Abstract
Aerobic ammonia-oxidizing archaea (AOA) play a crucial role in the global nitrogen cycle by oxidizing ammonia to nitrite, and nitric oxide (NO) is a key intermediate in AOA for sustaining aerobic ammonia oxidation activity. We herein heterologously expressed the NO-forming, copper-containing, dissimilatory nitrite reductase (NirK) from Nitrososphaera viennensis and investigated its enzymatic properties. The recombinant protein catalyzed the reduction of 15NO2− to 15NO, the oxidation of hydroxylamine (15NH2OH) to 15NO, and the production of 14–15N2O from 15NH2OH and 14NO2−. To the best of our knowledge, the present study is the first to document the enzymatic properties of AOA NirK.
Keywords: nitrite reduction, hydroxylamine oxidation, nitrous oxide production, ammonia oxidizing archaea, Nitrososphaera viennensis
Aerobic ammonia oxidation, a rate-limiting step of nitrification, drives the global nitrogen cycle (24, 40), which involves aerobic ammonia-oxidizing archaea and bacteria (AOA and AOB, respectively) and complete ammonia oxidizers (comammox) (9, 44). Of these, AOA primarily contribute to aerobic ammonia oxidation in natural environments including soil and open ocean (19, 31, 46). AOA are affiliated with the phylum Thaumarchaeota, which includes phylogenetically and physiologically diverse members (6) and the soil-inhabiting archaeon Nitrososphaera viennensis (41). The biochemistry of aerobic ammonia oxidation by AOA has received a great deal of interest because ammonia oxidation to nitrite (NO2−) proceeds in a different manner to that of AOB. AOA oxidize ammonia to hydroxylamine by ammonia monooxygenase (Amo) as well as AOB (43), while hydroxylamine is further oxidized to NO2− by an unidentified enzyme (17). All known AOA genomes lack the gene encoding hydroxylamine dehydrogenase (Hao), and the involvement of a copper-protein complex has been proposed (40, 45). In parallel with the oxidation of ammonia to NO2−, AOA produce nitric oxide (NO) (22). NO is a key intermediate in AOA cells because this highly reactive molecule is essential for sustaining aerobic ammonia oxidation activity (17, 33, 36, 47). To date, the following 2 pathways have been reported as a source of prokaryotic NO formation: NO2− reduction to NO by copper-containing and cytochrome cd1-type dissimilatory nitrite reductases (NirK and NirS, respectively) (38) and NH2OH oxidation to NO by hydroxylamine oxidoreductase (Hao) (4, 21). Although neither nirS nor hao are found in AOA genomes (6), AOA commonly possess nirK, which is transcribed and expressed during aerobic ammonia oxidation (8, 15, 20, 37). These findings suggest that NirK are involved in NO formation in AOA cells. However, NO2− reduction to NO by AOA NirK has never been demonstrated.
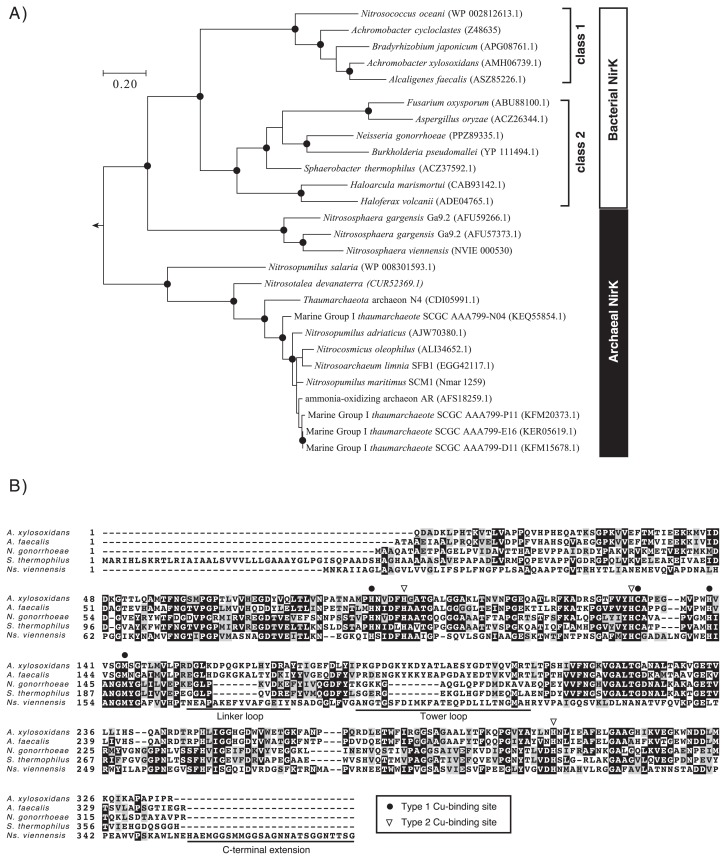
Bacterial NirK have been characterized as homotrimeric enzymes, and each subunit has 2 Cu-binding sites (Type 1 and 2 Cu-binding sites). Type 1 Cu-binding sites receive an electron from an electron donor, such as type 1 Cu proteins (single-domain cupredoxins) and/or cytochrome c, and the electron is then further transferred to a type 2 Cu-binding site that is the catalytic center of NirK (14, 25). Bacterial NirK have been classified into 2 phylogenetically distinct groups (class 1 and class 2 groups) based on sequence similarities, and the NirK of the class 1 group contains linker loop and tower loop regions in the amino acid sequence (3). AOA NirK, including Ns. viennensis NirK, are affiliated with a distinct clade of bacterial class 1 and 2 groups (Fig. 1A). Lund et al. (20) reported that AOA NirK may be further classified into several phylogenetic clades showing specific geographic distributions. Ns. viennensis NirK has amino acid residues consistent with those of type 1 and 2 Cu-binding sites (His106, His140, and His316 for type 1 Cu-binding sites and His101, Cys141, His152, and Met157 for type 2 Cu-binding sites) as well as the linker and tower loop regions, whereas the C terminus has unusual extensions of ~26 residues (Fig. 1B). These phylogenetic affiliations of and structural variations in Ns. viennensis NirK raise concerns regarding its enzymatic properties, such as specific enzymatic activity, affinity for NO2−, and products of NO2− reduction.
