Abstract
Accumulating evidence suggests the roles of glutamate metabotropic receptors (GRMs) in cancer, in addition to synaptic signalling. The present study assessed the associations of genetic variants in eight GRM genes with regard to risk and overall survival (OS) in 780 renal cell carcinoma (RCC) patients and controls. After adjustment for known risk factors, GRM5 rs7102764 T was associated with an increased risk of RCC (P = 0.006). Additional analysis has provided evidence that rs7102764 T was correlated with a higher expression of GRM5, which is consistently found to be upregulated in tumours, compared to normal tissues. Furthermore, the GRM3 rs701332 C, GRM4 rs2499707 T, and GRM4 rs4713742 T alleles were significantly associated with a poorer OS (P ≤ 0.030). The three loci were also observed to have strong cumulative effects on OS. Additional analysis has revealed a significant genotype‐expression correlation of rs2499707 T with increased GRM4 expression, which in turn leads to poorer OS in patients with RCC. GRMs might be involved in RCC development and progression, and genetic variants in GRMs might be promising biomarkers.
Keywords: glutamate metabotropic receptors, prognosis, renal cell carcinoma, single‐nucleotide polymorphisms, survival
1. INTRODUCTION
Renal cell carcinoma (RCC) is the most common type of kidney cancer, and is one of the most common cancers in the United States, with an estimate of 65 340 new cases and 14 970 deaths occurring in 2018.1 In Taiwan, kidney cancer was ranked as the 15th and 16th leading cause of cancer mortality in males and females, respectively, and accounted for 1.27% of all cancers in 2015. Multiple environmental risk factors, such as cigarette smoking and arsenic exposure, and genetic variations have been reported to be involved in the aetiology of RCC.2, 3 It is important to explore RCC‐related risk factors and its underlying mechanisms in order to improve therapeutic treatments.
Glutamate is an excitatory neurotransmitter in processes such as memory and learning,4 but recent studies have also implicated glutamate signalling in the development and progression of various cancers.5 Glutamate metabotropic receptors (GRMs) are G‐protein‐coupled receptors, which are activated by glutamate and stimulate secondary messengers such as phospholipase C/protein kinase C/calcium, phosphatidylinositol 3‐kinase/Akt/mammalian target of rapamycin, and mitogen‐activated protein kinase pathways, to generate the glutamate signalling cascades.6 The GRM family comprises eight members and is classified into three groups according to the difference in sequence similarity and downstream second messenger pathways.7 Group I (GRM1 and 5) initiates signalling via the phospholipase C/protein kinase C/calcium pathway, whereas group II (GRM2 and 3) and group III (GRM4, 6, 7 and 8) couple negatively with adenylyl cyclase to suppress the production of cyclic AMP and inhibit protein kinase A.
In addition to the central nervous system, a functional glutamatergic system has been reported in non‐neuronal peripheral cells.8 Furthermore, studies have suggested that glutamate signalling is dysregulated and may play roles in human malignancies.9 Here, we explored the associations of gene expression and genetic variants of GRMs with prognosis in patients with RCC.
2. MATERIALS AND METHODS
2.1. Patient population and clinical data collection
The study population consisting of 780 participants was recruited from three Taipei city hospitals: National Taiwan University Hospital, Taipei Medical University Hospital, and Taipei Municipal Wan Fang Hospital, as described previously.2, 10 A total of 390 patients with pathologically confirmed RCC were matched for age (±1 year) and gender with 390 cancer‐free controls. The demographic data were collected through in‐person interviews using a structured questionnaire, and the clinical and follow‐up information was obtained from medical records. Recurrence‐free survival (RFS) was defined as the time from surgery to the first date of recurrence. Overall survival (OS) was defined as the time from surgery to death due to any cause. This study was performed in accordance with the approval protocols by The Research Ethics Committee of National Taiwan University Hospital, and written informed consent was obtained from all participants before the questionnaire interview and specimen collection.
