Abstract
Introduction
Immunohistochemical staining for C4d in peritubular capillaries has been part of antibody-mediated rejection (AbMR) definition in the Banff Classification for Allograft Pathology since 2003. However, it has limited sensitivity and specificity, therefore the clinical significance of C4d-positive biopsies without evidence of rejection (C4d+ WER) is unknown. We investigated the transcript levels of genes associated with AbMR in C4d+ WER biopsies from both ABO-compatible and incompatible renal transplant patients.
Methods
RNA was extracted from formalin-fixed paraffin-embedded renal transplant biopsies (n = 125) and gene expression analysis of 35 AbMR-associated transcripts carried out using the NanoString nCounter system.
Results
AbMR-associated transcripts were significantly increased in samples with AbMR or suspicious AbMR. A subgroup of 17 of 35 transcripts that best distinguished AbMR from C4d-negative biopsies without evidence of rejection was used to study C4d+ WER samples. There was no differential expression between C4d-negative and C4d+ WER from both ABO-incompatible and -compatible transplants. The geometric mean of 17 differentially expressed genes was used to assign the C4d+ WER biopsies a high- or low-AbMR transcript score. Follow-up biopsies showed AbMR within 1 year of initial biopsy in 5 of 7 high-AbMR transcript patients but only 2 of 46 low-AbMR transcript patients. In multivariate logistic regression analysis, elevated transcript levels in a C4d+ WER biopsy were associated with increased odds for biopsy-proven AbMR on follow-up (P = 0.032, odds ratio 16.318), whereas factors including donor-specific antibody (DSA) status and time since transplantation were not.
Conclusion
Gene expression analysis in C4d+ WER samples has the potential to identify patients at higher risk of developing AbMR.
Keywords: antibody mediated rejection, C4d, kidney, molecular, transplant rejection
Complement split-product C4d was originally proposed as a marker for AbMR in renal transplant biopsies in the 1990s when Feucht et al.1, 2 identified a correlation between C4d deposition in peritubular capillaries and poor clinical outcome. The Banff Classification for Allograft Pathology incorporated immunohistochemistry for C4d in the definition of acute AbMR in 2003,3, 4 and chronic AbMR in 2005.5
In the context of AbMR, C4d positivity is thought to indicate activation of the classical pathway of complement by anti-donor antibody on the endothelial surface,6 leading to C4 enzymatic cleavage, producing C4b that covalently binds to nearby endothelial cell surfaces via a thioester bond. C4b is rapidly cleaved by factor I into iC4b then C4d, which remains covalently bound.7 This covalent bond makes C4d a better target for immunohistochemical detection than Igs or other complement fragments.
A positive immunohistochemical stain for C4d in peritubular capillaries is considered very specific for AbMR, although with limited sensitivity: not all cases of AbMR are C4d positive, with C4d-negative AbMR a well-recognized entity in the Banff classification.8 C4d-negative AbMR was discovered following gene expression analyses of transplant biopsies, which identified a set of endothelial transcripts that were elevated in cases of AbMR, whether they were C4d positive or negative.9 Since then, a wider set of transcripts elevated in AbMR has been described, which also includes natural killer cell transcripts.10, 11
C4d positivity is not entirely specific for AbMR. In biopsies from recipients of an ABO-incompatible (ABOi) graft, C4d-positivity is not associated with graft dysfunction, poor outcome, or ultrastructural evidence of endothelial injury.12, 13, 14, 15 In ABO-compatible (ABOc) grafts, occasional renal transplant biopsies show C4d+ WER (i.e., without inflammation or injury).
The significance of C4d-positivity without evidence of rejection is uncertain. The recent update to the Banff Classification of Allograft Pathology suggests that the presence of AbMR-associated transcripts in such samples can be taken as evidence that these cases represent AbMR, but data are lacking to support this statement, as acknowledged in Table 5 of the 2017 Banff classification.16, 17 We hypothesised that analysis of AbMR-associated transcripts would allow a better understanding of the biological significance of C4d+ WER biopsies. There is increasing evidence that RNA obtained from paraffin blocks can give reproducible results in gene expression analysis18 and the use of a novel technology applicable to paraffin blocks has allowed analysis of archival samples, increasing cases of this relatively uncommon phenotype, strengthening the observed results.
Methods
Sample Collection
Renal transplant tissue was obtained from the Imperial College Healthcare NHS Trust Tissue Bank, which has ethics approval to both collect human tissue and release material to researchers (MREC 07/MRE09/54). This study complies with the Declaration of Helsinki. All biopsies were graded using Banff 2015 criteria16; however, we did not use transcript analysis as a tool to classify our biopsies initially, as it has not yet been validated in our center.
C4d staining was carried out by immunoperoxidase on paraffin sections, using polyclonal rabbit anti-C4d antibody at 1/40 (BI-RC4D; Oxford Biosystems, Milton Park, UK). The slides were subjected to microwave antigen retrieval (in citrate buffer pH 6), then placed on the BioGenex i6000 autostainer (BioGenex, Fremont, CA). The BioGenex Non-Biotin detection kit was used. C4d staining in peritubular capillaries was classified as negative/minimal (C4d0/C4d1 <1% and 1%–10% of peritubular capillaries, respectively), focal (C4d2, 11%–50% of peritubular capillaries), or diffuse (C4d3, >50% of peritubular capillaries). Only cases with C4d2 or C4d3 were considered positive.
DSAs were assessed using LABScreen mixed beads (One Lambda, Inc., Canoga Park, CA) and, if positive, the anti-HLA antibody specificity was identified using LABScreen single antigen beads. Before transplantation, all donor-recipient pairs had a negative T- and B-cell complement-dependent cytotoxicity crossmatch and a negative T-cell flow cytometric crossmatch, defined as mean fluorescence intensity <300 pretransplantation. Posttransplantation, a mean fluorescence intensity >500 was considered DSA positive. Patients were typed for HLA -A, -B, -Cw, -DR, and -DQ antigens.
Formalin-Fixed Paraffin-Embedded RNA Isolation
Consecutive 10-μM sections were cut from each formalin-fixed paraffin-embedded (FFPE) block and between 4 and 6 sections were obtained. Microtome blades were replaced, and equipment was cleaned with RNaseZap (Life Technologies, Carlsbad, CA) between each block. Sections were immediately transferred to RNase-free 1.5-ml microcentrifuge tubes and placed on ice.
