ox-LDL is primarily taken up by the scavenger receptors through endocytosis. CE in LDL and ox-LDL can be hydrolyzed in lysosome; thus, a normal lysosomal function and autophagy is critical for CE hydrolysis and cholesterol efflux. Conversely, impaired lysosomal function and autophagy may lead to the accumulation of excessive CEs to form foam cells, an early hallmark of atherosclerosis. For what we believe is the first time, our studies identified that ALDH2 can be phosphorylated by AMPK at T356 and translocate into the nucleus, interact with HDAC3, and repress the expression of ATP6V0E2, a lysosomal proton pump protein that plays a critical role in the fusion of lysosome with endosome or autophagosome (Figure 9). Under normal circumstances, this process is blocked by LDLR to ensure normal endocytosis and autophagy and to avoid excessive lipid accumulation to form foam cells. However, importantly, when LDLR is knocked out (in the case of LDLR-KO) or interaction between LDLR and mutant ALDH2 (with human SNP rs671) is weakened, ALDH2 is phosphorylated by AMPK and translocated into the nucleus, which eventually leads to a dysfunctional lysosome and impaired CE hydrolysis. We provide evidence that LDLR most likely interacts with ALDH2 and AMPK with its cytosolic C-terminus to block the nuclear translocation of ALDH2. This serendipitous discovery was made from the unexpected observation of a phenotypic discrepancy in ALDH2-KO in LDLR and APOE backgrounds. More importantly, we have uncovered what we believe are a novel mechanism by which ALDH2 SNP rs671 increases atherosclerosis, and also a novel role of AMPK in macrophages for increasing this potentially detrimental effect of ALDH2 in the context of atherosclerosis.
Previous studies have provided evidence that ALDH2 plays an important role in cardiac protection against myocardial infarction, primarily based on the fact that ALDH2 is an important enzyme responsible for eliminating toxic aldehydes. Selective activation of ALDH2 improved the heart failure outcome by reducing toxic aldehyde overload and restoring mitochondrial function (24). Moreover, ALDH2 appears to protect against myocardial infarction–related cardiac fibrosis through modulation of the Wnt/β-catenin signaling pathway (25). Compared with cardiac protection, the role of ALDH2 in atherosclerosis remains poorly defined. A recent epidemiological study in East Asian populations using alcohol flushing status as an instrumental variable demonstrated that increased alcohol consumption was positively associated with an increased risk of subclinical coronary atherosclerosis, suggesting that ALDH2 might be involved in the pathogenesis of atherosclerosis (26). A previous study using APOE-KO mice infected with an ALDH2-expressing lentivirus or ALDH2 RNAi lentivirus found that ALDH2 had a protective role in atherosclerosis through attenuating the inflammation in endothelial cells (17). Likewise, we observed an increase in atherosclerotic plaques in ALDH2/APOE-DKO mice. However, ALDH2/LDLR-DKO mice had much fewer atherosclerotic lesions than the LDLR-KO mice. This unexpected observation led us to discover a nonenzymatic role of ALDH2, which promotes foam cell formation by translocating to the nucleus after phosphorylation by AMPK to repress ATP6V0E2 expression, a critical protein for lysosomal function and autophagy. The phenotypic discrepancy in DKO mice of ALDH2 with APOE and LDLR is most likely due to the different roles of inflammation and LDLR-regulated ALDH2 nuclear translocation. In APOE-KO mice, the mechanism identified in the present study does not operate due to the presence of LDLR, which blocks the nuclear translocation of ALDH2, whereas in APOE/ALDH2-DKO mice, increased atherosclerotic plaque formation compared with APOE-KO mice is primarily due to the elevated inflammation in endothelial cells (17). On the other hand, ALDH2/LDLR-DKO mice had a decrease in atherosclerosis compared with LDLR-KO mice, primarily through attenuated foam cell formation in macrophages, in addition to a reduction in inflammation. As one of the most important types of immune cells in atherogenesis, macrophages are critical for inflammatory responses and the formation of foam cells (27, 28). Macrophages tend to polarize to different phenotypes upon exposure to oxidized lipids (28). Consistent with the decreased number of atherosclerotic lesions in ALDH2/LDLR-DKO mice, the DKO macrophages exhibited an antiinflammatory M2 phenotype, whereas the markers for the M1 phenotype were markedly decreased despite the fact that other inflammation markers were increased in plasma and endothelial cells (Supplemental Figure 20). These data appear to suggest that the mechanisms identified in the present study play a predominant role in atherosclerosis in the context of foam cell formation.
