Abstract
Background and Aim
Human leukocyte antigen (HLA)‐DQ2 and/or ‐DQ8 is an essential risk factor for celiac disease (CD). About 90–95% of patients with CD carry HLA‐DQ2/‐DQ8 alleles, and HLA‐DQ typing is considered an additional diagnostic test. Conventional polymerase chain reaction (PCR)‐based HLA‐DQ typing methods are expensive, complex, and a time‐consuming process. We assessed the efficacy of a novel HLA‐DQ typing method, “Celiac Gene Screen,” for the detection of CD‐associated HLA haplotypes.
Methods
To assess the diagnostic performance of the Celiac Gene Screen test, 100 ethylenediaminetetraacetic acid (EDTA) blood samples, already characterized by the conventional HLA‐DQ typing method, that is, PCR sequence‐specific oligonucleotide probes (PCR‐SSOP), a concordance between both the methods were explored. For validity, a further 300 EDTA blood samples with unknown HLA‐DQ status were genotyped using the Celiac Gene Screen test, including 141 samples from CD, 56 first‐degree relatives (FDRs) of CD and 103 samples from controls.
Results
Of the 100 samples with known status of HLA‐DQ alleles, 79 samples were HLA‐DQ2 and/or ‐DQ8 positive, and 21 samples were HLA‐DQ2 and/or ‐DQ8 negative by conventional PCR. These 100 samples were re‐typed using the Celiac Gene screen kit; all 79 positives were typed positive, and 21 negatives were typed negative for HLA‐DQ alleles. Among 300 samples with unknown HLA‐DQ status, 118 of 141 (84%) patients with CD, 48 of 56 (86%) FDRs of CD, and 52 of 103 (50%) controls typed positive for HLA‐DQ alleles.
Conclusions
The Celiac Gene Screen HLA‐DQ typing method showed excellent concordance with the conventional HLA‐DQ typing method and could be a cost‐reducing and effective method for CD‐associated HLA screening.
Keywords: alleles, celiac disease, genotyping, HLA, polymerase chain reaction, susceptibility
Introduction
Celiac disease (CD) is a chronic inflammatory condition of the small intestine, initiated by the ingestion of dietary gluten peptides (contained in wheat, rye, and barley) in genetically susceptible individuals.1 The presence of HLA‐DQ2 and/or ‐DQ8 haplotypes, identified in the HLA class II histocompatibility system expressed on the surface of antigen‐presenting cells (APC), mainly macrophages, dendritic cells, and B cells, is considered as a necessary risk factor for the development of CD. Risk of CD is excluded if both genotypes are absent, with nearly 95% confidence.2
In a recent systematic review and meta‐analysis, we observed that 1.37% of the global population is seropositive for CD, and 0.68% of the global population is estimated to have biopsy‐confirmed CD.3 Furthermore, various landmark studies have reported a worldwide increase in the prevalence of CD in the past few decades.3, 4, 5, 6, 7 While CD was thought to be uncommon in India, in a pan‐India population‐based study involving more than 23 331 subjects, we observed that 0.68% of Indians have either potential CD or CD.8 While a large number of subjects is expected to have CD, the majority still remains undiagnosed.5
The HLA‐DQ2 heterodimer associated with the susceptibility to CD is formed by an alpha chain encoded by the HLA‐DQA1*05 allele and a beta chain encoded by the HLA‐DQB1*02 allele (now defined as HLA‐DQ 2.5).9 These two HLA alleles (in either cis or trans position) are present in at least 90% of patients with CD.10, 11 The HLA‐DQ8 heterodimer is found in 5% of CD patients and is formed by an alpha chain and beta chain encoded by HLA‐DQA1*03 and HLA‐DQB1*03:02, respectively.10, 12 However, HLA‐DQ2 or HLA‐DQ8 are expressed in almost 30–55% of the general population.13, 14 HLA‐DQ2/‐DQ8 typing is currently considered as an additional diagnostic test for CD, particularly useful when taking clinical decisions where there are discrepancies between celiac‐specific serological tests and intestinal mucosal biopsies and for the screening of “at‐risk” individuals, such as first‐degree relatives (FDRs) of patients with CD; patients with type 1 diabetes; and patients with chromosomal diseases such as William's syndrome, Turner syndrome, and Down's