2.2. Single‐nucleotide polymorphism (SNP) selection and genotyping
The candidate SNPs were identified across eight GRM genes (GRM1‐8), including 5 kb upstream and 1 kb downstream of each gene, using SNPinfo.11 TagSNPs were selected based on a minor allele frequency (MAF) of >0.05 in the HapMap CHB (Han Chinese in Beijing) population, a pairwise linkage disequilibrium squared correlation coefficient (r 2) of >0.8, and whether they were potentially functional; a maximum of five tagSNPs per gene was selected. A total of 35 tagSNPs were selected for genotyping. Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA). Genotyping was carried out as described previously,12 using Agena Bioscience iPLEX matrix‐assisted laser desorption/ionization time‐of‐flight mass‐spectrometry technology at the National Center for Genome Medicine, Taiwan. Any SNP that failed the assay design (N = 7), deviated from Hardy‐Weinberg equilibrium (P < 0.01, N = 2), or had an MAF of <0.01 (N = 1) was excluded. Finally, a total of 25 SNPs were included for further analysis, and the average genotype call rate was 98.8%.
2.3. Bioinformatics analysis
We used HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) to annotate the regulatory features of the region adjoining the risk SNPs.13 The association of selected SNP‐gene expression quantitative trait loci (eQTL) was evaluated using Genotype‐Tissue Expression (GTEx).14 The clinical significance of the expression of GRMs on RCC was analysed using The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA‐KIRC) data15 and DriverDB.16, 17
2.4. Statistical analysis
Chi‐square or Mann‐Whitney U test was used to compare the categorical or continuous variables, respectively, between the RCC cases and healthy controls. Univariate and multivariate logistic regression was used to estimate the crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) of the SNP genotypes and RCC risk. Multivariate logistic regression models for each SNP were adjusted for age, gender, alcohol consumption, and histories of hypertension and diabetes. Kaplan‐Meier analysis with the log‐rank test was used to assess the associations of SNPs or gene expression with OS. Multivariable Cox regression, after adjustment for age and gender, was performed to estimate the adjusted hazard ratios (HRs) and their 95% CIs for the association of SNP with OS. Spearman's rank correlation tests were used to determine the association between the expression of GRMs and clinical characteristics of RCC. All statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software version 19.0.0 (IBM, Armonk, NY, USA), and a two‐sided P value of <0.05 was considered nominally significant. False discovery rate (q value) was calculated using the R‐package to adjust for multiple testing.18 As previously suggested, all SNPs with q < 0.20 were reported to account for multiple testing while balancing the discovery nature of our study.19
3. RESULTS
The clinical characteristics of 390 RCC patients and 390 age‐ and gender‐matched healthy controls are shown in Table 1. Age, gender, body mass index (BMI), and cigarette smoking status were comparable between RCC patients and control subjects. However, significant differences between the cases and controls were noted in case of alcohol consumption and histories of hypertension or diabetes (P < 0.001). Most RCC cases had stage I‐II and grade I‐II of the disease, and the median follow‐up time was 19.6 months.
Table 1.
Clinical characteristics of the study population
| Characteristic | Cases (n = 390) | Controls (n = 390) | P |
|---|---|---|---|
| Age | |||
| Median, years (IQR) | 59 (50‐69) | 59 (50‐69) | 0.893 |
| Gender | |||
| Male | 261 (66.9) | 261 (66.9) | 1.000 |
| Female | 129 (33.1) | 129 (33.1) | |
| BMI | |||
| Median, kg/m2 (IQR) | 24.5 (22.3‐27.7) | 24.5 (22.6‐27.4) | 0.953 |
| Cigarette smoking status, n (%) | |||
| Never | 245 (63.0) | 254 (65.1) | 0.533 |
| Ever | 144 (37.0) | 136 (34.9) | |
| Alcohol consumption, n (%) | |||
| Never | 299 (76.9) | 220 (56.4) | <0.001 |
| Ever | 90 (23.1) | 170 (43.6) | |
| Hypertension, n (%) | |||
| No | 214 (54.9) | 280 (71.8) | <0.001 |
| Yes | 176 (45.1) | 110 (28.2) | |
| Diabetes, n (%) | |||
| No | 314 (80.7) | 352 (90.3) | <0.001 |
| Yes | 75 (19.3) | 38 (9.7) | |
| Stage, n (%) | |||
| I‐II | 300 (80.9) | ||
| III‐IV | 71 (19.1) | ||
| Grade, n (%) | |||
| I‐II | 243 (73.2) | ||
| III‐IV | 89 (26.8) | ||
| Follow‐upa, n (%) | |||
| Recurrence | 20 (7.2) | ||
| Deaths | 9 (3.2) | ||
BMI, body mass index; IQR, interquartile range.
P < 0.05 are in boldface.