The most exciting discovery of our current study is that AMPK-phosphorylated ALDH2 translocates to the nucleus and represses ATP6V0E2, a protein critical for lysosomal function, endocytosis, and autophagy. ATP6V0E2 belongs to the H+-ATPase family, which is important for the lysosomal proton pump to maintain the proper function of the lysosomes (29). Many proteins other than ATP6V0E2 belonging to this family (V-ATPase) have been reported to be closely involved in endocytosis and autophagy through maintenance of normal lysosomal function (30–33). Our study showed, for what we believe is the first time, that in the later stages of endocytosis and autophagy, ATP6V0E2 plays an important role for the fusion of lysosome with endosome or autophagosome. Furthermore, normal lysosomal function and autophagic flux in macrophages enable the degradation of ox-LDL and thereby decrease the formation of foam cells (7). In ox-LDL–loaded LDLR-KO macrophages, the formation of autophagosome was intact, as indicated by the similar expression levels of ATG5. Increased expression of LC3 II and P62 in LDLR-KO macrophages compared with ALDH2/LDLR-DKO macrophages signaled a decreased autophagic flux due to an impaired lysosomal function caused by decreased expression of ATP6V0E2 (Figure 3B). Intriguingly, however, ALDH2 deficiency alone appears to decrease autophagic flux without affecting endocytosis compared with WT (Supplemental Figure 5, B and C), suggesting that other unknown mechanisms are most likely involved in autophagic regulation in the presence of LDLR. It remains to be studied how AMPK-phosphorylated ALDH2 is translocated into the nucleus. We performed a preliminary proteomic study on the ALDH2-immunoprecipitated nuclear fraction of LDLR-KO BMDMs treated with metformin and identified a list of interacting proteins with ALDH2 in the nucleus. Among them, DDX5 (known as P68) was reported as a shuttle between the nucleus and the cytoplasm, consistent with the function of the protein in transcriptional regulation and pre-mRNA splicing (data not shown) (34). These data suggest that ALDH2 is likely translocated to the nucleus by binding to nuclear transporters such as P68.
AMPK is a key factor that enables the nuclear translocation of ALDH2 in the absence of LDLR or ALDH2 rs671 mutant. The role of AMPK in CVD has been widely recognized, including its inflammatory signaling suppression, monocyte differentiation, and foam cell formation (35). Our data, presented here, strongly support the notion that AMPK plays an important role in regulating macrophage metabolism of ox-LDL through phosphorylating ALDH2. AMPK activators metformin and AICAR inhibited monocyte-to-macrophage differentiation through AMPK-mediated inhibition of STAT3 activation (36). Ma et al. demonstrated that ALDH2 exerted cardio-protection against myocardial ischemia/reperfusion injury through an AMPK-dependent induction of autophagy during ischemia and a paradoxical Akt-dependent reduction in autophagy during reperfusion (37). This elegant study appeared to suggest a role for detoxification of toxic aldehydes in ALDH2-elicited regulation of the AMPK and AKT pathways via LKB1 and PTEN signaling. Intriguingly, however, the regulation of AMPK in macrophages appears to be different from that in cardiomyocytes, in which we did not observe an alteration of the upstream kinases of AMPK (Supplemental Figure 9A). The discrepancy in mouse phenotypes between APOE and LDLR backgrounds prompted us to study the unknown role of LDLR in the binding of ALDH2 and AMPK. Data shown in Figure 5 and Figure 6 strongly support a direct protein-protein interaction of AMPK and ALDH2, which is important in regulating lysosome function and autophagy in macrophages.