RNA was extracted using the RNeasy FFPE kit (Qiagen, Hilden, Germany) and deparaffinization solution (Qiagen) according to the manufacturer’s large-volume protocol and RNA eluted in the minimum volume. RNA concentration and purity were measured with a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and 2100 Bioanalyzer with an RNA 6000 Nano chip (Agilent, Santa Clara, CA).
NanoString Gene Expression Analysis
A literature search was carried out to identify genes associated with AbMR.9, 10, 19, 20 A custom nCounter XT CodeSet (NanoString Technologies, Seattle, WA) was used to analyze gene expression in the FFPE samples. Quality control and normalization of raw gene expression counts were performed with nSolver Analysis Software Version 4.0 (NanoString Technologies). Default parameters for quality control flagging were used for imaging (field of view registration >75%), binding density (0.05–2.25), positive control linearity (R2 value >0.95), and positive control limit of detection (0.5 fM positive control ≥2 SDs above the mean of the negative controls). Background subtraction was performed for each sample by subtracting the mean of the negative controls from all data points. nSolver Analysis Software was also used to generate agglomerative cluster heat map using the Z-Score of each gene and calculating Euclidian distance between samples.
Data Analysis
GraphPad Prism 7.02 (GraphPad, La Jolla, CA) was used to analyze results. Gene expression between 2 groups was compared using a 2-tailed Mann-Whitney test. Gene expression among 3 or more groups was compared using analysis of variance and a Dunn’s test. Correction for multiple testing was done using the Benjamini-Hochberg procedure with a false discovery rate of 0.05.
Receiver operator characteristic (ROC), survival analysis and logistic regression were performed using SPSS (IBM Corp, Chicago, IL).
Results
C4d+ WER
A retrospective analysis of renal transplant biopsies obtained at our institution between January 2008 and December 2016 showed that among 5265 renal transplant biopsies, 108 (2.16%) of 5001 from ABOc transplants and 104 (39.4%) of 264 from ABOi transplants were C4d+ WER. Cases were selected to include only biopsies showing no or mild tubular atrophy/interstitial fibrosis (Banff ct and ci 0 or 1), no tubulointerstitial or vascular inflammation (Banff scores t0, v0, i0 and ti0), and no microcirculation inflammation (Banff scores g0 and ptc0) or injury (Banff score cg0). C4d immunohistochemistry was reviewed and only cases with C4d positivity in >10% of peritubular capillaries were retained (Banff scores C4d 2 or 3).
NanoString Analysis
RNA was extracted from 157 FFPE samples, representing multiple diagnostic categories; C4d+ WER (n = 67, including 17 ABOi), C4d-negative WER (C4d− WER) (n = 36), AbMR or suspicious for AbMR (n = 27, including 3 ABOi), T-cell–mediated rejection (TCMR) (n = 16, including 4 borderline cases), and mixed rejection (n = 11). Diagnostic category was assigned according to the Banff 2015 classification, although not including results of transcript analysis in defining cases of AbMR.16 The AbMR and suspicious for AbMR groups were combined for analysis and included both C4d-positive and C4d-negative cases, as well as active and chronic active cases. In addition, 6 of the 157 samples had RNA extracted from a portion of biopsy stored in RNAlater (C4d− WER [n = 2], mixed rejection [n = 1], TCMR [n = 1], C4d+ WER [n = 1], AbMR [n = 1]).
A custom NanoString nCounter CodeSet (Supplementary Table S1) containing 35 genes associated with AbMR,10, 11, 19, 20 including endothelial-associated transcripts, DSA-selective transcripts, and natural killer cell transcripts, was used to assess gene expression levels from 157 FFPE samples and 6 samples stored in RNAlater.
Fifteen samples failed to pass NanoString quality control (5 C4d− WER, 6 C4d+ WER [3 ABOi], 1 mixed, 2 AbMR and 1 TCMR). A further 13 samples (4 C4d− WER, 6 C4d+ WER [1 ABOi], 1 TCMR, 2 AbMR) were removed after normalization quality control. Four samples were analyzed twice for batch control across code sets, and the second sample results were not included (1 C4d− WER, 1 C4d+ WER ABOc, 1 C4d+ WER ABOi and 1 TCMR).
Normalized counts from the NanoString analysis were compared for 6 samples between the FFPE and RNAlater stored biopsies and there was good correlation between the 2 sample storage methods (Supplementary Figure S1). RNAlater samples were excluded from further analysis. The remaining 125 samples were included in analysis and patient information is given in Table 1.
Table 1.
Patient demographics
| C4d− WER (n = 26) | C4d+ WER (n = 53) | AbMR/suspicious AbMR (n = 23) | TCMR (n = 13) | Mixed (n = 10) | |
|---|---|---|---|---|---|
| Recipient sex: males (%) | 19 (73) | 42 (79.2) | 10 (43.5) | 11 (84.6) | 4 (40) |
| Recipient age at transplant, median (IQR) | 49.1 (40.6–55.0) | 44.7 (38.5–52.6) | 38.6 (26.2–47.5) | 51.0 (38.0–57.4) | 42.2 (37.2–56.3) |
| Recipient ethnicity | |||||
| Afro-Caribbean | 5 | 6 | 4 | 2 | 1 |
| Asian | 9 | 13 | 3 | 2 | 2 |
| White | 10 | 29 | 13 | 8 | 6 |
| Other | 1 | 5 | 1 | 1 | 1 |
| Unknown | 1 | 0 | 1 | 0 | 0 |
| Transplant type | |||||
| Deceased donor | 15 | 22 | 7 | 5 | 5 |
| Live related donor | 4 | 7 | 3 | 4 | 2 |
| Live unrelated donor | 6 | 8 | 2 | 3 | 1 |
| Simultaneous pancreas and kidney | 0 | 3 | 2 | 1 | 1 |
| Pancreas after kidney | 0 | 1 | 4 | 0 | 1 |
| Unknowna | 1 | 0 | 2 | 0 | 0 |
| ABO-incompatible transplants | 0 | 12 | 3 | 0 | 0 |
| Anti-HLA donor-specific antibody (at time of biopsy) | |||||
| Positive | 5 | 18 | 15 | 1 | 8 |
| Negative | 21 | 35 | 8 | 12 | 2 |
| C4d positivity | 0 | 53 | 7 | 4 | 5 |
| Time from transplant to biopsy, d | 941.5 (181.5–1232.5) | 107 (23–660) | 637.5 (213–1823) | 455 (154–816) | 151.5 (82.5–257) |
| Induction regimen | |||||
| Campath | 20 | 49 | 16 | 11 | 9 |
| Anti-IL2 receptor inhibition | 5 | 0 | 31 | 10 | 00 |
| Rituximab + Anti-IL2 receptor | 0 | 4 | 1 | 0 | 0 |
| Campath + Eculizumab | 0 | 0 | 2 | 0 | 0 |
| None | 0 | 0 | 0 | 1 | 0 |
| Unknowna | 1 | 0 | 1 | 0 | 1 |
AbMR, antibody-mediated rejection; C4d− WER, C4d-negative without evidence of rejection; C4d+ WER, C4d-positive without evidence of rejection; IL, interleukin; IQR, interquartile range; TCMR, T-cell–mediated rejection.