With median follow‐up of 19.6 mo.
Of the 25 GRM SNPs evaluated, GRM5 rs7102764 and GRM7 rs756084 were associated with RCC risk (nominal P ≤ 0.049, Table S1). However, only GRM5 rs7102764 attained significance after adjustment for the false discovery rate (q value) at a level of <0.20 (q = 0.140, Table 2). In addition, this association persisted after controlling for age, gender, alcohol consumption, and histories of hypertension and diabetes (P = 0.006).
Table 2.
Association between GRM SNPs and RCC risk
| Gene | SNP | Genotype | Cases, n (%) | Controls, n (%) | OR (95% CI) | P | q | OR (95% CI)a | P a |
|---|---|---|---|---|---|---|---|---|---|
| GRM5 | rs7102764 | AA | 209 (54.3) | 249 (63.8) | |||||
| AT | 151 (39.2) | 125 (32.1) | |||||||
| TT | 25 (6.5) | 16 (4.1) | |||||||
| Trend | 1.41 (1.11‐1.79) | 0.005 | 0.140 | 1.42 (1.11‐1.83) | 0.006 | ||||
| GRM7 | rs756084 | CC | 100 (26.0) | 119 (30.5) | |||||
| CA | 191 (49.6) | 197 (50.5) | |||||||
| AA | 94 (24.4) | 74 (19.0) | |||||||
| Trend | 1.22 (1.00‐1.50) | 0.049 | 0.403 |
95% CI, 95% confidence interval; OR, odds ratio; RCC, renal cell carcinoma; SNP, single‐nucleotide polymorphism.
q < 0.20 are in boldface.
ORs were adjusted for age, gender, alcohol consumption, and histories of hypertension and diabetes.
Two SNPs, GRM3 rs701332 and GRM4 rs2499707, showed a nominal correlation with RFS, but none of them passed the q value threshold (Tables S1 and S2). GRM3 rs701332, GRM4 rs2499707, and GRM4 rs4713742 were associated with OS (nominal P ≤ 0.018, Table S1), and all had a q value of ≤0.133 (Table 3). A strong gene‐dosage effect on OS was observed when these three SNPs were analysed in combination, and the HRs increased as the number of risk alleles increased (P = 0.001, Table 3 and Figure 1).
Table 3.
Association between GRM SNPs and overall survival in patients with RCC
| Gene | SNP | Genotype | n of patients | n of events | 5‐y survival rate (%) | P a | q | HR (95% CI)b | P b |
|---|---|---|---|---|---|---|---|---|---|
| GRM3 | rs701332 | TT | 235 | 5 | 96.6 | ||||
| TC | 42 | 4 | 81.6 | ||||||
| Trend | 0.015 | 0.133 | 4.28 (1.15‐16.0) | 0.030 | |||||
| GRM4 | rs2499707 | CC | 192 | 3 | 96.7 | ||||
| CT | 70 | 4 | 89.9 | ||||||
| TT | 11 | 2 | 70.0 | ||||||
| Trend | 0.001 | 0.028 | 3.55 (1.46‐8.65) | 0.005 | |||||
| GRM4 | rs4713742 | CC | 117 | 2 | 96.1 | ||||
| CT | 117 | 2 | 97.6 | ||||||
| TT | 35 | 4 | 82.1 | ||||||
| Trend | 0.018 | 0.133 | 3.11 (1.17‐8.27) | 0.023 | |||||
| n of risk alleles | |||||||||
| 0 | 69 | 0 | 100.0 | ||||||
| 1‐2 | 149 | 3 | 95.8 | ||||||
| >2 | 28 | 5 | 70.6 | ||||||
| Trend | <0.001 | 10.4 (2.73‐39.8) | 0.001 | ||||||
95% CI, 95% confidence interval; HR, hazard ratio; RCC, renal cell carcinoma; SNP, single‐nucleotide polymorphism.
q < 0.20 are in boldface.
P values were calculated using the log‐rank test.
HRs were adjusted for age and gender.
Figure 1.