Fig. 1.
Phylogeny (A) and sequence alignments (B) of prokaryotic NirK. A) A phylogenetic tree of prokaryotic NirK was constructed by the maximum likelihood method with the Jones-Taylor-Thornton model using the protein sequence of multicopper oxidase type 3 of Nitrososphaera viennensis (accession number; AIC14243.1) as an outgroup. Branching points that support a probability >80% in bootstrap analyses (based on 500 replicates) are shown as filled circles. The scale bar represents 10% sequence divergence. Sequence accession numbers are indicated in parentheses. B) Protein sequence alignment of nirK. NirK sequences were aligned using ClustalW software. Circles and triangles correspond to the amino acid residues of type 1 and 2 Cu-binding sites, respectively. Linker, Tower loop (3), and C-terminal extension regions are underlined. Abbreviations of microorganisms are as follows: Nitrosomonas europaea is N. europaea, A. xylosoxidans is Achromobacter xylosoxidans, A. faecalis is Alcaligenes faecalis, N. gonorrhoeae is Neisseria gonorrhoeae, S. thermophilus is Sphaerobacter thermophilus, and Ns. viennensis is Nitrososphaera viennensis.
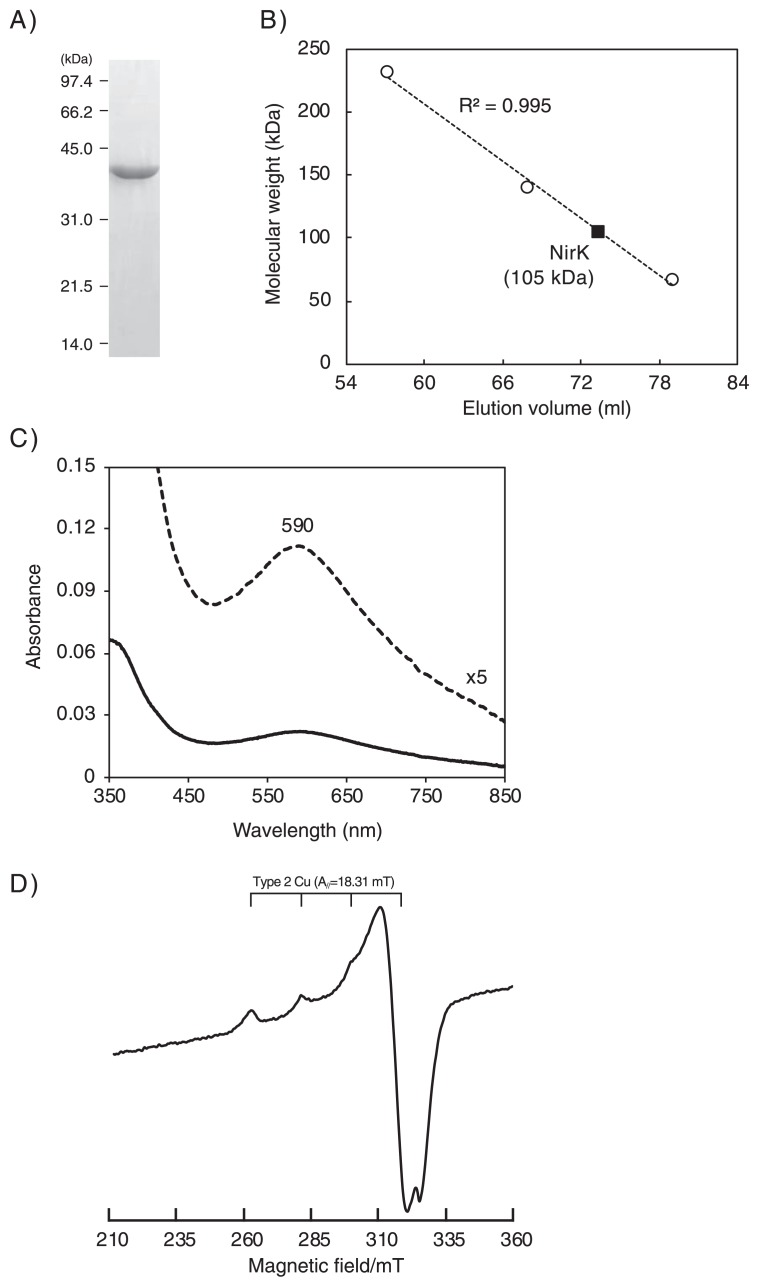
Based on its unique sequence and lack of biochemical information, the purpose of the present study was to characterize Ns. viennensis NirK. Prior to the present study, we aimed to isolate Ns. viennensis NirK from a batch culture of Ns. viennensis as a native enzyme. However, the activity of aerobic ammonia oxidation often disappeared when we scaled up the cultures (data not shown). Additionally, a slow growth rate (μmax 0.024 h−1) (41) and low biomass concentration in the culture (ca. 107~8 cells mL−1) further precluded the preparation of the biomass required for protein purification. Since recombinant NirK proteins have been successfully used to previously examine several enzymatic properties (7, 16, 32), the authors decided to heterologously express Ns. viennensis NirK in Escherichia coli, and investigate its enzymatic properties. The nirK gene located in the Ns. viennensis genome (accession number; CP007536.1) was cloned into the expression vector pCold I (Takara Bio, Shiga, Japan) with the 6×His tag using the Mighty cloning reagent set (Takara Bio), and transformed into E. coli strain BL21(DE3) (Takara Bio). The N-terminal region of Ns. viennensis NirK was predicted to be the signal peptide sequence (Met1 to Ala24), and nirK without the signal peptide sequence was amplified by PCR using ExTaq polymerase (Takara Bio) and specific forward (5′-GGCATATGGCCCCGACTGGTGTCACTAGACACTAT-3′) and reverse (5′-GGAAGCTTAACCAGAGGTGGTGTTGC CACCGGAGG-3′) oligonucleotide primers. The restriction sites of NdeI and HindIII in the forward and reverse primers above are underlined. Genomic DNA extracted from Ns. viennensis cells (JCM19564) was used as the DNA template for PCR. The constructed plasmid was subjected to Sanger sequencing, and no