Transplantation took place abroad and information is unavailable.
Expression of AbMR-Associated Transcripts
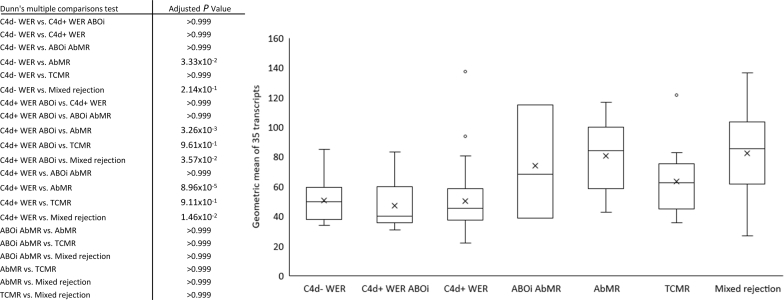
Transcript analysis in AbMR produces varying lists of “top targets” (the most highly expressed in and most strongly associated with AbMR), depending on samples included.21 To assess the 35-gene set as a whole, the geometric mean of the 35 AbMR transcripts was calculated for each sample with the results for each diagnostic category summarized in Figure 1. There were significant differences between the groups (P = 8.05 × 10–6, Kruskal-Wallis). The transcripts were elevated in the AbMR group compared with the C4d− WER (P = 0.033, Dunn’s multiple comparison test) and C4d+ WER groups (P = 8.96 × 10–5). In contrast to previous studies,22 the AbMR-associated genes were not elevated in the TCMR group (vs. C4d− WER, P = 0.999). There was no significant difference between C4d− WER and C4d+ WER groups (P = 0.999).
Figure 1.
Antibody-mediated rejection (AbMR) gene transcript levels in different diagnostic categories. The geometric mean of the 35 AbMR-associated gene transcripts was calculated for each of the 125 samples. Median and first and third quartiles are plotted. Transcript geometric mean is elevated in AbMR samples compared with samples without evidence of rejection. ABOi, ABO-incompatible; C4d+ WER, C4d-positive biopsies without evidence of rejection; TCMR, T-cell–mediated rejection.
For the purpose of this study, we aimed to select top transcripts for distinguishing C4d− WER biopsies from AbMR biopsies. We therefore assessed the 35 AbMR-associated transcripts individually to confirm differential gene expression between AbMR (n = 20) and C4d− WER biopsies (n = 26). ABOi patients (n = 3) were excluded from this analysis. Seventeen transcripts demonstrated increased expression in the AbMR group compared with C4d− WER biopsies (Table 2) and these genes were used in subsequent analyses. These included endothelial11 (n = 5), natural killer–associated10 (n = 6), DSA-selective10 (n = 3), and AbMR19 (n = 3) transcripts.
Table 2.
Comparison of AbMR-associated transcripts in C4d− WER and AbMR biopsies
| C4d− WER vs. AbMR | |
|---|---|
| GNLY | 1.00E-09 |
| FGFBP2 | 8.1E-09 |
| CCL4 | 2.981E-07 |
| SH2D1b | 7.831E-07 |
| CXCL10 | 1.0651E-06 |
| KLRF1 | 4.51E-06 |
| TRD | 4.30622E-05 |
| HLA-DRB3 | 0.000055 |
| DARC | 0.0004 |
| MYBL1 | 0.0005 |
| PLA1A | 0.001 |
| ICAM2 | 0.0014 |
| PECAM1 | 0.0029 |
| KLF4 | 0.005 |
| SELE | 0.0058 |
| CX3CR1 | 0.0083 |
| TM4SF18 | 0.01 |
| COL13A1 | 0.0374a |
| SOST | 0.0442a |
| CDH5 | 0.078 |
| RAMP3 | 0.078 |
| CDH13 | 0.087 |
| GNG11 | 0.1244 |
| VWF | 0.1358 |
| EVA1C | 0.1609 |
| PLAT | 0.2297 |
| CAV1 | 0.3851 |
| CRHBP | 0.4224 |
| PGM5 | 0.4483 |
| MEOX1 | 0.5717 |
| SOX7 | 0.5755 |
| CETP | 0.7009 |
| APOBEC | 0.7949 |
| PALMD | 0.8866 |
| TEK | 0.8866 |
AbMR samples (n = 20) were compared with C4d− WER samples (n = 26) using the Mann-Whitney test and Benjamini-Hochberg correction for multiple testing with a significance value set to 0.05. Significant values are shown in bold.
AbMR, antibody-mediated rejection; C4d− WER, C4d-negative without evidence of rejection.
Not significant after correction for multiple testing.
AbMR-Associated Transcript Expression in C4d+ WER Samples
ABO and DSA Status
Transcript expression in C4d+ WER ABOc samples (n = 41) was compared with transcript expression in C4d+ WER ABOi samples (n = 12) to determine if ABO status affected gene expression in C4d-positive samples. The geometric mean of 17 gene transcripts was compared between the 2 groups and there was no significant difference (Supplementary Figure S2, P = 0.1532).
A comparison was next made between gene expression from DSA-negative (n = 35) and DSA-positive (n = 18) patients. There was no significant difference between the geometric means of the 2 groups (Supplementary Figure S3, P = 0.7307). In subsequent analysis, the C4d+ WER consisted of ABOi and ABOc samples that were either DSA positive or negative (n = 53).