syndrome.15, 16, 17, 18, 19, 20 Due to its high negative predictive value, it is helpful in excluding the subjects requiring further investigations and may find application in both mass screening or case finding strategies. A combination of different serological tests (e.g. IgA anti‐transglutaminase, anti‐endomysial ab) with HLA DQ‐2/‐DQ8 genotyping can strengthen the serological test result and could minimize the burden of unnecessary duodenal biopsies.21 In the recent European pediatric guidelines (European Society for Pediatric Gastroenterology Hepatology and Nutrition [ESPGHAN]) for the diagnosis of CD, HLA typing is indicated as an adjunctive and an appropriate test to avoid the mucosal biopsies in patients demonstrating symptoms and a high titer of celiac autoantibodies (i.e. IgA anti‐transglutaminase).22
There are several HLA‐DQ typing approaches currently available, such as polymerase chain reaction‐sequence‐specific primers (PCR‐SSP), PCR‐sequence‐specific oligonucleotide probes (PCR‐SSOP), real‐time PCR (RT‐PCR), and microarray.2 Commercially available kits used to perform these tests are expensive and time consuming, and their methodologies are complex, requiring a well‐trained technician to perform the test. Duration of analysis, costs, and availability of these tests limit their widespread use in the clinical practice. There is an unmet need for an accurate, quick, easy, and affordable method to perform HLA‐DQ typing.
To make HLA‐DQ typing easy and inexpensive, we proposed a new‐fangled, reliable, and unambiguous sequence‐specific primer‐based rapid, single PCR HLA‐DQ typing method, “Celiac Gene Screen,” developed by BioDiagene S.R.L (Palermo, Italy). This quick HLA‐DQ typing test isolates DNA in approximately 1 min and runs total PCR (amplification of HLA‐DQ alleles) in 90 min. The Celiac Gene Screen HLA‐DQ typing kit provides information on the presence/absence of HLA‐DQ2 and/or ‐DQ8 alleles (yes/no), without identification of all critical HLA alleles necessary to identify the CD risk heterodimers.
The aim of this study was to validate the accuracy of the “Celiac Gene Screen” through a comparison of this new test with the conventional HLA‐DQ alleles typing test (SSOP‐PCR method).
Methods
During June 2017, the rapid HLA‐DQ typing method, “Celiac Gene Screen,” was applied to the stored EDTA blood samples collected from the biorepository of the Celiac Clinic of the Department of Gastroenterology and Human Nutrition, All India Institute of Medical Sciences, New Delhi, India. This study was approved by the Ethics Committee of the institution.
Selection of blood samples
In the present study, we included a total of 400 blood samples that had been collected in EDTA blood collection vials on earlier occasions and were stored in the biorepository of our institution. These samples had been collected during a community prevalence study that was conducted earlier and also from patients with CD being followed up at our center. Written and informed consent was requested from all the participants during the respective studies. To determine the diagnostic performance of the Celiac Gene Screen kit, we tested 100 blood samples in which HLA genotyping had already been characterized by the conventional SSOP‐PCR with the Luminex‐based One Lambda (LAB Type SSO Class II DQA1/DQB1 typing kit, CA, USA). In the remaining 300 EDTA blood samples where HLA‐DQ status had not been assessed previously, 141 samples were collected from patients with CD (having IgA anti‐tTG ab positivity and villous abnormality of modified Marsh grade 2 or more), 56 samples from FDRs of patients with CD and 103 IgA anti‐tTG negative samples (controls).
The procedure of the Celiac Gene Screen kit
The Celiac Gene Screen kit involves three major steps, including preliminary lysis of EDTA blood sample, DNA amplification (one reaction/test), and fluorescence detection using a BioRun Reader. The total procedure time of this test to obtain the final result is less than 2 h. During the HLA‐DQ typing procedure, manufacturer's guidelines were strictly followed. Major steps of the procedure are described below.