ALDH2 SNP rs671, the inactive ALDH2*2 variant (E487K), affects around 40% of East Asians. This ALDH2 SNP has been associated with increased CVDs, such as heart failure and stroke (38), and gastrointestinal cancers (39). However, previous studies have exclusively focused on the role of alcohol consumption, and it remains elusive whether other molecular mechanisms are involved in diseases associated with this SNP. Our data clearly showed that SNP rs671 mutant attenuated the binding of ALDH2 to LDLR and increased its binding to AMPK, which enables more mutant ALDH2 to translocate into the nucleus to suppress the expression of ATP6V0E2 and cause impaired lysosomal function and autophagy. The net outcome of these effects is an excessive accumulation of ox-LDL and enhanced foam cell formation. The molecular mechanism identified in the present study raises an interesting concern — that activation of AMPK by the widely used antidiabetic drug metformin may potentially exert an increased risk in atherosclerosis for humans with ALDH2 SNP rs671. Even though a substantial amount of data support metformin as protective against CVD, metformin’s role in the patients with ALDH2 SNP rs671 remains to be studied.
In conclusion, our study discovered what we believe is a new pathway through which ALDH2 rs671 SNP promotes macrophage foam cell formation in atherosclerosis. It does so by attenuating the interaction with LDLR but increasing the interaction with AMPK to promote the translocation of ALDH2 into the nucleus, suppressing the transcription of ATP6V0E2 and thereby impairing a lysosomal function in macrophages. This finding represents a novel molecular mechanism by which ALDH2 rs671 carriers are prone to CVDs, implying that AMPK activation increases atherosclerosis for humans with ALDH2 rs671 mutant.
Methods
Further information can be found in the Supplemental Methods.
Animals.
Mice were housed in a pathogen-free, temperature- and light-controlled animal facility under a 12-hour light/dark cycle. ALDH2–/– LDLR–/– mice were obtained by crossing ALDH2–/– mice (a gift from Jun Ren and Aijun Sun, Zhongshan Hospital affiliated with Fudan University, Shanghai, China) (37, 40) and LDLR–/– mice (Jackson Laboratory). ALDH2–/–APOE–/– mice were obtained by crossing ALDH2–/– mice and APOE–/– mice (Shanghai Model Organisms). All mice used in the present study were on a C57BL/6 background. For atherosclerotic study, 6-week-old mice were fed Western Diet (Research Diets, catalog D12079B) for 12 weeks or 26 weeks (26-week experiment was done in duplicate). The number of mice studied in each experiment is shown in the figure legends.
Cell culture.
HEK 293T cell lines were purchased from the CAS Cell Bank and cultured in DMEM containing 10% FBS. Isolation of bone marrow–derived macrophages (BMDMs) followed a previously published protocol (41). All BMDMs or human macrophages for detecting autophagy were pretreated with 50 μg/ml ox-LDL.
Cholesterol depletion.
To deplete cholesterol, BMDMs were treated with RPMI-1640 supplemented with 5% lipoprotein-deficient serum (LPDS), 1 μM lovastatin, and 10 μM mevalonate for 16 hours.
Bone marrow transplantation.
BMT experiments were performed according to a published protocol (42). After recovery, chimeric mice were fed WD for 12 weeks.
Analysis of aortic lesions.
A previously published protocol was followed to analyze aortic lesions (43).
Histology and immunohistochemistry.
The mice hearts were fixed with 4% formaldehyde overnight at 4°C and embedded in paraffin. Then, the heart samples were divided into 8-μm sections from the brachiocephalic trunk through the aortic root. The sections were then stained with Masson blue for collagen components, and IHC staining was used to analyze macrophages and α-SMCs with specific primary antibodies and corresponding secondary antibodies (ZSGB-Biotech). For IHC, the sections were washed with dimethylbenzene twice for 15 minutes, ethanol twice for 5 minutes, 95% ethanol for 5 minutes, 70% ethanol for 5 minutes, ddH2O twice for 5 minutes, and PBS for 5 minutes. Antigen retrieval was conducted using 10 mM sodium citrate. Slides in sodium citrate were boiled 3 times for 2 minutes and cooled for 2 minutes. Slides were then blocked in 10% normal serum with 1% BSA in TBS for 30 minutes at room temperature, and incubated with specific primary antibodies (anti-CD68, catalog MCA1957, Bio-Rad; anti–α-SMC-actin, catalog ab5694, Abcam) overnight at 4°C. After recovering to room temperature, slides were incubated with secondary antibody and stained with DAB buffer (ZSGB-Biotech, catalog ZLI-9017). All sections were analyzed with ImageJ for quantitative measurements.
Phagocytotic assay.