Agglomerative Heat Map With Hierarchical Clustering Analysis
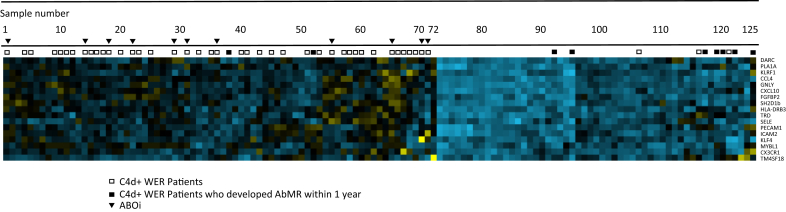
To visualize the 17-gene AbMR-associated expression profile (determined in Table 2) of C4d+ WER samples, an agglomerative heat map with hierarchical clustering was generated with all 125 samples (Figure 2). This clustered 72 samples into a mainly non-AbMR group and 53 into a mainly AbMR group. Most C4d+ WER samples clustered with the non-AbMR group (43/53), whereas 10 of 53 clustered with AbMR. Within 1 year of the initial biopsy, 2 of 43 and 6 of 10 C4d+ WER patients, respectively, went on to develop biopsy-proven AbMR. Further details of these samples are given in the analysis of discrepancies between gene expression and histology section and in Table 4.
Figure 2.
Heat map of 17 significant genes in 125 samples analyzed. Gene expression levels are given by color, with bluer squares indicating higher expression and yellower squares indicating lower expression. Two broad categories were created. The left, containing 72 samples, was mainly non–antibody-mediated rejection (AbMR), and the right, containing 53 samples, was mainly AbMR. The position of each C4d-positive biopsy without evidence of rejection (C4d+ WER) sample is represented by squares above the heat map, with open squares representing samples without AbMR within 1 year of follow-up and filled squares representing samples that developed AbMR within 1 year of follow-up. Individual transcript names are indicated on the right. ABOi, ABO-incompatible.
Table 4.
C4d+ WER sample details
| Patient (based on heat map cluster location, Figure 2) | Biopsy type | ABO status | DSA status (at time of biopsy) | Time from transplant to biopsy, d | Transcript geometric mean | AbMR transcripts | Heat map group | Follow-up biopsy | Follow-up biopsy type | Time to follow-up, d |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P | i | Negative | 97 | 22.81 | Low | No Rejection | No AbMR | FC | 665 |
| 4 | FC | c | Negative | 9 | 23.19 | Low | No Rejection | None | N/A | 0 |
| 5 | FC | c | Negative | 11 | 27.16 | Low | No Rejection | No AbMR | FC | 112 |
| 9 | FC | c | Negative | 40 | 28.67 | Low | No Rejection | AbMR | FC | 1792 |
| 10 | FC | c | Negative | 115 | 21.38 | Low | No Rejection | None | N/A | 0 |
| 11 | FC | c | Positive | 12 | 22.97 | Low | No Rejection | No AbMR | FC | 44 |
| 12 | FC | c | Negative | 362 | 28.64 | Low | No Rejection | No AbMR | FC | 1386 |
| 14 | FC | i | Negative | 1169 | 34.67 | Low | No Rejection | No AbMR | FC | 755 |
| 15 | FC | c | Negative | 1530 | 27.19 | Low | No Rejection | None | N/A | 0 |
| 16 | FC | c | Negative | 59 | 31.63 | Low | No Rejection | No AbMR | P | 1075 |
| 17 | FC | c | Negative | 179 | 33.43 | Low | No Rejection | No AbMR | FC | 1687 |
| 18 | P | i | Negative | 1914 | 31.85 | Low | No Rejection | None | N/A | 0 |
| 20 | FC | c | Negative | 2853 | 32.22 | Low | No Rejection | None | N/A | 0 |
| 22 | P | i | Negative | 1827 | 28.30 | Low | No Rejection | None | N/A | 0 |
| 23 | FC | c | Positive | 62 | 31.13 | Low | No Rejection | No AbMR | P | 252 |
| 25 | P | c | Positive | 495 | 24.78 | Low | No Rejection | None | N/A | 0 |
| 29 | P | i | Negative | 1201 | 25.13 | Low | No Rejection | None | N/A | 0 |
| 31 | P | i | Negative | 260 | 23.82 | Low | No Rejection | None | N/A | 0 |
| 33 | FC | c | Negative | 132 | 44.26 | Low | No Rejection | AbMR | FC | 2164 |
| 35 | FC | c | Negative | 1282 | 32.55 | Low | No Rejection | No AbMR | FC | 1083 |
| 36 | P | i | Negative | 1886 | 31.87 | Low | No Rejection | None | N/A | 0 |
| 38 | FC | c | Positive | 41 | 37.88 | Low | No Rejection | AbMR | FC | 170 |
| 40 | FC | c | Negative | 44 | 44.52 | Low | No Rejection | No AbMR | FC | 1133 |
| 41 | FC | c | Negative | 48 | 33.80 | Low | No Rejection | None | N/A | 0 |
| 43 | FC | c | Positive | 23 | 33.62 | Low | No Rejection | No AbMR | FC | 2483 |
| 45 | FC | c | Positive | 17 | 29.25 | Low | No Rejection | AbMR | FC | 2370 |
| 47 | FC | c | Positive | 123 | 31.98 | Low | No Rejection | AbMR | FC | 1262 |
| 51 | FC | c | Positive | 1822 | 44.81 | Low | No Rejection | None | N/A | 0 |
| 52 | FC | c | Positive | 16 | 42.89 | Low | No Rejection | AbMR | FC | 28 |
| 53 | FC | c | Positive | 17 | 26.55 | Low | No Rejection | None | N/A | 0 |