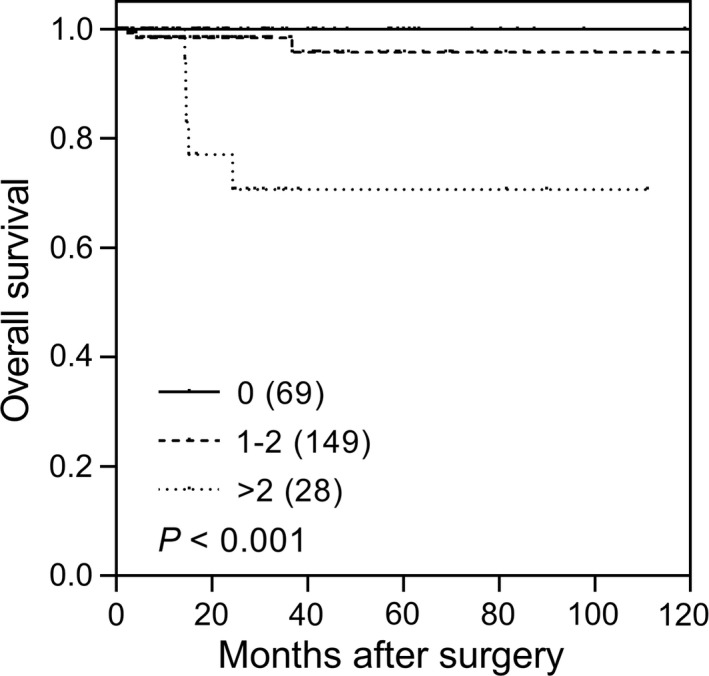
Impact of genetic variants of GRM3 and GRM4 on renal cell carcinoma (RCC) survival. Kaplan‐Meier curves of overall survival for RCC patients with 0, 1‐2, or >2 risk alleles (GRM3 rs701332 C, GRM4 rs2499707 T, and GRM4 rs4713742 T). The numbers in parentheses indicate the number of patients
We investigated the functional significance of all genetic variants in linkage disequilibrium with the prognostic SNPs identified in this study using HaploReg (Tables S3‐S6). GRM5 rs7102764, GRM3 rs701332, GRM4 rs2499707, and GRM4 rs4713742 all coincided with enhancer histone marks in multiple tissues, suggesting that these SNPs might influence the gene expression of GRMs. In the eQTL analysis from the GTEx dataset, the risk allele T of rs7102764 showed an increased GRM5 expression (P = 0.016, Figure 2A), and the risk allele T of rs2499707 showed an increased GRM4 expression (P = 0.001, Figure 2C) in most GRMs abundantly expressed brain cerebellum tissues.
Figure 2.
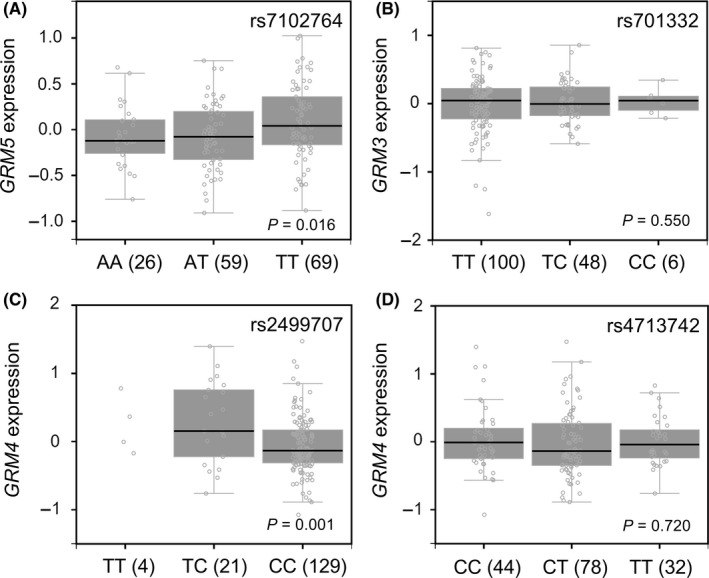
The expression quantitative trait loci analysis of GRM SNPs and gene expression levels. Correlations of (A) rs7102764 genotypes with GRM5 expression, (B) rs701332 genotypes with GRM3 expression, (C) rs2499707 genotypes with GRM4 expression, and (D) rs4713742 genotypes with GRM4 expression. The numbers in parentheses indicate the number of patients
We further determined the clinical relevance of GRM3, GRM4, and GRM5 expression in RCC using the TCGA KIRC dataset. GRM3 gene expression was significantly lower in tumour, late‐stage, and high‐grade tissues (P < 0.001, Figure 3A‐C), and low expression of GRM3 was associated with poor OS in patients with RCC (P = 0.004, Figure 3D). GRM4 was highly expressed in tumour, late‐stage, and high‐grade tissues (P ≤ 0.003), and a high expression of GRM4 was associated with poor OS (P < 0.001). However, there was only a slight trend towards higher GRM5 expression levels in cancer tissues than in adjacent normal tissues (P = 0.079); GRM5 expression was not associated with survival.