BMDMs were incubated at 37°C for detecting ox-LDL content or 4°C for binding with 10 μg/ml Dil–ox-LDL (catalog YB-0010, Yiyuan Biotechnologies) for 4 hours or 50 μg/ml ox-LDL (catalog YB-002, Yiyuan Biotechnologies) for 6 hours. Staining and analyses were performed as previously described (41). AICAR (catalog S1802, Selleck) or Bafilomycin-A1 (catalog S1413, Selleck) was added to cell medium to make the final concentrations of 1 mM AICAR and 10 nM Baf-A1, respectively, 1 hour before adding Dil–ox-LDL or ox-LDL.
Western blotting.
SDS-PAGE was used to separate total cell lysate, and proteins were transfered to PVDF membranes (Millipore). The immune-reactive protein bands were visualized by ECL kit (Proteintech). Antibodies against ALDH2 (catalog 15310-1-AP), LDLR (catalog 10785-1-AP), ATG5 (catalog 10181-2-AP), HDAC1 (catalog 10197-1-AP), HDAC2 (catalog 12922-3-AP), HDAC3 (catalog 10255-1-AP), Lamin B1 (catalog 66095-1-Ig), ACAT1 (catalog 16215-1-AP), His-Tag (catalog 66005-1-Ig), and GAPDH (catalog 60004-1-Ig) were purchased from Proteintech. Antibodies against P-AMPK (Thr172; catalog 2535S), T-AMPK (catalog 5832T), LC3B (catalog 2775S), Myc-Tag (catalog 2276S), RAB11 (catalog 5589T), EEA1 (catalog 3288T), RAB7 (catalog 9367T), RAB5 (catalog 3547T), LAMP1 (catalog 90917), Clathrin (catalog 4796T), and normal rabbit IgG (catalog 2729S) were purchased from Cell Signaling Technology. Antibodies against ATP6V0E2 (catalog ab178934), LOX1 (catalog ab60178), H3 (catalog ab4729), P62 (catalog ab91526), and SRA (catalog ab183725) were purchased from Abcam. ABCA1 (catalog NB400-105) and CD36 (catalog NB400-144SS) were purchased from Novus. Flag-Tag (catalog F1804) was purchased from MilliporeSigma.
Detection of phosphorylated ALDH2 using iTRAQ proteomics.
A 10% SDS-PAGE was used to separate total cell lysate for silver stain (catalog P0017S, Beyotime). ALDH2 was extracted by cutting the gel at approximately 53 kDa, and ALDH2 phosphorylation was measured using iTRAQ proteomics carried out by Shanghai Applied Protein Technology.
ATP, ADP, and AMP detection by LC–MS.
BMDMs (107) with or without 50 μg/ml ox-LDL treatment were collected by Trypsin-EDTA (catalog 25200056, Gibco) and washed twice using 1× PBS. BMDMs were pretreated according to previously established procedures (44). Methods for detecting ATP, ADP, and AMP levels were as previously published (45).
Detection of phosphorylated ALDH2 in SuperSepPhos-tag SDS-PAGE.
SuperSepPhos-tag SDS-PAGE (catalog 198-17981, Wako) was used for detection of phosphorylated ALDH2 in BMDMs.
Nuclear and cytoplasmic fractionation.
Separation of the cytoplasmic and nuclear proteins was conducted using the NE-PER nuclear and cytoplasmic extraction kit (catalog P0027, Beyotime) according to the manufacturer’s instructions.
Cholesterol efflux assay.
BMDMs were treated with 15 μg/ml cholesterol-d7 (chol-d7) for 24 hours. After washing the cells thoroughly with PBS, 50 μg/ml HDL was added to induce cholesterol efflux. After a 24-hour incubation, medium was collected for gas chromatography–mass spectrometry (GC-MS) and 10% of the BMDM proteins was collected for quantification. Free cholesterol was extracted using 2 ml hexane/isopropyl alcohol (IPA) (3:2, vol/vol) supplemented with 1% acetic acid and 6 μg 5α-cholestane as internal standard. The upper organic phase was dried under nitrogen, and 40 μl pyridine and 40 μl derivatizing agent (BSTFA + TMCS) were added for GC-MS analysis. GC-MS was performed in the EI mode using Shimadzu GC-MS QP2010. Peak areas were integrated using the instrument software. Calibration curves were generated using chol-d7 and internal standard. The final results were normalized to cell lysate protein.