| 55 | P | i | Negative | 1897 | 19.84 | Low | No Rejection | None | N/A | 0 |
| 57 | FC | c | Positive | 13 | 22.75 | Low | No Rejection | No AbMR | FC | 694 |
| 58 | P | c | Negative | 1918 | 28.40 | Low | No Rejection | None | N/A | 0 |
| 59 | FC | i | Negative | 107 | 26.78 | Low | No Rejection | None | N/A | 0 |
| 60 | FC | c | Positive | 18 | 22.42 | Low | No Rejection | No AbMR | FC | 1250 |
| 62 | FC | c | Negative | 7 | 20.09 | Low | No Rejection | AbMR | FC | 1984 |
| 65 | P | i | Negative | 238 | 13.59 | Low | No Rejection | AbMR | P | 2932 |
| 66 | FC | c | Positive | 2422 | 13.09 | Low | No Rejection | None | N/A | 0 |
| 67 | FC | c | Positive | 14 | 21.01 | Low | No Rejection | None | P | 0 |
| 68 | FC | c | Negative | 565 | 24.38 | Low | No Rejection | No AbMR | FC | 1183 |
| 69 | FC | c | Negative | 660 | 34.42 | Low | No Rejection | No AbMR | FC | 1289 |
| 70 | P | i | Negative | 92 | 36.03 | Low | No Rejection | No AbMR | P | 301 |
| 71 | P | i | Negative | 262 | 47.18 | Low | No Rejection | None | N/A | 0 |
| 92 | FC | c | Positive | 37 | 84.80 | High | Rejection | AbMR | FC | 120 |
| 95 | FC | c | Positive | 348 | 148.11 | High | Rejection | AbMR | FC | 198 |
| 106 | P | c | Negative | 1111 | 66.44 | High | Rejection | AbMR | FC | 2234 |
| 116 | FC | c | Negative | 23 | 61.94 | High | Rejection | No AbMR | C | 16 |
| 117 | FC | c | Negative | 40 | 72.78 | High | Rejection | AbMR | P | 160 |
| 119 | FC | c | Negative | 37 | 58.63 | Low | Rejection | AbMR | P | 319 |
| 120 | FC | c | Positive | 20 | 59.51 | High | Rejection | AbMR | FC | 9 |
| 121 | FC | c | Negative | 434 | 56.26 | Low | Rejection | No AbMR | FC | 3108 |
| 122 | FC | c | Negative | 16 | 74.30 | High | Rejection | AbMR | P | 340 |
| 125 | FC | c | Positive | 0 | 25.20 | Low | Rejection | AbMR | P | 206 |
AbMR, antibody-mediated rejection; c, compatible; C4d+ WER, C4d-positive without evidence of rejection; DSA, donor-specific antibody; FC, for cause biopsy; I, incompatible; N/A, not applicable; P, protocol biopsy.
ROC and Survival Analysis
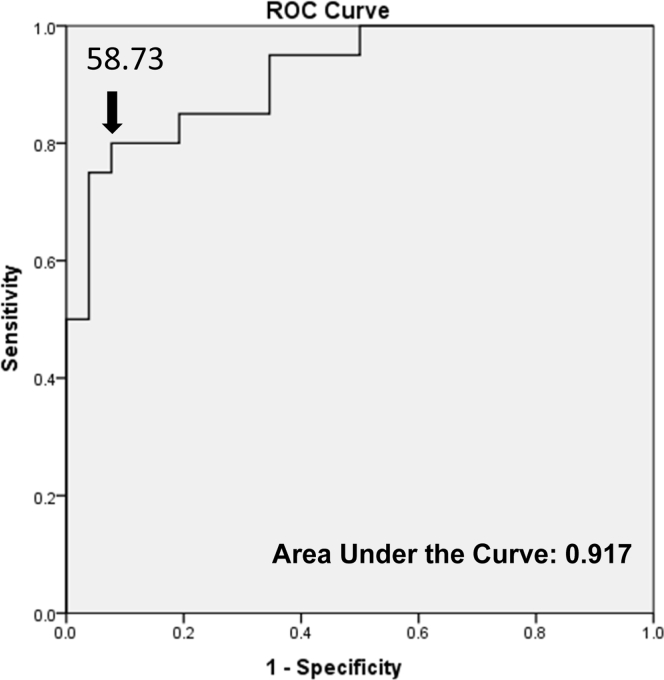
The geometric mean of the 17 AbMR-associated transcripts was calculated for the C4d− WER and AbMR samples and used in the generation of an ROC curve. A cutoff was selected where samples with a geometric mean greater than or equal to 58.7309 were considered high-risk for AbMR in the biopsy, maximizing sensitivity (0.80) and specificity (0.923) (Figure 3).
Figure 3.
Receiver operator characteristic (ROC) curve analysis. The geometric mean of the 17 transcripts significantly associated with antibody-mediated rejection (AbMR) in our analysis was calculated for AbMR and C4d-positive biopsy without evidence of rejection (C4d+ WER) samples. A cutoff (58.7309) was selected for AbMR risk, maximizing sensitivity (0.80) and specificity (0.923), indicated by the arrow. The transcripts were a good predictor of AbMR with an area under the curve of 0.917.
The 17-gene geometric mean of each of the 53 C4d+ WER samples was used to assign each case to a low-AbMR transcripts or high-AbMR transcripts group based on the 58.7309 threshold. Forty-six C4d+ WER samples were classified as low-AbMR transcripts (including all 12 ABOi patients) and 7 as high-AbMR transcripts. Further details of these samples are given the “Analysis of Discrepancies Between Gene Expression and Histology” section and in Table 4.
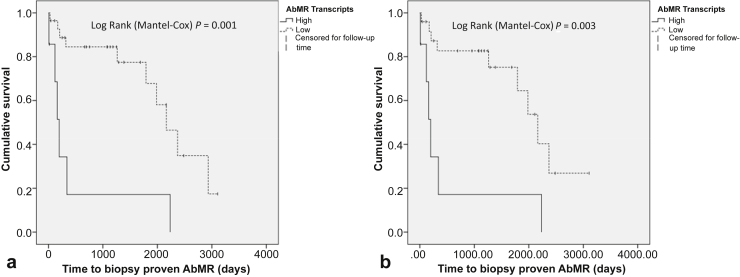
Data from follow-up biopsies were available for 34 of 53 C4d+ WER patients. These were used as the basis for Kaplan-Meier survival plots with biopsy-proven AbMR used as the outcome measure (Figure 4). AbMR-free survival was significantly worse in the high-AbMR transcripts group (P = 0.001). This remained the case when ABOi samples were excluded from the analysis (P = 0.003).
Figure 4.
Survival analysis. Kaplan-Meier survival analysis was performed on the C4d-positive biopsy without evidence of rejection (C4d+WER) as a whole (a) and additionally only on the ABO-compatible (ABOc) C4d+ WER samples (b). The outcome measure was biopsy-proven antibody-mediated rejection (AbMR). The high-AbMR transcripts group had a significantly worse outcome than the low-AbMR transcripts group.