Step 1: lysis of blood samples collected in EDTA tubes
Tubes containing EDTA blood samples were allowed to thaw at room temperature and vortexed gently. For each blood sample, 200 μl of extraction buffer (provided with the Celiac Gene Screen kit) was poured in a 1.5‐ml Eppendorf tube. Ten microliter of EDTA blood was mixed with the extraction buffer and incubated at room temperature for 1 min. This step facilitated quick DNA isolation. Of this lysate, 2 μl was used further for the DNA amplification.
Step 2: amplification of HLA‐DQ alleles
In 0.2‐ml PCR tubes provided with the Celiac Gene Screen kit, containing dried primers/probes for a housekeeping gene (internal control) and the target alleles, 18 μl of ready‐to‐use Taq mix (provided with the kit) and 2 μl of isolated DNA were mixed by pipetting. PCR tubes were vortexed for about 15 s and were then placed into a thermocycler. After 90 min of PCR run, tubes were ready for use in the analysis.
Step 3: detection of HLA‐DQ alleles
PCR tubes were placed in the BioRun Reader. In‐built software analyzed all the data at once and gave the interpretation in 10–20 s. The Celiac Gene Screen identifies the alleles DQB1*02 codifying for the beta chain of the DQ2 antigen and the DQB1*03:02 alleles codifying for the DQ8 beta chain. In each BioDiagene PCR tube, two fluorescence probes corresponding to primers were also coated, one that recognized the housekeeping gene and the second one that was perfectly complementary to that of the target DNA. The tubes were excited with two well‐defined wavelengths. The increase in the reported fluorescent signal was directly proportional to the number of amplicons generated. Therefore, depending on the fluorescence value in the tube, the BioRun Reader reported the presence or absence of CD‐associated alleles in the sample.
Statistical analysis
Quantitative variables were summarized as the mean ± SD; 95% confidence interval (CI) was calculated using the Software Program Stata System (SPSS) IBM version 25, Chicago, USA.
Results
Concordance of Celiac Gene Screen Kit with conventional HLA‐DQ2 and ‐DQ8 typing method
Of 100 blood samples (mean age ± SD: 25.3 ± 11.1 years; 40 males) with a known status of HLA‐DQ2 and ‐DQ8, 79 samples (mean age ± SD: 24.5 ± 10.7 years, 31 males) were HLA‐DQ2 and/or ‐DQ8 positive, and 21 samples (mean age ± SD: 28.4 ± 12.5 years; 9 males) were HLA‐DQ2 and ‐DQ8 negative by the conventional SSOP HLA typing method. These 100 samples were regenotyped using the Celiac Gene Screen kit, and all of the 79 HLA‐DQ2 and/or ‐DQ8 positive samples were positive, and all the 21 HLA‐DQ2 and ‐DQ8 negative samples were typed negative by the Celiac Gene Screen Kit as well. This shows an excellent concordance (100%) rate between HLA testing using the SSOP HLA typing method and the Celiac Gene Screen method (Fig. 1).
Figure 1.