Hydrolysis of cholesteryl esters.
Macrophages were incubated with 50 μg/ml ox-LDL with or without 5 nM Baf-A1 for 20 hours. CEs were extracted by adding 4 ml hexane/IPA (3:2, vol/vol), and 3 μg CE 19:0 as an internal standard. The upper organic phase was dried under nitrogen and 50 μl hexane was added for liquid chromatography–mass spectrometry (LC-MS) analysis. Normal phase LC was carried out with the Agilent 1260 Quat pump VL and Accela 1250 pump. The MS was performed on a TSQ Vantage (Thermo Fisher Scientific) in positive ion and multiple reaction monitoring mode. Chromatographic peaks were integrated and area ratios (samples vs. internal standard) were generated using Xcalibur (Thermo Fisher Scientific). CE changes (hydrolysis) were expressed as fold change relative to control, calculated as follow: % hydrolysis = (CE1 – CE2)/(CE1) × 100, where CE1 represented CE after treating with ox-LDL for 20 hours, and CE2 represents CE after the macrophages were incubated for 20 hours with 50 μg/ml HDL. Fold change = % hydrolysis sample/% hydrolysis control.
Real-time PCR.
BMDMs were lysed with TRIzol reagent (catalog 9109, Takara). RNA was purified and reverse transcribed into cDNA. Gene expression was normalized to L32 expression. The primer sequences are shown in Supplemental Table 2. The fold change was calculated using the comparative ΔCt method and is shown as relative transcript levels.
Retrovirus (TRV).
LDLR-KO BMDMs were incubated with 5 μg/ml polybrene and 100 μl 5 × 107 TU/ml HBhTRV-m-LDLR-3xflag-GFP in 900 μl cell medium (RPMI-1640 plus 10% FBS) at 37°C for 4 hours. We then added 1 ml cell medium with 5 μg/ml polybrene. Cell medium was changed after 24-hour incubation at 37°C, and BMDMs were incubated for 48 hours.
Immunoprecipitation.
For BMDMs, target protein was precipitated with respective antibodies and protein A/G beads (catalog B23202, Biotool). 293T cells were transfected with specifically tagged protein using Attractene Transfection Reagent (catalog 301005, Qiagen), lysed and incubated with myc-beads or flag-beads (catalog B26301 for myc-beads, catalog B23102 for flag-beads, Biotool).
Fluorescence microscopy.
BMDMs were grown on glass coverslips in 6-well dishes. Immunofluorescence staining was performed as previously described (42). The percentage of ALDH2 translocation was calculated according to areas of ALDH2 signal in nucleus/nucleus area (quantified by Image J).
RNA-Seq analysis.
BMDMs were lysed with TRIzol reagent. The library preparation and sequencing were carried out by Beijing Genomics Institute BGI (Wuhan, China). We measured the expression of the transcript isoforms using RSEM, and used the NOISeq method to screen differentially expressed genes (DEGs) between 2 groups. The top 3 gene ontology functional classifications on overlapping DEGs are shown in Supplemental Table 3. Heatmaps were generated using the R software. Microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE121310).
ChIP assay.
BMDMs were cultured on 10-cm plates and pretreated with or without 1 mM AICAR for 1 hour. ChIP assay was conducted using a ChIP Assay kit (catalog 17-295, Millipore). Eluted DNA was further purified using a PCR purification kit (Qiagen).
Detection of autophagic flux.
BMDMs were plated and grown overnight on a 20-mm, glass-bottom cell-culture dish (catalog 801001, NEST). The formation of autolysosomes in BMDMs in control and in cells treated with Baf-A1 (1 nM), chloroquine (10 μM), or leupeptin (100 μM) was detected using the Premo Autophagy Tandem Sensor RFP-GFP-LC3B Kit (catalog P36239, Thermo Fisher Scientific). Fluorescent images were taken using confocal microscopy (Carl Zeiss Meditec, Inc.) and quantified using Image J software. The quantified mean number of autolysosomes per cell was according to the dots of RFP signal in the merge picture.
Luciferase assay.