Follow-up data were available for 15 of 26 C4d− WER biopsies. None had developed biopsy-proven AbMR.
These results support the hypothesis that a high-risk score is an indication of potential development of AbMR in C4d+ WER samples. Although some patients with low-AbMR transcripts developed AbMR, this was mostly several years after the initial biopsy (see the “Comparison of ROC and Heat Map Analyses” section) and it is likely the C4d positivity in the initial biopsy did not indicate current antibody-mediated injury.
Comparison of ROC and Heat Map Analyses
Seven patients had high-AbMR transcripts by ROC analysis and were in the AbMR portion of the heat map analysis (Table 4: patients 92, 95, 106, 116, 117, 120, and 122). All patients had a subsequent follow-up biopsy, with 5 of 7 developing AbMR within 1 year. The C4d+ WER biopsies were all taken within 1 year of transplantation in these patients. Patient 106 from this group developed AbMR 5 years after biopsy (8 years posttransplantation). Patient 116 had only 1 follow-up biopsy available 16 days after the initial biopsy (23 days posttransplantation), but has now had 3 years of follow-up with no further biopsy and no graft loss.
Three C4d+ WER samples were assigned a low-AbMR transcripts score (ROC analysis) but were assigned to the AbMR group in the heat map (Table 4 patients 119, 121, and 125). Patient 119 developed AbMR within 1 year and had a transcript geometric mean of 58.63, very close to the high-AbMR transcripts cutoff of 58.7309. Patient 125 also developed AbMR within 1 year, although the transcript geometric mean was only 25.2. Patient 121 had not developed AbMR over 8 years after the initial C4d+ WER biopsy.
Forty-three patients were assigned to both the non-AbMR (heat map analysis) and low-AbMR transcripts (ROC analysis) groups. Follow-up biopsies were available for 24 of these patients and 8 had subsequently developed AbMR, although in 6 of 8 patients the biopsy-proven AbMR episode was at least 3 years after the initial biopsy. It is likely the absence of an AbMR transcript signature in the initial biopsy indicates true absence of AbMR at that time.
Patients 38 and 52 developed AbMR less than 6 months after the initial biopsy. Both patients were DSA positive at the time of biopsy.
Sixteen of 24 were rejection-free in follow-up biopsies, with 12 having a biopsy 1 year or beyond after the initial biopsy. The 19 patients with no follow-up biopsy may not have had a clinical indication for biopsy, which suggests that they have not progressed to graft rejection; however, subclinical AbMR cannot be ruled out.
Of the 34 patients with follow-up biopsies, 10 had a change in treatment between C4d+ WER biopsy and follow-up biopsy. In 4 of the 10, this was an increase in baseline immunosuppression, from tacrolimus only to tacrolimus, mycophenolate mofetil, and prednisolone (patients 1, 12, 47, and 95 in Table 4). Six received anti-AbMR treatment: plasma exchange/i.v. Ig in 5 (patients 9, 11, 23, 45, 92) and rituximab in 1 (patient 60).
Analysis of Discrepancies Between Gene Expression and Histology
AbMR Samples
Among samples classified by histology as AbMR or suspicious for AbMR, 3 of 23 were assigned to the non-AbMR (heat map analysis) and low-AbMR transcript (ROC analysis) groups. In 2 of these 3 samples, patients had received plasma exchange and i.v. Ig for presumed AbMR before the biopsy, raising the possibility that the low-level transcript expression represented a therapeutic response. In the first case, a subsequent biopsy also showed no rejection. The second patient has not had a further biopsy, but has not lost the graft 5 years on. In the final case of the 3, there was mild peritubular capillary inflammation only in the biopsy (peritubular capillaritis score 2, C4d negative). The patient was sensitized (with anti-class II antibodies with a mean fluorescence intensity of approximately 4500), but there were no data on the donor type. She has not lost her graft yet, 6 years after the biopsy showing histological features of chronic active AbMR using conventional criteria.
There were 2 more patients who were classified as AbMR by histology and heat map rejection cluster but low-AbMR transcripts by ROC analysis.
C4d− WER Samples
Two of the 26 C4d− WER samples were assigned to the AbMR (heat map analysis) and high-AbMR transcripts (ROC analysis) groups. There is no clear explanation; both patients have only ever had surveillance biopsies with minor nonrejection changes and are 9 and 10 years posttransplantation with a functioning graft.
A further 4 C4d− WER samples were assigned to the non-AbMR (heat map analysis) group but high-risk (ROC analysis) group. All 4 had a transcript score close to the cutoff (50.88–55.72).
Follow-up biopsies were available for 15 of 26 C4d− WER patients and all have been AbMR-free for between 7 months and 6.5 years.
The 17-gene set was not specifically chosen to distinguish among AbMR, TCMR, and mixed rejection, so these samples are not further discussed.
Logistic Regression Analysis
Transcripts demonstrated a predictive value for progression to AbMR in the C4d+ WER patient group. To test for the effect of other risk factors, ABO status, C4d score (2 or 3), DSA status at biopsy, time from transplantation to biopsy, and the 17-gene geometric mean were included in multivariate logistic regression analysis, with biopsy-proven AbMR as the dependent variable. High-AbMR transcripts were the only factor with a significant odds ratio (16.3; confidence interval 1.265–210.6) for progression to AbMR (P = 0.032, Table 3).
Table 3.
Multivariate logistic regression analysis
| Variable | Odds ratio (confidence interval) | Significance |
|---|---|---|
| ABO status | ||
| Compatible | (1) | |
| Incompatible | 0.532 (0.023–12.585) | 0.696 |
| C4d score | ||
| 2 | (1) | |
| 3 | 0.645 (0.117–3.551) | 0.614 |
| DSA | ||
| Negative | (1) | |
| Positive | 1.578 (0.268–9.306) | 0.614 |
| Time from transplant to biopsy, mo | ||
| <3 | 0.287 (0.011–7.831) | 0.459 |
| 3–6 | 0.492 (0.018–13.697) | 0.676 |
| 6–12 | (1) | |
| >12 | 0.063 (0.001–3.664) | 0.182 |
| AbMR transcript group | ||
| Low | (1) | |
| High | 16.318 (1.265–210.556) | 0.032 |
Significant values are shown in bold.