Figure 3.
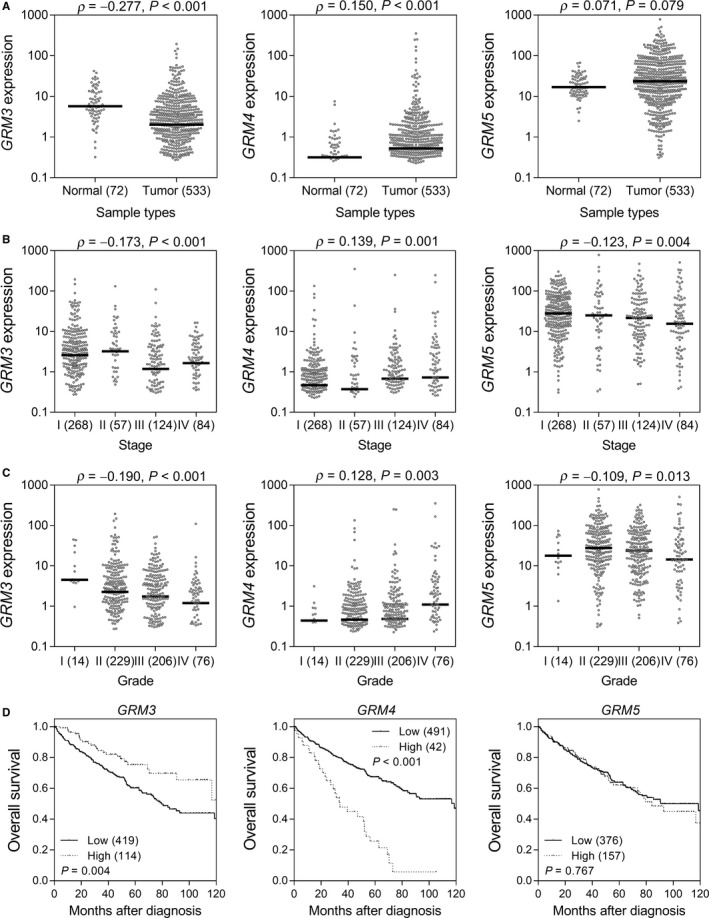
Roles of GRM3, GRM4, and GRM5 expression in renal cell carcinoma (RCC) progression. GRM3, GRM4, and GRM5 expression levels in (A) normal and tumour tissues, (B) different stages, and (C) different grades of RCC. (D) Kaplan‐Meier curves of overall survival according to GRM3, GRM4, and GRM5 expression levels. Patients were dichotomised at the mean gene expression level into the low and high groups. The numbers in parentheses indicate the number of patients
4. DISCUSSION
In the present study, we explored the effects of genetic variants in GRMs on the risk and the prognosis of RCC patients. Several significant associations of GRM3, GRM4, and GRM5 with RCC susceptibility and survival were identified. These findings highlight the importance of GRMs in RCC and might have the potential to guide the selection of patients at a high risk of poor prognosis.