mutations were found in the sequence. Regarding the expression of the recombinant protein in E. coli cells, the expression culture was aerobically cultivated at 37°C in Luria-Bertani media containing 100 ng μL−1 ampicillin. When the OD600 of the culture increased to 0.4, the culture was transferred to 15°C and held for 30 min, and protein expression was then induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM. After being incubated at 15°C for 24 h, cells were harvested by centrifugation at 8,500×g at 4°C for 10 min. The harvested cells were suspended in buffer containing 20 mM Tris HCl (pH 8), 200 mM NaCl, and 10% glycerol. The cells were disrupted using a sonifier 250 (Branson) (output 20, duty 20% for 60 s, 6 cycles), and centrifuged at 13,000×g at 4°C for 1 h. The supernatant was recovered as a soluble protein fraction, and the recombinant protein was purified using His-tag affinity chromatography. The recombinant protein was bound to His60 Ni Superflow resin (Takara Bio), and washed with washing buffer containing 20 mM Tris HCl (pH 8), 200 mM NaCl, 10% glycerol, and 20 mM imidazole. The bound recombinant protein was eluted with elution buffer containing 20 mM Tris HCl (pH 8), 200 mM NaCl, 10% glycerol, and 300 mM imidazole. Protein concentrations were measured using the DC-protein assay kit (Bio-Rad, Hercules, CA, USA) with bovine serum albumin as previously described (26), and purity was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% polyacrylamide gel as previously described (28). As shown in Fig. 2A, a single protein band appeared at a molecular mass of 40 kDa, which closely matched the molecular mass deduced from amino acid sequences of the recombinant protein (i.e., 39.7 kDa). The protein band was excised from the gel, and subjected to a matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis after in-gel tryptic digestion for protein identification (The detailed methodology is described in the Supplementary text). The MALDI-TOF MS analysis confirmed that the protein band corresponded to Ns. viennensis NirK (Fig. S1). Regarding the reconstitution of Cu-binding sites of the recombinant protein, the purified recombinant protein was dialyzed against buffer containing 20 mM Tris HCl (pH 8), 300 mM NaCl, and 0.5 mM CuSO4 at 4°C for 57 h. The protein solution was dialyzed again using the above Tris buffer without CuSO4 at 4°C for 6 h. The dialyzed recombinant protein was concentrated using a Vivaspin column (MWCO; 30 kDa) (GE Healthcare Japan, Tokyo, Japan). The recombinant protein was loaded onto a gel-filtration HiLoad 16/600 Superdex 200 pg column (GE Healthcare) to assess the molecular mass of the recombinant protein, which was 105±1.3 kDa (Fig. 2B). Since the deduced molecular mass of Ns. viennensis NirK was 39.7 kDa, the molecular mass obtained by gel filtration indicated that the recombinant protein forms a homotrimeric structure, similar to canonical NirK.