AbMR, antibody-mediated rejection; DSA, donor-specific antibody.
Discussion
Molecular analysis of transplant biopsies is yielding novel biomarkers with the potential to improve diagnosis, predict graft outcome, or predict response to therapy. They may also add significantly to existing knowledge, in cases in which traditional histopathological assessment provides ambiguous results. One such area is the significance of C4d-positive biopsies that show no other histological features of rejection. We sought to determine whether transcripts indicative of antibody-mediated injury were elevated in these cases. The geometric mean of AbMR-associated gene expression levels split 53 C4d+ WER samples into 46 cases with low-AbMR transcripts, similar to C4d− WER and including 12 C4d+ WER in ABOi transplants, and 7 cases with high-AbMR transcripts, similar to that seen in AbMR. A heat map and hierarchical clustering assigned 10 of C4d+ WER cases to a rejection group. Six of 7 of the high-AbMR transcript samples or 8 of 10 of those in the AbMR cluster went on to develop AbMR, mostly within 1 year of the initial biopsy. Patients with low-AbMR transcripts who developed AbMR tended to do so at a later time point.
This evidence suggests that most cases of C4d+ WER in ABOc transplant biopsies do not represent AbMR, akin to ABOi C4d+ WER samples, but that in occasional cases, it may represent the earliest sign of AbMR, warranting treatment or close monitoring. Conventional histology is not able to distinguish these 2 groups from each other, resulting in the suggestion that all C4d+ WER should be treated as AbMR.13 This study gives evidence that individualized gene expression analysis could be a useful tool to guide therapeutic options in this situation.
We also confirmed that NanoString nCounter analysis could detect the presence of an AbMR gene set signature in samples with AbMR, as has been previously described using microarrays23 or reverse transcriptase–polymerase chain reaction,24 in paraffin-embedded material up to 10 years old.
Few previous studies have addressed the significance of C4d+ WER biopsies in ABO-compatible transplantation, partly because it is a rare lesion. We found that such biopsies represented approximately 2% of transplant biopsies in our center, comparable with figures found by Kikic et al.25 (approximately 2%, when considering only C4d2 and C4d3 Banff scores). In sensitized patients, and in particular in post-reperfusion or early biopsies, C4d+ WER is associated with subsequent AbMR.26, 27, 28 In later biopsies, with or without allograft dysfunction, the significance of C4d+ WER yielded conflicting results, with variable responses to antirejection therapy and variable outcomes in terms of progressive functional deterioration and graft failure.29, 30, 31, 32, 33
Nickeleit et al.29 proposed 2 groups of C4d+ biopsies: a large group associated with allograft dysfunction or histologic signs of rejection, and a smaller group with only mild allograft dysfunction and no histological evidence of rejection. It was only patients from the first, larger group, who showed benefit from antirejection therapy. However, a follow-up report from the same group30 found improved graft function in C4d+ WER patients after antirejection therapy, and concluded that this lesion was highly suspicious for smoldering rejection.26, 30 Of note, many of these studies precede the most recent updates to the Banff classification, including the formal recommended grading for peritubular capillaritis,5 and might include cases with low levels of microcirculation inflammation. A more recent study by Djamali et al.27 looked at post-reperfusion biopsies in moderately sensitized patients; those with C4d positivity most often did not have microcirculation inflammation (23/28 patients) and yet C4d was found to a risk factor for rejection. Kikic et al.25 found C4d to be associated with graft loss independently of the presence of histological features of AbMR; 42% of patients in the C4d+ group as a whole were reported as presensitized and the mean time to biopsy in the group with C4d+ WER was 0.75 months. In our analysis, the 9 C4d+ WER patients who subsequently developed AbMR within 1 year of the initial biopsy were also within 1 year posttransplantation at the time of the initial biopsy. Overall, the literature is in keeping with our finding that C4d+ WER biopsies are a heterogeneous group, and that in early biopsies, particularly in presensitized patients, this finding may indicate AbMR.
Why might C4d immunohistochemistry represent a heterogeneous biological footprint? C4d is typically used to establish the pathogenetic link between a circulating DSA and injury noted in the graft, on the assumption that injury is triggered by local complement activation by antibody fixed to the endothelial cell surface, via the classical pathway. Although not conclusively proven, C4d deposition could also occur via local activation of the lectin pathway,34 which could explain C4d positivity in DSA-negative patients. Because Igs, C1q, or mannose-binding lectin are not consistently detected in the C4d+ biopsies, it remains unclear to what extent the classical and/or lectin pathways of complement activation contribute to C4d deposition.34 In addition, C4d positivity indicates activation of complement only as far as the C3 convertase, and C4d itself is thought to be biologically inactive with no identified receptor.35 It is possible that tight regulation of complement activation means that only partial complement cascade activation occurs in C4d+ WER, with generation of C4d but severe limitation of formation of C5 convertase and membrane attack complex.15 One study found negative immunohistochemical staining for membrane attack complex36 in C4d-positive renal biopsies with rejection. Such a dissociation might provide support for a local mechanism of accommodation in a subset of patients. In a recent meta-analysis, C4d was found to have only a modest agreement with histological features of AbMR and presence of DSA.37 We also know that in ABOi transplants, C4d positivity is not associated with graft dysfunction, poor outcome, or ultrastructural evidence of endothelial injury.13, 38
There are competing biomarkers to establish the pathogenetic link between antibody and graft injury, such as the molecular analysis of AbMR-associated transcripts used here, and ultrastructural examination for features of endothelial activation.39 C4d will need to be reassessed in light of these developments for its effectiveness as a biomarker.
This study has limitations. It is a retrospective, single-center study using small numbers of samples, with no validation set. Optimal gene sets for AbMR diagnosis are still evolving and will need to be confirmed in a large multicenter study. Additionally, we cannot formally exclude the possibility that subclinical AbMR may be occurring at the follow-up stage of C4d+ WER patients with low-AbMR gene signature, as we do not have a consistent protocol biopsy program for these patients. In some cases, AbMR did occur in patients that were C4d+ WER with low-AbMR gene signature, but much later after the C4d+ WER biopsy.