293T cells were transfected with pGL3.1/pCMV-ATP6V0E2 (20:1) for 24 hours. Luciferase activity was measured using the Dual Luciferase Reporter Assay System (catalog E1960, Promega). The GL (firefly luciferase) activity was normalized to coexpressed RL (renilla luciferase) activity. 293T cells were transfected with Lipofectamine 2000 Reagent (catalog 1854311, Invitrogen).
Human macrophages.
Human monocytes were isolated from PBMCs. PBMCs were isolated using a Ficoll (catalog 17-1440-02, BD Biosciences) density gradient, and then were isolated from PBMCs using anti-CD14 magnetic beads (catalog 11149D, Thermo Fisher Scientific) according to the manufacturers’ protocols. Then, human monocytes were differentiated into human macrophages by treatment with differentiated medium (RPMI -1640, 5% AB human serum, 10% FBS, penicillin/streptomycin, 25 ng/ml M-CSF) for 7 days.
Plasmids.
Flag-tagged human ALDH2, ALDH2 (rs671) mutant clone, Myc, and 6xHis-tagged human LDLR were constructed into a pcDNA3.0 vector. Clones were picked and amplified to be sequenced to verify the final results.
Statistics.
Results are mean ± SD or mean ± SEM from at least 3 independent experiments using GraphPad Prism 5. Statistical analysis was conducted using an unpaired Student’s t test or 2-way ANOVA. A 2-tailed P value of less than 0.05 was considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences, Shanghai, China (approval number 2015-AN-2). All human samples were collected by Shanghai General Hospital and human experiments were approved by the Ethics Committee of Shanghai General Hospital, affiliated with Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Author contributions
HY and SZ designed the research studies and experiments. SZ carried out a majority of the experiments. LL helped perform bone marrow transplant experiments and measured aorta lesions. YLZ and LZ helped with the clinical study. JL and SG developed the LC-MS method and helped measure cholesteryl esters and cholesterol efflux. NL performed BMDM primary culture experiments. JG performed RNA-seq analysis. MZ performed phosphoproteomics data analysis. YT developed the GC-MS method and helped detect cholesterol. YCW and HY designed the clinical study. HY and SZ wrote and approved this manuscript.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Jun Ren and Aijun Sun from Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital affiliated with Fudan University, Shanghai, China, for providing ALDH2–/– mice. We acknowledge the help from Zhimin Hu at SIBS in bone marrow transplant experiments. A constructive discussion with Dawei Zhang at University of Alberta, Canada, Ming-Hui Zou from Georgia State University, USA, and Bao-liang Song from Wuhan University, China, is also greatly appreciated. This work was financially supported by the National Natural Science Foundation of China (91857112, 31470831, 91439103, 91539127, and 31401015) and the National Key R&D Program of China, administered by the Chinese Ministry of Science and Technology (MOST) (2016YFD0400205, 2016YFC0903403), and a grant from the Chinese Academy of Sciences (ZDBS-SSW-DQC-02).
Version 1. 10/30/2018
In-Press Preview
Version 2. 12/03/2018
Electronic publication
Version 3. 01/02/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(1):252–267.https://doi.org/10.1172/JCI122064.
See the related Commentary at Not just correlative: a new pathway defines how an ALDH2 SNP contributes to atherosclerosis.
Contributor Information
Shanshan Zhong, Email: sszhong@sibs.ac.cn.
Luxiao Li, Email: lilx@shanghaitech.edu.cn.
Yu-Lei Zhang, Email: 1195485885@qq.com.
Lili Zhang, Email: zhanglili2018@sibs.ac.cn.
Jianhong Lu, Email: lujianhong@sibs.ac.cn.
Shuyuan Guo, Email: guoshy@shanghaitech.edu.cn.
Ningning Liang, Email: liangningning2017@sibs.ac.cn.
Jing Ge, Email: gejing@sibs.ac.cn.
Mingjiang Zhu, Email: mjzhu@sibs.ac.cn.
Yongzhen Tao, Email: yztao01@sibs.ac.cn.
Yun-Cheng Wu, Email: drwu2006@163.com.
Huiyong Yin, Email: hyyin@sibs.ac.cn.