Fig. 2.
Characterization of recombinant Nitrososphaera viennensis NirK. A) SDS-PAGE of the recombinant protein purified by His-tag affinity chromatography. B) Assessment of the molecular mass of the recombinant protein by gel filtration chromatography. Catalase from bovine liver (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (66 kDa) were used to prepare a standard calibration curve. C) UV-VIS absorption spectra. The measurement was performed in a 20 mM Tris buffer (pH 8) containing 300 mM NaCl at 25°C. The solid line indicates the recombinant protein (1 mL mL−1) oxidized with air. A 5×enlarged spectrum is also shown as a dashed line. D) ESR spectra. The measurement was performed using the recombinant protein (4.9 mg mL−1) at −253°C.
NirK have been characterized as metalloproteins showing a blue or green color spectrum, and exhibit absorption peaks at approximately 450 and/or 600 nm (3). Bacterial NirK, which belong to the class 1 group, often show a maximum absorption peak at approximately 450 nm, although an exception (Achromobacter xylosoxidans NirK) that shows a peak at 593 nm has been previously reported (16). The purified recombinant protein was pale blue in color, and showed an absorption peak at 590 nm (Fig. 2C). This feature indicated that Ns. viennensis NirK is affiliated with the subgroup of NirK showing a blue color spectrum. The blue or green color spectrum of NirK is derived from a copper atom in the type 1 Cu-binding site (14), while the type 2 Cu-binding site does not contribute to the UV or visible spectrum. The type 2 Cu-binding site shows a characteristic electron spin resonance (ESR) spectrum (7, 16); therefore, an ESR analysis was performed using a JES-FA200 spectrometer (JEOL, Tokyo, Japan) to test for the presence of the type 2 Cu-binding site in the recombinant protein. An axial type 2 Cu signal (g//=2.24, A//=18.31 mT, and g⊥=2.06) was found in the ESR measurement (Fig. 2D), indicating that the recombinant protein has a type 2 Cu-binding site coordinating with a copper atom. Additionally, we assessed the copper content of the recombinant protein by inductively coupled plasma mass spectrometry (ICP-MS). The copper content was found to be 2.9 atoms per subunit of the recombinant protein, indicating that Cu was fully incorporated into the recombinant protein. Overall, the recombinant protein shared the structural and spectroscopic features of class 1 and 2 bacterial NirK, which is consistent with sequencing information.
The kinetics of NO2− reduction were examined by anoxically incubating the recombinant protein at 25°C and pH 6.5 with 15NO2− and artificial electron donors as previously described (7). All of the buffers and stock solutions were prepared anoxically as previously described (27). Two milliliters of reaction buffer (20 mM phosphate buffer, 0.1 to 1.6 mM Na15NO2−, 0.5 mM benzyl viologen (BV), and 0.24 mM sodium dithionite) was dispensed into a 1-cm sealable quartz cuvette and placed in an anaerobic chamber in which the O2 concentration was maintained at lower than 1 ppm. BV was used as an artificial electron donor because it has been employed to examine the kinetics of the NO2− reduction of bacterial NirK (7, 13). The cuvette was set in a UV-VIS spectrometer UV-2700 (Shimadzu, Kyoto, Japan), and the initial absorbance of the prepared reaction mixture at a wavelength of 550 nm was approximately 2.0. The reaction was initiated by adding the recombinant protein (50 μL containing 250 μg of protein) using a gastight syringe, and the oxidation rate of reduced BV (molecular extinction coefficient, 10.4 mM−1 cm−1) (13) was monitored at 550 nm. The recombinant protein reduced NO2− by oxidizing BV, whereas no significant BV oxidation was found in the cuvette without the recombinant protein. The turnover number and Km value for NO2− reduction by the recombinant protein were 3.1 s−1 and 287 μM, respectively (Table 1), and the turnover number and affinity constant were markedly lower and higher, respectively, than those of other canonical NirK proteins, including those from AOB. The product of NO2− reduction by the recombinant protein was examined using phenazine methosulfate (PMS) as the electron donor instead of BV. When BV was used as the electron donor, NO2− was reduced to NO, and further reduced to ammonia (approx. 60% of consumed 15NO2−) as observed in a previous study in which the NO2− reduction activity of A. xylosoxidans NirK was examined using methyl viologen (MV) as the electron donor (1). BV and MV have low redox potentials (−350 and −440 mV, respectively) (23), resulting in the reduction of NO to NH3; therefore, PMS with a higher redox potential (+80 mV) was used in the present study. The recombinant protein was incubated as described above in a 1.8-mL gas-tight vial with the addition of 0.5 mM PMS and 5 mM ascorbic acid instead of BV and dithionite, and the production of 15N-labeled gaseous compounds (i.e., N2, NO, and N2O) in the headspace was examined by gas chromatography mass spectrometry (GC/MS) as previously described (27). The diluted gases of 15-15N2 (Cambridge Isotope Laboratories, Tewksbury, MA, USA), 14NO, and 14-14N2O (GL Science, Tokyo, Japan) were also analyzed to prepare standard curves for quantification. The recombinant protein reduced 15NO2− with the oxidation of PMS, and 38 and 48% of consumed 15NO2− were converted to 15NO and 15-15N2O, respectively. This is direct evidence to show that the recombinant protein is a NO-forming nitrite reductase. We found that the production of 15-15N2O was equal to the production of 15NO, which likely results from the reduction of 15NO2− to H15NO (i.e., NO2−+2e−+3H+ → HNO+H2O) and the chemical formation of 15-15N2O from the formed H15NO (i.e., 2HNO → N2O+H2O) (35), as previously observed for a sulfide-linked nitrite reductase (34).