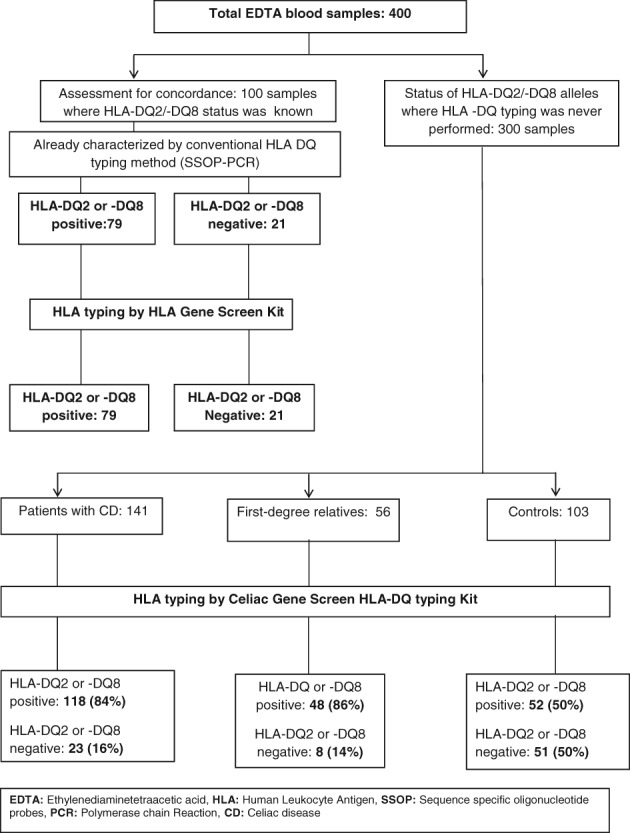
HLA‐DQ2/‐DQ8 status in the samples selected in the study. CD, celiac disease; EDTA, ethylenediaminetetraacetic acid; HLA, human leukocyte antigen; PCR, polymerase chain reaction; SSOP, sequence‐specific oligonucleotide probes.
HLA DQ status in CD samples
Of 300 samples with unknown HLA‐DQ status (mean age ± SD: 30 ± 14.7 years; 145 males), 141 blood samples (mean age ± SD: 30 ± 14 years; 68 males) were from patients with CD. Of these, 118 (84%, 95% CI: 78–90) samples (mean age ± SD: 26.2 ± 13.5 years; 57 males) showed HLA‐DQ2 and/or ‐DQ8 positivity, and 23 (16%, 95% CI: 10–22) samples (mean age ± SD: 30.7 ± 14.1 years; 11 males) were HLA‐DQ2 and ‐DQ8 negative (Fig. 1).
HLA DQ status in FDRs of patients with CD
Of 56 samples (mean age ± SD: 27.4 ± 13.9 years; 33 males) from FDRs, 48 (86%, 95% CI: 85–87%) samples (mean age ± SD: 28.1 ± 14.4 years; 32 males) tested positive for HLA‐DQ2 and/or ‐DQ8. On the other hand, eight (14%, 95% CI: 5–23) samples (mean age ± SD: 23.2 ± 10.8 years; 1 male) were HLA‐DQ2 and/or DQ8 negative (Fig. 1).
HLA‐DQ status in control samples
In 103 samples (mean age ± SD: 38.4 ± 13.5 years; 44 males) from control subjects, 52 (50%, 95% CI: 40–60) samples (mean age 36.7 ± 13.2 years; 18 males) were HLA‐DQ2 and/or ‐DQ8 positive, whereas the remaining 51 (50%, 95% CI: 40–60) samples (mean age ± SD: 40.3 ± 13.9 years; 26 males) were reported to be HLA‐DQ2 and ‐DQ8 negative (Fig. 1).