Our results indicated that GRM5 rs7102764, an intronic variant, was associated with RCC risk. Functional prediction implicated this variant as an eQTL regulating the expression of GRM5, potentially through the modulation of the enhancer activities and transcription‐factor binding affinities. A tendency of GRM5 gene upregulation was observed in tumour tissues, suggesting that this gene may play a role in RCC carcinogenesis. Studies have shown that GRM5 is upregulated in lung and glial cancers,20, 21 and inactivation of GRM5 suppresses liver and bone cancer cell proliferation by blocking mitogen‐activated protein kinase pathways.22, 23 The intronic variants rs701332 located in GRM3, and rs2499707 and rs4713742 located in GRM4, were associated with the survival of RCC patients. According to the GTEx dataset, rs2499707 is an eQTL that affects the expression of GRM4. In addition, downregulation of GRM3 was observed in tumour tissues and correlated with a shorter survival of RCC patients, whereas GRM4 overexpression was found in tumours and correlated with poor survival. Consistent with our findings, it has been shown that GRM3 mediates B‐cell‐related tumour cell apoptosis via forkhead box O1, and GRM3 deficiency promotes tumour progression.24 Mounting evidence has also revealed that GRM4 overexpression is correlated with poor prognosis in various cancers such as osteosarcoma,25 colorectal cancer,9 and glioma.26 Further mechanistic studies have demonstrated that transcriptional misregulation in cancers and peroxisome proliferator‐activated receptor signalling pathways might participate in GRM4‐mediated osteosarcoma progression.27 Taken together, the genetic variants identified in this study might affect the gene expression of GRMs, which in turn influence the progression of RCC through modulating tumour cell apoptosis by GRM3, as well as misregulating peroxisome proliferator‐activated receptor signalling pathway by GRM4, and mitogen‐activated protein kinase pathway by GRM5. However, the mechanisms underlying the effects of GRMs in RCC remain undetermined and warrant further investigations.
Several limitations should be noted with regard to interpreting the results of our study. First, the sample size was relatively small, the follow‐up time was limited, and the frequencies of recurrent/death events and some homozygous variants were low in subgroups, which could limit the accuracy and reliability of our results. Second, our findings may not be generalized to other ethnicities as the study population was mainly Taiwanese; however, similar genetic backgrounds can minimise the potential confounding of population heterogeneity. Third, the SNPs genotyped in this study were haplotype‐tagging SNPs to capture most of the genomic diversity; however, the linked causal SNPs and exact molecular mechanisms need to be further identified. Fourth, due to the low number (N = 39) of kidney tissue samples in the GTEx dataset, our eQTL analyses were limited to the brain cerebellum, in which most GRMs are abundantly expressed, but not the target tissues. Finally, although false discovery rates were reported to account for multiple hypothesis testing, we still cannot rule out the possibility of false‐positive findings. However, functional studies support the clinical significance of GRMs in RCC. To our knowledge, this study is the first attempt to discover the effect of GRMs on RCC development and progression. The results are intriguing and worth replicating in further independent studies with larger sample size, as well as performing functional experiments to confirm our findings.
In conclusion, we identified multiple novel associations of genetic variants in GRM3, GRM4, and GRM5 with the risk and the survival of RCC. Furthermore, these relationships were supported by gene expression profiles obtained using bioinformatics analysis. Specifically, the expression of GRM4 and GRM5 showed an increasing trend in RCC tissues, compared to that in normal tissue, whereas GRM3 was downregulated in RCC. Consistently, the increased expression of GRM4 and decreased expression of GRM3 were associated with poorer survival in patients with RCC. Collectively, these results provide evidence in support of the hypothesis that GRMs modulate the development and progression of RCC.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank Chao‐Shih Chen for data analysis, and the National Center for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support. The results published here are based in part on data generated by the HapMap, HaploReg, and TCGA projects.
Huang C‐Y, Hsueh Y‐M, Chen L‐C, et al. Clinical significance of glutamate metabotropic receptors in renal cell carcinoma risk and survival. Cancer Med. 2018;7:6104–6111. 10.1002/cam4.1901
Chao‐Yuan Huang, Yu‐Mei Hsueh, Lih‐Chyang Chen, and Wei‐Chung Cheng contributed equally to this work.
Funding information
This work was supported by the Ministry of Science and Technology of Taiwan (grant nos: 106‐2314‐B‐039‐018, 106‐2221‐E‐039‐011‐MY3, and 107‐2320‐B‐039‐010), the Kaohsiung Medical University Hospital (grant no: KMUH106‐6R57), and the China Medical University (grant no: CMU106‐S‐24, CMU106‐N‐05, CMU107‐S‐24, and CMU107‐S‐42). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Shu‐Pin Huang, Email: shpihu@yahoo.com.tw.