Table 1.
Enzymatic properties of archaeal and bacterial copper-containing nitrite reductase (NirK). ND; not determined.
| Organisms | MW* (kDa) | Cu content† (atom per subunit) | Absorption (nm) | Activity‡ Turnover (s−1) |
Km (μM) | Reference |
|---|---|---|---|---|---|---|
| Archaeal NirK | ||||||
| Nitrososphaera viennensis | 105±1.5 | 2.9 | 590 | |||
| NO2− reduction | 3.1 | 287 | This study | |||
| NH2OH oxidation | 0.039 | 97 | This study | |||
| Bacterial NirK (NO2− reduction) | ||||||
| Nitrosomonas europaea | 96 | ND | 450, 597 | 288 | ND | 18 |
| Nitrosococcus oceani | 114 | 1.67 | 455, 575 | 1,600 | 52 | 16 |
| Achromobacter xylosoxidans | 110 | 1.99 | 595 | 172 | 35 | 14, 32 |
| Candidatus Jettenia caeni | 101 | ND | 449, 598 | 319 | 250 | 7 |
Molecular weight (MW) of a trimeric NirK. The MW of Ca. Jettenia caeni NirK was calculated from amino acid sequences without a signal peptide sequence.
Copper contents previously assessed by chemical analyses were shown.
The following electron donors were used to evaluate the turnover number of NO2− reduction; methyl viologen for N. europaea and Nc. oceani, pseudoazurine for A. xylosoxidans, and benzyl viologen for Ns. viennensis and Ca. Jettenia caeni NirK.
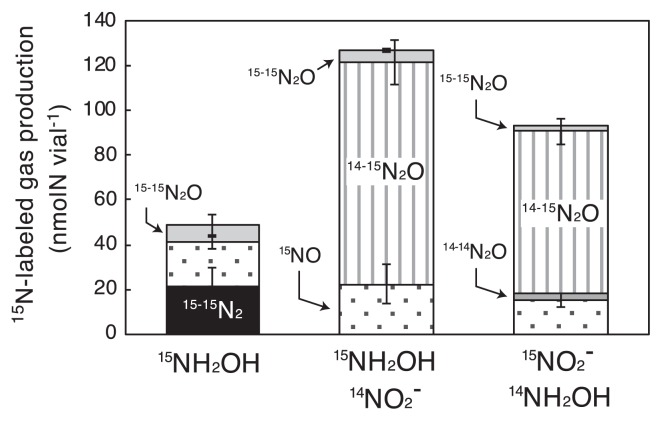
Aside from NO2− reduction, NH2OH oxidation was also investigated using the recombinant protein because NH2OH is produced as an intermediate during aerobic ammonia oxidation by AOA. The kinetics of NH2OH oxidation were examined by aerobically incubating the recombinant protein (245 μg mL−1) at 30°C and pH 7.5 with 0.5 mM NH2OH, with dissolved oxygen being available as an oxidant. The reaction was initiated by the addition of NH2OH solution, and the concentration of NH2OH was assessed colorimetrically (5). The concentration of H2O2, which may be produced by the oxidase activity of NirK (12), was also evaluated colorimetrically using horseradish peroxidase (Wako, Osaka, Japan) and 3,3′,5,5′-tetramethylbenzidine (TMBZ) (Dojindo, Kumamoto, Japan) (2). As shown in Fig. S2, the recombinant protein oxidized NH2OH with the production of H2O2. No NH2OH oxidation or H2O2 production was observed when the incubation was repeated without the addition of the recombinant protein. The values for the turnover number and affinity constant for NH2OH oxidation were 0.039 s−1 and 97 μM (Table 1), respectively, and the value for the turnover number was two orders of magnitude lower than that observed for NO2− reduction; therefore, the recombinant protein catalyzed NO2− reduction more efficiently. The addition of cytochrome c from equine heart (1 mg mL−1) or BV (0.5 mM) did not result in an increase in the reaction rate or affinity for NH2OH oxidation. The product of NH2OH oxidation by the recombinant protein was examined in a 15NH2OH tracer experiment (29). The recombinant protein was incubated in a 1.8-mL gas-tight vial with the addition of 0.5 mM 15NH2OH (Cambridge Isotope Laboratories) instead of 14NH2OH. After a 2-h incubation, the concentrations of the 15N-labeled gaseous products were assessed by GC/MS. The recombinant protein oxidized 15NH2OH and produced 15NO, 15-15N2O, and 15-15N2 gases quantitatively (Fig. 3), whereas the production of NO2− and NH3 was not detectable (detection limits: 50 and 100 μM, respectively). The oxidation of NH2OH to NO has been described in bacterial Hao (21); however, to the best of our knowledge, this is the first description of NH2OH oxidation by NirK. We also observed 15-15N2O production from 15NH2OH oxidation, which likely resulted from the oxidation of 15NH2OH to H15NO and abiotic coupling of H15NO, as previously described. Notably, 15-15N2 was the major product of 15NH2OH oxidation by the recombinant protein. Hydroxylamine disproportionation (30) may not be responsible for 15-15N2 production because NH3 production was not detectable in the liquid phase. The molecular mechanisms underlying the oxidation of 15NH2OH to 15-15N2 by the recombinant protein warrant further studies.