References
- 1.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17(11):1410–1422. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 2.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao B, Song J, Chow WN, St Clair RW, Rudel LL, Ghosh S. Macrophage-specific transgenic expression of cholesteryl ester hydrolase significantly reduces atherosclerosis and lesion necrosis in Ldlr mice. J Clin Invest. 2007;117(10):2983–2992. doi: 10.1172/JCI30485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouimet M, et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37(6):1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sergin I, Evans TD, Razani B. Degradation and beyond: the macrophage lysosome as a nexus for nutrient sensing and processing in atherosclerosis. Curr Opin Lipidol. 2015;26(5):394–404. doi: 10.1097/MOL.0000000000000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sergin I, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8:15750. doi: 10.1038/ncomms15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Getz GS, Reardon CA. Do the Apoe-/- and Ldlr-/- mice yield the same insight on atherogenesis? Arterioscler Thromb Vasc Biol. 2016;36(9):1734–1741. doi: 10.1161/ATVBAHA.116.306874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defesche JC, Gidding SS, Harada-Shiba M, Hegele RA, Santos RD, Wierzbicki AS. Familial hypercholesterolaemia. Nat Rev Dis Primers. 2017;3:17093. doi: 10.1038/nrdp.2017.93. [DOI] [PubMed] [Google Scholar]
- 10.Somanathan S, Jacobs F, Wang Q, Hanlon AL, Wilson JM, Rader DJ. AAV vectors expressing LDLR gain-of-function variants demonstrate increased efficacy in mouse models of familial hypercholesterolemia. Circ Res. 2014;115(6):591–599. doi: 10.1161/CIRCRESAHA.115.304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc Natl Acad Sci U S A. 1992;89(10):4471–4475. doi: 10.1073/pnas.89.10.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pradhan AD, Aday AW, Rose LM, Ridker PM. Residual inflammatory risk on treatment with PCSK9 inhibition and statin therapy. Circulation. 2018;138(2):141–149. doi: 10.1161/CIRCULATIONAHA.118.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raal FJ, et al. Inhibition of PCSK9 with evolocumab in homozygous familial hypercholesterolaemia (TESLA Part B): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;385(9965):341–350. doi: 10.1016/S0140-6736(14)61374-X. [DOI] [PubMed] [Google Scholar]
- 15.Chen CH, Sun L, Mochly-Rosen D. Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc Res. 2010;88(1):51–57. doi: 10.1093/cvr/cvq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JM, et al. ALDH2 protects against stroke by clearing 4-HNE. Cell Res. 2013;23(7):915–930. doi: 10.1038/cr.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan C, et al. Aldehyde dehydrogenase 2 inhibits inflammatory response and regulates atherosclerotic plaque. Oncotarget. 2016;7(24):35562–35576. doi: 10.18632/oncotarget.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Q, Weiner H, Crabb DW. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest. 1996;98(9):2027–2032. doi: 10.1172/JCI119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han H, Wang H, Yin Z, Jiang H, Fang M, Han J. Association of genetic polymorphisms in ADH and ALDH2 with risk of coronary artery disease and myocardial infarction: a meta-analysis. Gene. 2013;526(2):134–141. doi: 10.1016/j.gene.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Choi JW, et al. Malondialdehyde inhibits an AMPK-mediated nuclear translocation and repression activity of ALDH2 in transcription. Biochem Biophys Res Commun. 2011;404(1):400–406. doi: 10.1016/j.bbrc.2010.11.131. [DOI] [PubMed] [Google Scholar]
- 22.Tang WH, Hazen SL. Atherosclerosis in 2016: advances in new therapeutic targets for atherosclerosis. Nat Rev Cardiol. 2017;14(2):71–72. doi: 10.1038/nrcardio.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, et al. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet. 2009;73(Pt 3):335–345. doi: 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes KM, et al. Aldehyde dehydrogenase 2 activation in heart failure restores mitochondrial function and improves ventricular function and remodelling. Cardiovasc Res. 2014;103(4):498–508. doi: 10.1093/cvr/cvu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, et al. Aldehyde dehydrogenase-2 protects against myocardial infarction-related cardiac fibrosis through modulation of the Wnt/β-catenin signaling pathway. Ther Clin Risk Manag. 2015;11:1371–1381. doi: 10.2147/TCRM.S88297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun KE, et al. Alcohol and coronary artery calcification: an investigation using alcohol flushing as an instrumental variable. Int J Epidemiol. 2017;46(3):950–962. doi: 10.1093/ije/dyw237. [DOI] [PubMed] [Google Scholar]