Discussion
HLA‐DQ genes (HLA ‐DQ2/‐DQ8) play a primary role in the development of CD. Clinically, HLA‐DQ genotyping does not provide a final diagnosis of the CD but indicates the necessary predisposition required for the development of CD.23, 24 Due to its high negative predictive value, the absence of HLA‐DQ predisposing alleles makes the diagnosis of CD unlikely. Different combinations of HLA‐DQ CD predisposing alleles also determine the level of risk of CD development. For instance, individuals presenting a double dose of the HLA‐DQ B1*02 variant remain at high risk of developing CD in comparison to those expressing a single dose of the DQB1*02 allele.25, 26 At‐risk subjects, such as FDRs of patients with CD; patients with other autoimmune diseases (type 1 diabetes); and individuals with Williams, Turner, and Down's syndrome showing HLA‐DQ2 and ‐DQ8 negativity, particularly in Western countries, remain out of risk of developing CD on a lifelong basis.16, 17, 18, 19, 20 A single HLA determination in such subjects may set them free from future surveillance and unnecessary follow‐up in the clinic (repetitive serology, duodenal endoscopy/biopsy).27
Furthermore, a duodenal biopsy may be avoided in some subjects if one demonstrates that the patient has a genetic susceptibility to CD by showing the presence of HLA‐DQ2/‐DQ8 haplotype if they have a positive antiendomysial antibody (EMA) and high titer of antitissue transglutaminase antibody (IgA anti‐tTG ab). However, the latest ESPGHAN guideline specifically recommends avoiding histological assessment in children and adolescents with signs or symptoms suggestive of CD with high anti‐tTG ab titers with levels >10 times the upper limits of normal (ULN), supported by a positive EMA test, also with positive HLA‐DQ2 and/or HLA‐DQ8 alleles.22 The ESPGHAN guideline also recommends HLA‐DQ‐2.5/‐DQ8 typing in all type 1 diabetes mellitus patients for screening of CD and encourages the assessment of HLA‐DQ2.5/‐DQ8 genotyping as the first screening tool in asymptomatic individuals with a family risk of CD.22
Conventional HLA‐DQ typing is an expensive method. Due to its high cost, HLA‐DQ typing is not performed in many studies, and a considerable number of at‐risk individuals do not undergo this test.28 At our center (AIIMS) in New Delhi, HLA‐DQ2/‐DQ8 typing (high resolution) costs approximately 3000 Indian rupees (INR)/test (approximately US Dollar $46/test), while private laboratories in New Delhi offer this test at about 9000 INR/test (approximately USD138/test). In Italy, the cost of HLA genotyping varies from €46 to €180/test. In other parts of Europe, HLA‐DQ typing costs €167–€326/test, and within the United States, in the nonprofit private medical center, HLA‐DQ typing costs approximately USD 350/test.29, 30 The cost of HLA‐DQ genotyping in our study was much lower (per test price by manufacturer, about €15, excluding all other laboratory costs). However, we wish to emphasize that the Celiac Gene Screen kit is a low‐resolution test most suitable for CD‐associated screening studies. It covers the set of critical HLA alleles necessary to identify the CD risk heterodimers (DQB1*02 and DQB1*03:02 alleles). An affordable price for HLA‐DQ typing may increase the use of this test in developing countries, especially for CD screening studies, and may help physicians resolve many clinical dilemmas related to CD diagnosis.
In the present study, we compared the efficacy of a novel rapid HLA‐DQ2 and/or ‐DQ8 typing kit (“Celiac Gene Screen Kit”) with that of the conventional SSOP HLA‐DQ typing method. We observed an excellent (100%) concordance rate in the reporting of two methods. We also determined the HLA‐DQ status in blood samples collected from different groups (CD, FDRs of CD patients and controls) where HLA‐DQ status was not known previously. In the present study, we observed that 84% of patients with CD were HLA‐DQ2/‐DQ8 positive compared with approximately 90–95% Caucasian CD patients in Europe and the United States.10, 11 The results of this study match the observations in our previous population‐based study, where we observed that about 76% of patients from the northern part of India detected to have CD were HLA‐DQ2/‐DQ8 positive.8 There may be another HLA genotype in the rest of North Indian patients with CD. A high throughput HLA DQ allele characterization may identify the risk alleles for these 23 (16%) negative HLA but confirmed CD samples. However, these 23 CD patients who were typed negative for HLA‐DQ alleles had a high titer of anti‐tTG ab and had villous abnormality of Modified Marsh grade between 3a and 3c (Appendix S1, Supporting Information). One pediatric patient was exempted from duodenal endoscopy biopsy test as he or she had a significantly higher level of anti‐tTG ab. In FDRs of CD patients and control subjects, the percentage of HLA‐DQ2 and/or ‐DQ8 positivity was in line with other well‐designed CD prevalence studies.31, 32, 33
Small sample size was a limitation of this study. A higher number of samples could have provided a more robust result. This test does not provide homo‐ or heterozygous status; instead, it only reveals the presence or absence of CD‐associated alleles (HLA‐DQ2/‐DQ8). Nevertheless, this method can be useful in screening both individuals at risk of CD and the general population. The advantage of this test is to provide HLA‐DQ2 and/or‐DQ8 status quickly. Setting up this machinery in standard‐level laboratories is feasible and quite easy. Unlike the conventional HLA‐DQ typing method, this method does not involve complex steps and is not restricted to deeply trained and experienced laboratory personnel.