Fig. 3.
NH2OH oxidation by recombinant Nitrososphaera viennensis NirK. The recombinant protein was incubated at 30°C and pH 7.5 in 1.8-mL vials (volume of the headspace: 1.5 mL), with i) 0.5 mM 15NH2OH, ii) 15NH2OH and 14NO2− (each 0.5 mM), or iii) 14NH2OH and 15NO2−. The production of N2, NO, and N2O in the headspace was examined by gas chromatography mass spectrometry (GC/MS). NH3 and NO2− concentrations were also measured; however, they were not detectable during the incubation. During a 2-h incubation, i) 63±35 (mean±SD), ii) 149±1, and iii) 120±1 nmol N of NH2OH were consumed in the liquid phase, resulting in 75–137% of the 15N-labeled nitrogen mass balance in the vials. Error bars represent the SD derived from triplicate incubations, and the graph bars represent the mean values. NH2OH oxidation was not found in the vials without the addition of the recombinant protein.
We repeated the above incubation with the addition of NH2OH and NO2− because both compounds are available in AOA cells during aerobic ammonia oxidation. Therefore, the above incubation was repeated with the addition of 15NH2OH and 14NO2− (each 0.5 mM) or 14NH2OH and 15NO2− (Cambridge Isotope Laboratories) (each 0.5 mM). In both cases, 14–15N2O was the major product (Fig. 3), indicating that the recombinant protein produces N2O by oxidizing NH2OH using NO2− as an electron acceptor. N2O production by the denitrifier NirK from NH2OH and NO2− has been previously described (10), and the N-nitrosation reaction is involved in N2O production (39). Notably, Ns. viennensis cells produce N2O when they are incubated aerobically with NH3 and NO2− (42), although the Ns. viennensis genome lacks the gene encoding nitric oxide reductase (nor) that is involved in N2O production from nitrifier-denitrification. Stieglmeier et al. (42) suggested the involvement of Ns. viennensis NirK in the production of N2O in an Ns. viennensis culture, and our results support this hypothesis. Although the catalytic efficiency of Ns. viennensis NirK for NH2OH oxidation was markedly lower than that of NO2− reduction (Table 1), Ns. viennensis NirK may act as an NH2OH oxidase in Ns. viennensis cells and produce N2O under oxic growth conditions. Aside from 14–15N2O production, the production of 15NO and 15-15N2O was also observed when the recombinant protein was incubated with 14NH2OH and 15NO2− (Fig. 3).
Although the recombinant protein catalyzes NO2− reduction and NH2OH oxidation, the catalytic efficiency of both reactions was low, as shown in Table 1. AOA nirK transcripts are abundant in the transcriptome (8, 11, 20, 37), suggesting the strong expression of AOA NirK in cells. NirK was the 225th most abundant protein of the 1,503 proteins detected in the proteome of the late exponential phase of Ns. viennensis cells aerobically oxidizing ammonia (15). The strong expression of NirK appears to support the activity of NO2− reduction to NO as well as NH2OH oxidation to NO by the low efficiency catalytic enzyme. Ns. viennensis NirK may function as a bifunctional enzyme that supplies NO molecules from 2 different sources (i.e., NH2OH and NO2−), which provides Ns. viennensis cells with a competitive advantage. In the present study, the enzymatic kinetics of recombinant Ns. viennensis NirK for NO2− reduction were examined using artificial electron donors; further studies are needed to identify physiological electron donors in Ns. viennensis cells. Bacterial NirK may accept electrons supplied from single-domain cupredoxin and cytochrome c (14, 25). A number of genes encoding single-domain cupredoxin were found in the Ns. viennensis genome (Table S1), whereas the ortholog of the gene encoding cytochrome c was not. To date, the biochemistry of AOA cupredoxin has not been investigated using natural enzymes and recombinant proteins, and our study provides basic information that furthers our understanding of the biochemistry of AOA.
Supplementary text
Acknowledgements
This work was supported by JSPS KAKENHI Grant numbers 17K15305, 16H02371, and 16H04442 to M.O., T.Y., and N.A., respectively. The authors acknowledge Ko Furukawa, Akihiro Suzuki, and Hisashi Satoh for their technical assistance with the ESR, UV-VIS, and ICP-MS measurements, respectively.