- 27.Chan L, et al. Role of Rab5 in the formation of macrophage-derived foam cell. Lipids Health Dis. 2017;16(1):170. doi: 10.1186/s12944-017-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chistiakov DA, Melnichenko AA, Myasoedova VA, Grechko AV, Orekhov AN. Mechanisms of foam cell formation in atherosclerosis. J Mol Med. 2017;95(11):1153–1165. doi: 10.1007/s00109-017-1575-8. [DOI] [PubMed] [Google Scholar]
- 29.Höhn A, Sittig A, Jung T, Grimm S, Grune T. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radic Biol Med. 2012;53(9):1760–1769. doi: 10.1016/j.freeradbiomed.2012.08.591. [DOI] [PubMed] [Google Scholar]
- 30.Mijaljica D, Prescott M, Devenish RJ. V-ATPase engagement in autophagic processes. Autophagy. 2011;7(6):666–668. doi: 10.4161/auto.7.6.15812. [DOI] [PubMed] [Google Scholar]
- 31.Florey O, Gammoh N, Kim SE, Jiang X, Overholtzer M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11(1):88–99. doi: 10.4161/15548627.2014.984277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forgac M, Cantley L, Wiedenmann B, Altstiel L, Branton D. Clathrin-coated vesicles contain an ATP-dependent proton pump. Proc Natl Acad Sci U S A. 1983;80(5):1300–1303. doi: 10.1073/pnas.80.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake-Palmer KG, Su Y, Smith AN, Karet FE. Molecular cloning and characterization of a novel form of the human vacuolar H+-ATPase e-subunit: an essential proton pump component. Gene. 2007;393(1-2):94–100. doi: 10.1016/j.gene.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Gao X, Huang Y, Yang J, Liu ZR. P68 RNA helicase is a nucleocytoplasmic shuttling protein. Cell Res. 2009;19(12):1388–1400. doi: 10.1038/cr.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salt IP, Hardie DG. AMP-activated protein kinase: an ubiquitous signaling pathway with key roles in the cardiovascular system. Circ Res. 2017;120(11):1825–1841. doi: 10.1161/CIRCRESAHA.117.309633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasamsetti SB, Karnewar S, Kanugula AK, Thatipalli AR, Kumar JM, Kotamraju S. Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: potential role in atherosclerosis. Diabetes. 2015;64(6):2028–2041. doi: 10.2337/db14-1225. [DOI] [PubMed] [Google Scholar]
- 37.Ma H, Guo R, Yu L, Zhang Y, Ren J. Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J. 2011;32(8):1025–1038. doi: 10.1093/eurheartj/ehq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai Q, Wu J, Cai Q, Chen EZ, Jiang ZY. Association between Glu504Lys polymorphism of ALDH2 gene and cancer risk: a meta-analysis. PLoS ONE. 2015;10(2):e0117173. doi: 10.1371/journal.pone.0117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin S, et al. ALDH2(E487K) mutation increases protein turnover and promotes murine hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2015;112(29):9088–9093. doi: 10.1073/pnas.1510757112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun A, Ren J. ALDH2, a novel protector against stroke? Cell Res. 2013;23(7):874–875. doi: 10.1038/cr.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, et al. Identification of a novel series of anti-inflammatory and anti-oxidative phospholipid oxidation products containing the cyclopentenone moiety in vitro and in vivo: implication in atherosclerosis. J Biol Chem. 2017;292(13):5378–5391. doi: 10.1074/jbc.M116.751909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J, et al. Activation of E-prostanoid 3 receptor in macrophages facilitates cardiac healing after myocardial infarction. Nat Commun. 2017;8:14656. doi: 10.1038/ncomms14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan S, et al. Prostaglandin E2 promotes hepatic bile acid synthesis by an E prostanoid receptor 3-mediated hepatocyte nuclear receptor 4α/cholesterol 7α-hydroxylase pathway in mice. Hepatology. 2017;65(3):999–1014. doi: 10.1002/hep.28928. [DOI] [PubMed] [Google Scholar]
- 44.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang CS, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548(7665):112–116. doi: 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.