To make HLA‐DQ typing easy, more than a few sincere efforts have been made to deliver a simpler but reliable HLA‐DQ typing test. In recent times, several easy‐to‐perform commercial HLA‐DQ typing kits have been introduced, claiming to be able to characterize celiac‐associated genes (HLA‐DQ genes) specifically, unambiguously, and exactly by using conventional HLA‐DQ typing methods with overlapping sensitivity to the conventional ones. While these tests are easier than the conventional method, they require critical steps to be performed as DNA isolation has to be performed either manually (that takes several hours) or using a rapid kit‐based method for DNA amplification (PCR run), and the amplicons have to be run on an agarose gel to read for interpretation of HLA‐DQ status.13, 24, 34, 35 The celiac gene screen HLA‐DQ typing test makes every step easier, from DNA isolation (few minutes) to PCR run (90 min), and results in interpretation (within minutes). Furthermore, to the best of our current knowledge, in comparison to conventional and other commercially available HLA‐DQ typing methods we have explored, the Celiac Gene Screen HLA‐DQ typing method costs less and is probably the easiest, quickest, and most economical and reliable method to identify celiac‐associated alleles.24, 29, 30
In conclusion, the Celiac Gene Screen method showed excellent concordance with the conventional HLA‐DQ typing test (SSOP‐PCR). This is a rapid, easy‐to‐conduct, and relatively less expensive method for the specific and unambiguous detection of CD‐associated HLA alleles. The Celiac Gene Screen kit can be an effective tool for CD‐associated HLA screening, which could be particularly useful whenever problems of budget limitations hamper the possibility of applying this important diagnostic method.
Supporting information
Appendix S1 Clinical features of celiac disease patients, typed HLA‐DQ2/‐DQ8 negative using the Celiac Gene Screen method.
Acknowledgments
We acknowledge the support of BioDiagene S.R.L (Palermo, Italy) in providing the Celiac Gene Screen kit. We thank Emanuela Maria Mariani (Department of Pediatrics, Università Politecnica delle Marche, Ancona Italy) for the execution of administrative work. We appreciate and thank the Università Politecnica delle Marche (Ancona, Italy) for approving funds for travel. We acknowledge and recognize the support and the help of Anu Verma (Ancona, Italy) in assisting with the study of data entry. We appreciate the contribution of students and staff of the Department of Gastroenterology and Human Nutrition, AIIMS, New Delhi, India, for their support.
Declaration of conflict of interest: Elena Lionetti and Carlo Catassi are scientific consultants of Dr. Schär's. All other authors declare no conflict of interest.
Author contribution: Anil K Verma did the interpretation of data and statistical analysis. Anil K Verma and Alka Singh did the selection of biological material and performance of HLA‐DQ typing. Carlo Catassi, Vineet Ahuja, Govind K Makharia designed the overall concept and supervised the study. Anil K Verma, Carlo Catassi, Govind K Makharia drafted the manuscript. Elena Lionetti, Simona Gatti, Tiziana Galeazzi, Chiara Monachesi, Elisa Franceschini, and Vineet Ahuja provided critical analysis of final outcome of the results and its analysis, and review of the manuscript. Carlo Catassi and Govind K Makharia provided administrative and financial support and final approval of the version to be published. All authors read and approved the final version of this manuscript.
Financial support: This study has not received any external grant support.