References
- 1.Abraham Z.H.L., Lowe D.J., Smith B.E. Purification and characterization of the dissimilatory nitrite reductase from Alcaligenes xylosoxidans subsp. xylosoxidans (N.C.I.M.B. 11015): Evidence for the presence of both type 1 and type 2 copper centres. Biochem J. 1993;296:587–593. doi: 10.1042/bj2950587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos E.S., van der Doelen A.A., van Rooy N., Schuurs A.H.W.M. 3,3′,5,5′-Tetramethylbenzidine as an Ames test negative chromogen for horse-radish peroxidase in enzyme-immunoassay. J Immunoassay. 1981;2:187–204. doi: 10.1080/15321818108056977. [DOI] [PubMed] [Google Scholar]
- 3.Boulanger M.J., Murphy M.E. Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper-containing nitrite reductases. J Mol Biol. 2002;315:1111–1127. doi: 10.1006/jmbi.2001.5251. [DOI] [PubMed] [Google Scholar]
- 4.Carantoa J.D., Lancaster K.M. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc Natl Acad Sci USA. 2017;114:8217–8222. doi: 10.1073/pnas.1704504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frear D.S., Burrell R.C. Spectrophotometric method for determining hydroxylamine reductase activity in higher plants. Anal Chem. 1955;27:1664–1665. [Google Scholar]
- 6.Hatzenpichler R. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol. 2012;78:7501–7510. doi: 10.1128/AEM.01960-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hira D., Toh H., Migita C.T., Okubo H., Nishiyama T., Hattori M., Furukawa K., Fujii T. Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd1. FEBS Lett. 2012;586:1658–1663. doi: 10.1016/j.febslet.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 8.Hollibaugh J.T., Gifford S., Sharma S., Bano N., Moran M.A. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2011;5:866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isobe K., Ohte N. Ecological perspectives on microbes involved in N-cycling. Microbes Environ. 2014;29:4–16. doi: 10.1264/jsme2.ME13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki H., Matsubara T. A nitrite reductase from Achromobacter cycloclast. J Biochem. 1972;71:645–652. [PubMed] [Google Scholar]
- 11.Jung M.Y., Park S.J., Min D., Kim J.S., Rijpstra W.I.C., Sinninghe Damsté J.S., Kim G.J., Madsen E.L., Rhee S.K. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic Crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol. 2011;77:8635–8647. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakutani T., Watanabe H., Arima K., Beppu T. A blue protein as an inactivating factor for nitrite reductase from Alcaligenes faecalis strain S-6. J Biochem. 1981;89:463–472. doi: 10.1093/oxfordjournals.jbchem.a133221. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka K., Furusawa H., Takagi K., Yamaguchi K., Suzuki S. Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J Biochem. 2000;127:345–350. doi: 10.1093/oxfordjournals.jbchem.a022613. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka K., Yamaguchi K., Kobayashi M., Mori T., Bokui N., Suzuki S. Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances intermolecular electron transfer reaction with pseudoazurin. J Biol Chem. 2004;279:53374–53378. doi: 10.1074/jbc.M410198200. [DOI] [PubMed] [Google Scholar]
- 15.Kerou M., Offre P., Valledor L., Abby S.S., Melcher M., Nagler M., Weckwerth W., Schleper C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2016;113:E7937–E7946. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo K., Yoshimatsu K., Fujiwara T. Expression, and molecular and enzymatic characterization of Cu-containing nitrite reductase from a marine ammonia-oxidizing gammaproteobacterium, Nitrosococcus oceani. Microbes Environ. 2012;27:407–412. doi: 10.1264/jsme2.ME11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozlowski J.A., Stieglmeier M., Schleper C., Klotz M.G., Stein L.Y. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawton T.J., Bowen K.E., Sayavedra-Soto L.A., Arp D.J., Rosenzweig A.C. Characterization of a nitrite reductase involved in nitrifier denitrification. J Biol Chem. 2013;288:25575–25583. doi: 10.1074/jbc.M113.484543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G.W., Prosser J.I., Schuster S.C., Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 20.Lund M.B., Smith J.M., Francis C.A. Diversity, abundance and expression of nitrite reductase (nirK)-like genes in marine thaumarchaea. ISME J. 2012;6:1966–1977. doi: 10.1038/ismej.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maalcke W.J., Dietl A., Marritt S.J., Butt J.N., Jetten M.S.M., Keltjens J.T., Barends T.R.M., Kartal B. Structural basis of biological NO generation by octaheme oxidoreductases. J Biol Chem. 2014;289:1228–1242. doi: 10.1074/jbc.M113.525147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martens-Habbena W., Qin W., Horak R.E., et al. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol. 2015;17:2261–2274. doi: 10.1111/1462-2920.12677. [DOI] [PubMed] [Google Scholar]
- 23.Nagashima K.V. Redox titration for electron transfer proteins. Low Temp Sci. 2009;67:545–550. (in Japanese) [Google Scholar]