Bo‐Ying Bao, Email: bao@mail.cmu.edu.tw.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Huang CY, Chu JS, Pu YS, et al. Effect of urinary total arsenic level and estimated glomerular filtration rate on the risk of renal cell carcinoma in a low arsenic exposure area. J Urol. 2011;185(6):2040‐2044. [DOI] [PubMed] [Google Scholar]
- 3. Zbar B, Brauch H, Talmadge C, Linehan M. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature. 1987;327(6124):721‐724. [DOI] [PubMed] [Google Scholar]
- 4. Fairman WA, Amara SG. Functional diversity of excitatory amino acid transporters: ion channel and transport modes. Am J Physiol. 1999;277(4 Pt 2):F481‐F486. [DOI] [PubMed] [Google Scholar]
- 5. Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18(16):4240‐4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205‐237. [DOI] [PubMed] [Google Scholar]
- 7. Tanabe Y, Masu M, Ishii T, Shigemoto R, Nakanishi S. A family of metabotropic glutamate receptors. Neuron. 1992;8(1):169‐179. [DOI] [PubMed] [Google Scholar]
- 8. Tong Q, Ouedraogo R, Kirchgessner AL. Localization and function of group III metabotropic glutamate receptors in rat pancreatic islets. Am J Physiol Endocrinol Metab. 2002;282(6):E1324‐E1333. [DOI] [PubMed] [Google Scholar]
- 9. Chang HJ, Yoo BC, Lim SB, Jeong SY, Kim WH, Park JG. Metabotropic glutamate receptor 4 expression in colorectal carcinoma and its prognostic significance. Clin Cancer Res. 2005;11(9):3288‐3295. [DOI] [PubMed] [Google Scholar]
- 10. Huang CY, Su CT, Chu JS, et al. The polymorphisms of P53 codon 72 and MDM2 SNP309 and renal cell carcinoma risk in a low arsenic exposure area. Toxicol Appl Pharmacol. 2011;257(3):349‐355. [DOI] [PubMed] [Google Scholar]
- 11. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37(Web Server issue):W600‐W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang CN, Huang SP, Pao JB, et al. Genetic polymorphisms in oestrogen receptor‐binding sites affect clinical outcomes in patients with prostate cancer receiving androgen‐deprivation therapy. J Intern Med. 2012;271(5):499‐509. [DOI] [PubMed] [Google Scholar]
- 13. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877‐D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Consortium GT . The Genotype‐Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas Research N . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061‐1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng WC, Chung IF, Chen CY, et al. DriverDB: an exome sequencing database for cancer driver gene identification. Nucleic Acids Res. 2014;42(Database issue):D1048‐D1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chung IF, Chen CY, Su SC, et al. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016;44(D1):D975‐D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440‐9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith NL, Hindorff LA, Heckbert SR, et al. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007;297(5):489‐498. [DOI] [PubMed] [Google Scholar]
- 20. Boer K, Troost D, Timmermans W, et al. Cellular localization of metabotropic glutamate receptors in cortical tubers and subependymal giant cell tumors of tuberous sclerosis complex. Neuroscience. 2008;156(1):203‐215. [DOI] [PubMed] [Google Scholar]
- 21. Li S, Huang S, Peng SB. Overexpression of G protein‐coupled receptors in cancer cells: involvement in tumor progression. Int J Oncol. 2005;27(5):1329‐1339. [PubMed] [Google Scholar]
- 22. Liao S, Ruiz Y, Gulzar H, et al. Osteosarcoma cell proliferation and survival requires mGluR5 receptor activity and is blocked by Riluzole. PLoS ONE. 2017;12(2):e0171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu YL, Wang NN, Gu L, Yang HM, Xia N, Zhang H. The suppressive effect of metabotropic glutamate receptor 5 (mGlu5) inhibition on hepatocarcinogenesis. Biochimie. 2012;94(11):2366‐2375. [DOI] [PubMed] [Google Scholar]
- 24. Liu X, Zhang Y, Wang Z, et al. Metabotropic glutamate receptor 3 is involved in B‐cell‐related tumor apoptosis. Int J Oncol. 2016;49(4):1469‐1478. [DOI] [PubMed] [Google Scholar]
- 25. Yang W, Maolin H, Jinmin Z, Zhe W. High expression of metabotropic glutamate receptor 4: correlation with clinicopathologic characteristics and prognosis of osteosarcoma. J Cancer Res Clin Oncol. 2014;140(3):419‐426. [DOI] [PubMed] [Google Scholar]
- 26. Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7(9):1010‐1015. [DOI] [PubMed] [Google Scholar]
- 27. Pang Y, Zhao J, Fowdur M, Liu Y, Wu H, He M. To explore the mechanism of the GRM4 gene in osteosarcoma by RNA sequencing and bioinformatics approach. Med Sci Monit Basic Res. 2018;24:16‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