References
- 1. Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2002; 2: 647–55. [DOI] [PubMed] [Google Scholar]
- 2. Fasano ME, Dametto E, D'Alfonso S. HLA Genotyping: Methods for the Identification of the HLA‐DQ2,‐DQ8 Heterodimers Implicated in Celiac Disease (CD) Susceptibility. Methods Mol. Biol. 2015; 1326: 79–92. [DOI] [PubMed] [Google Scholar]
- 3. Singh P, Arora A, Strand TA et al Global Prevalence of Celiac Disease: Systematic Review and Meta‐analysis. Clin. Gastroenterol. Hepatol. 2018; 16: 823–836.e2. [DOI] [PubMed] [Google Scholar]
- 4. Mustalahti K, Catassi C, Reunanen A et al The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann. Med. 2010; 42: 587–95. [DOI] [PubMed] [Google Scholar]
- 5. Catassi C, Gatti S, Lionetti E. World perspective and celiac disease epidemiology. Dig. Dis. 2015; 33: 141–6. [DOI] [PubMed] [Google Scholar]
- 6. Catassi C, Gatti S, Fasano A. The new epidemiology of celiac disease. J. Pediatr. Gastroenterol. Nutr. 2014; 59(Suppl. 1): S7–9. [DOI] [PubMed] [Google Scholar]
- 7. Kang JY, Kang AHY, Green A, Gwee KA, Ho KY. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment. Pharmacol. Ther. 2013; 38: 226–45. [DOI] [PubMed] [Google Scholar]
- 8. Ramakrishna BS, Makharia GK, Chetri K et al Prevalence of Adult Celiac Disease in India: Regional Variations and Associations. Am. J. Gastroenterol. 2016; 111: 115–23. [DOI] [PubMed] [Google Scholar]
- 9. Tye‐Din JA, Cameron DJS, Daveson AJ et al Appropriate clinical use of human leukocyte antigen typing for coeliac disease: an Australasian perspective. Intern. Med. J. 2015; 45: 441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA‐DQ alpha/beta heterodimer. J. Exp. Med. 1989; 169: 345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sollid LM, Thorsby E. The primary association of celiac disease to a given HLA‐DQ alpha/beta heterodimer explains the divergent HLA‐DR associations observed in various Caucasian populations. Tissue Antigens. 1990; 36: 136–7. [DOI] [PubMed] [Google Scholar]
- 12. Margaritte‐Jeannin P, Babron MC, Bourgey M et al HLA‐DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens. 2004; 63: 562–7. [DOI] [PubMed] [Google Scholar]
- 13. Megiorni F, Mora B, Bonamico M et al A rapid and sensitive method to detect specific human lymphocyte antigen (HLA) class II alleles associated with celiac disease. Clin. Chem. Lab. Med. 2008; 46: 193–6. [DOI] [PubMed] [Google Scholar]
- 14. Cecilio LA, Bonatto MW. The prevalence of HLA DQ2 and DQ8 in patients with celiac disease, in family and in general population. Arq. Bras. Cir. Dig. 2015; 28: 183–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hill ID, Horvath K. Nonbiopsy diagnosis of celiac disease: are we nearly there yet? J. Pediatr. Gastroenterol. Nutr. 2012; 54: 310–11. [DOI] [PubMed] [Google Scholar]
- 16. Vitoria JC, Castaño L, Rica I, Bilbao JR, Arrieta A, García‐Masdevall MD. Association of insulin‐dependent diabetes mellitus and celiac disease: a study based on serologic markers. J. Pediatr. Gastroenterol. Nutr. 1998; 27: 47–52. [DOI] [PubMed] [Google Scholar]
- 17. Barera G, Bonfanti R, Viscardi M et al Occurrence of celiac disease after onset of type 1 diabetes: a 6‐year prospective longitudinal study. Pediatrics. 2002; 109: 833–8. [DOI] [PubMed] [Google Scholar]
- 18. Giannotti A, Tiberio G, Castro M et al Coeliac disease in Williams syndrome. J. Med. Genet. 2001; 38: 767–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonamico M, Pasquino AM, Mariani P et al Prevalence and clinical picture of celiac disease in Turner syndrome. J. Clin. Endocrinol. Metab. 2002; 87: 5495–8. [DOI] [PubMed] [Google Scholar]