- 24.Nelson M.B., Martiny A.C., Martiny J.B.H. Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci USA. 2016;113:8033–8040. doi: 10.1073/pnas.1601070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nojiri M., Koteishi H., Nakagami T., Kobayashi K., Inoue T., Suzuki K.Y.S. Structural basis of inter-protein electron transfer for nitrite reduction in denitrification. Nature. 2009;462:117–120. doi: 10.1038/nature08507. [DOI] [PubMed] [Google Scholar]
- 26.Oshiki M., Awata T., Kindaichi T., Satoh H., Okabe S. Cultivation of planktonic anaerobic ammonium oxidation (anammox) bacteria by using membrane bioreactor. Microbes Environ. 2013;28:436–443. doi: 10.1264/jsme2.ME13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshiki M., Ishii S., Yoshida K., Fujii N., Ishiguro M., Satoh H., Okabe S. Nitrate-dependent ferrous iron oxidation by anaerobic ammonium oxidation (anammox) bacteria. Appl Environ Microbiol. 2013;79:4087–4093. doi: 10.1128/AEM.00743-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshiki M., Takagi R., Hatamoto M., Yamaguchi T., Araki N. High-cell-density cultivation of Nitrosomonas europaea in a membrane bioreactor for performing protein purification and characterization studies. J Gen Appl Microbiol. 2016;62:330–333. doi: 10.2323/jgam.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Oshiki M., Ali M., Shinyako-Hata K., Satoh H., Okabe S. Hydroxylamine-dependent anaerobic ammonium oxidation (anammox) by “Candidatus Brocadia sinica”. Environ Microbiol. 2016;18:3133–3143. doi: 10.1111/1462-2920.13355. [DOI] [PubMed] [Google Scholar]
- 30.Pacheco A.A., McGarry J., Kostera J., Corona A. Techniques for investigating hydroxylamine disproportionation by hydroxylamine oxidoreductases. Methods Enzymol. 2011;486:447–463. doi: 10.1016/B978-0-12-381294-0.00020-1. [DOI] [PubMed] [Google Scholar]
- 31.Prosser J.I., Nicol G.W. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- 32.Prudêncio M., Eady R.R., Sawers G. The blue copper-containing nitrite reductase from Alcaligenes xylosoxidans: cloning of the nirA gene and characterization of the recombinant enzyme. J Bacteriol. 1999;181:2323–2329. doi: 10.1128/jb.181.8.2323-2329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauder L.A., Ross A.A., Neufeld J.D. Nitric oxide scavengers differentially inhibit ammonia oxidation in ammonia-oxidizing archaea and bacteria. FEMS Microbiol Lett. 2016;363:fnw052. doi: 10.1093/femsle/fnw052. [DOI] [PubMed] [Google Scholar]
- 34.Sawhney V., Nicholas D.J.D. Sulphide-linked nitrite reductase from Thiobacillus denitrificans with cytochrome oxidase activity: purification and properties. J Gen Microbiol. 1978;106:119–128. [Google Scholar]
- 35.Shafirovich V., Lymar S.V. Nitroxyl and its anion in aqueous solutions: Spin states, protic equilibria, and reactivities toward oxygen and nitric oxide. Proc Natl Acad Sci USA. 2002;99:7340–7345. doi: 10.1073/pnas.112202099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen T., Stieglmeier M., Dai J., Urich T., Schleper C. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett. 2013;344:121–129. doi: 10.1111/1574-6968.12164. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y., Tyson G.W., Eppley J.M., DeLong E.F. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon J., Klotz M.G. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta. 2013;1827:114–135. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Spott O., Russow R., Stange C.F. Formation of hybrid N2O and hybrid N2 due to codenitrification: First review of a barely considered process of microbially mediated N-nitrosation. Soil Biol Biochem. 2011;43:1995–2011. [Google Scholar]
- 40.Stahl D.A., de la Torre J.R. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 41.Stieglmeier M., Klingl A., Alves R.J.E., Rittmann S.K.-M.R., Melcher M., Leisch N., Schleper C. Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2014;64:2738–2752. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stieglmeier M., Mooshammer M., Kitzler B., Wanek W., Zechmeister-Boltenstern S., Richter A., Schleper C. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J. 2014;8:1135–1146. doi: 10.1038/ismej.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vajrala N., Martens-Habbena W., Sayavedra-Soto L.A., Schauer A., Bottomley P.J., Stahl D.A., Arp D.J. Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc Natl Acad Sci USA. 2013;110:1006–1011. doi: 10.1073/pnas.1214272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Kessel M.A.H.J., Speth D.R., Albertsen M., Nielsen P.H., Op den Camp H.J.M., Kartal B., Jetten M.S.M. Complete nitrification by a single microorganism. Nature. 2015;528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker C.B., de la Torre J.R., Klotz M.G., et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuchter C., Abbas B., Coolen M.J.L., et al. Archaea nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan J., Haaijer S.C.M., Op den Camp H.J.M., et al. Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ Microbiol. 2012;14:3146–3158. doi: 10.1111/j.1462-2920.2012.02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.