- 20. Bonamico M, Mariani P, Danesi HM et al Prevalence and clinical picture of celiac disease in Italian down syndrome patients: a multicenter study. J. Pediatr. Gastroenterol. Nutr. 2001; 33: 139–43. [DOI] [PubMed] [Google Scholar]
- 21. Anderson RP, Henry MJ, Taylor R et al A novel serogenetic approach determines the community prevalence of celiac disease and informs improved diagnostic pathways. BMC Med. 2013; 11: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Husby S, Koletzko S, Korponay‐Szabó IR et al European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012; 54: 136–60. [DOI] [PubMed] [Google Scholar]
- 23. Megiorni F, Mora B, Bonamico M et al HLA‐DQ and risk gradient for celiac disease. Hum. Immunol. 2009; 70: 55–9. [DOI] [PubMed] [Google Scholar]
- 24. Kaukinen K, Partanen J, Mäki M, Collin P. HLA‐DQ typing in the diagnosis of celiac disease. Am. J. Gastroenterol. 2002; 97: 695–9. [DOI] [PubMed] [Google Scholar]
- 25. Megiorni F, Pizzuti A. HLA‐DQA1 and HLA‐DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J. Biomed. Sci. 2012; 19: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vader W, Stepniak D, Kooy Y et al The HLA‐DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten‐specific T cell responses. Proc. Natl. Acad. Sci. USA. 2003; 100: 12390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubio‐Tapia A, Van Dyke CT, Lahr BD et al Predictors of family risk for celiac disease: a population‐based study. Clin. Gastroenterol. Hepatol. 2008; 6: 983–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martins R de CA, Gandolfi L, Modelli IC, Almeida RC, Castro LC, Pratesi R. Serologic screening and genetic testing among Brazilian patients with celiac disease and their first degree relatives. Arq. Gastroenterol. 2010; 47: 257–62. [DOI] [PubMed] [Google Scholar]
- 29. Elias J, Hoorweg‐Nijman JJG, Balemans WA. Clinical relevance and cost‐effectiveness of HLA genotyping in children with Type 1 diabetes mellitus in screening for coeliac disease in The Netherlands. Diabet. Med. 2015; 32: 834–8. [DOI] [PubMed] [Google Scholar]
- 30. Pallav K, Kabbani T, Tariq S, Vanga R, Kelly CP, Leffler DA. Clinical utility of celiac disease‐associated HLA testing. Dig. Dis. Sci. 2014; 59: 2199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Srivastava A, Yachha SK, Mathias A, Parveen F, Poddar U, Agrawal S. Prevalence, human leukocyte antigen typing and strategy for screening among Asian first‐degree relatives of children with celiac disease. J. Gastroenterol. Hepatol. 2010; 25: 319–24. [DOI] [PubMed] [Google Scholar]
- 32. Mishra A, Prakash S, Kaur G et al Prevalence of celiac disease among first‐degree relatives of Indian celiac disease patients. Dig. Liver Dis. 2016; 48: 255–9. [DOI] [PubMed] [Google Scholar]
- 33. Castro‐Antunes MM, Crovella S, Brandão LAC, Guimaraes RL, Motta MEFA, Silva GAP. Frequency distribution of HLA DQ2 and DQ8 in celiac patients and first‐degree relatives in Recife, northeastern Brazil. Clinics (Sao Paulo). 2011; 66: 227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piancatelli D, Ben El Barhdadi I, Oumhani K, Sebastiani P, Colanardi A, Essaid A. HLA Typing and Celiac Disease in Moroccans. Med. Sci. 2017; 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joda H, Beni V, Curnane D et al Low–medium resolution HLA‐DQ2/DQ8 typing for coeliac disease predisposition analysis by colorimetric assay. Anal. Bioanal. Chem. 2012; 403: 807–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Clinical features of celiac disease patients, typed HLA‐DQ2/‐DQ8 negative using the Celiac Gene Screen method.