Despite these limitations, we present novel evidence that many biopsies showing C4d+ WER do not show upregulation of AbMR-related transcripts. Occasional cases that did show a gene expression profile similar to AbMR often developed AbMR within a few months. This study provides the basis for a targeted use of gene expression analysis on routine histological samples, as a complement to standard histological biopsy assessment, in cases in which other routinely used biomarkers, in this case C4d immunohistochemistry, have reached their limitations.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We thank the UCL NanoString Facility for providing the nCounter system and related services. Tissue samples were provided by the Imperial College Healthcare NHS Trust Tissue Bank.
The research is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Figure S1. Formalin-fixed paraffin-embedded (FFPE) and RNAlater comparison. A comparison of NanoString counts obtained from samples from the same patient and stored by different methods.
Figure S2. Comparison of geometric mean from ABO-incompatible (ABOi) and ABO-compatible (ABOc) C4d-positive biopsies without evidence of rejection (C4d+ WER) biopsies. There was no difference in geometric mean transcript expression between ABOi and ABOc C4d+ WER biopsies (Mann-Whitney test, P = 0.1532).
Figure S3. Comparison of geometric mean from donor-specific antibody (DSA)-positive and DSA-negative C4d-positive biopsies without evidence of rejection (C4d+ WER). There was no difference in geometric mean transcript expression between DSA-positive and DSA-negative C4d+ WER biopsies (Mann-Whitney test, P = 0.7307).
Supplementary material is linked to the online version of the paper at http://www.kireports.org/.
Supplementary Material
Formalin-fixed paraffin-embedded (FFPE) and RNAlater comparison. A comparison of NanoString counts obtained from samples from the same patient and stored by different methods.
Comparison of geometric mean from ABO-incompatible (ABOi) and ABO-compatible (ABOc) C4d-positive biopsies without evidence of rejection (C4d+ WER) biopsies. There was no difference in geometric mean transcript expression between ABOi and ABOc C4d+ WER biopsies (Mann-Whitney test, P = 0.1532).
Comparison of geometric mean from donor-specific antibody (DSA)-positive and DSA-negative C4d-positive biopsies without evidence of rejection (C4d+ WER). There was no difference in geometric mean transcript expression between DSA-positive and DSA-negative C4d+ WER biopsies (Mann-Whitney test, P = 0.7307).
References
- 1.Feucht H.E., Felber E., Gokel M.J. Vascular deposition of complement-split products in kidney allografts with cell-mediated rejection. Clin Exp Immunol. 1991;86:464–470. doi: 10.1111/j.1365-2249.1991.tb02954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feucht H.E., Schneeberger H., Hillebrand G. Capillary deposition of C4d complement fragment and early renal graft loss. Kidney Int. 1993;43:1333–1338. doi: 10.1038/ki.1993.187. [DOI] [PubMed] [Google Scholar]
- 3.Racusen L.C., Halloran P.F., Solez K. Banff 2003 meeting report: new diagnostic insights and standards. Am J Transplant. 2004;4:1562–1566. doi: 10.1111/j.1600-6143.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- 4.Racusen L.C., Colvin R.B., Solez K. Antibody-mediated rejection criteria—an addition to the Banff 97 classification of renal allograft rejection. Am J Transplant. 2003;3:708–714. doi: 10.1034/j.1600-6143.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 5.Solez K., Colvin R.B., Racusen L.C. Banff '05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy ('CAN') Am J Transplant. 2007;7:518–526. doi: 10.1111/j.1600-6143.2006.01688.x. [DOI] [PubMed] [Google Scholar]
- 6.Feucht H.E., Opelz G. The humoral immune response towards HLA class II determinants in renal transplantation. Kidney Int. 1996;50:1464–1475. doi: 10.1038/ki.1996.460. [DOI] [PubMed] [Google Scholar]
- 7.Murata K., Baldwin W.M. Mechanisms of complement activation, C4d deposition, and their contribution to the pathogenesis of antibody-mediated rejection. Transplant Rev (Orlando) 2009;23:139–150. doi: 10.1016/j.trre.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas M., Sis B., Racusen L.C. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant. 2014;14:272–283. doi: 10.1111/ajt.12590. [DOI] [PubMed] [Google Scholar]
- 9.Sis B., Halloran P.F. Endothelial transcripts uncover a previously unknown phenotype: C4d-negative antibody-mediated rejection. Curr Opin Organ Transplant. 2010;15:42–48. doi: 10.1097/MOT.0b013e3283352a50. [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo L.G., Sis B., Sellares J. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant. 2010;10:1812–1822. doi: 10.1111/j.1600-6143.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 11.Sis B., Jhangri G.S., Bunnag S. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9:2312–2323. doi: 10.1111/j.1600-6143.2009.02761.x. [DOI] [PubMed] [Google Scholar]
- 12.Park W.D., Grande J.P., Ninova D. Accommodation in ABO-incompatible kidney allografts, a novel mechanism of self-protection against antibody-mediated injury. Am J Transplant. 2003;3:952–960. doi: 10.1034/j.1600-6143.2003.00179.x. [DOI] [PubMed] [Google Scholar]
- 13.Haas M. The significance of C4d staining with minimal histologic abnormalities. Curr Opin Organ Transplant. 2010;15:21–27. doi: 10.1097/MOT.0b013e3283342ebd. [DOI] [PubMed] [Google Scholar]
- 14.Haas M., Rahman M.H., Racusen L.C. C4d and C3d staining in biopsies of ABO- and HLA-incompatible renal allografts: correlation with histologic findings. Am J Transplant. 2006;6:1829–1840. doi: 10.1111/j.1600-6143.2006.01356.x. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin W.M., Ota H., Rodriguez E.R. Complement in transplant rejection: diagnostic and mechanistic considerations. Springer Semin Immunopathol. 2003;25:181–197. doi: 10.1007/s00281-003-0133-3. [DOI] [PubMed] [Google Scholar]
- 16.Loupy A., Haas M., Solez K. The Banff 2015 Kidney Meeting Report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17:28–41. doi: 10.1111/ajt.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas M., Loupy A., Lefaucheur C. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307. doi: 10.1111/ajt.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigdel T.K., Nguyen M., Dobi D. Targeted transcriptional profiling of kidney transplant biopsies. Kidney Int Rep. 2018;3:722–731. doi: 10.1016/j.ekir.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellares J., Reeve J., Loupy A. Molecular diagnosis of antibody-mediated rejection in human kidney transplants. Am J Transplant. 2013;13:971–983. doi: 10.1111/ajt.12150. [DOI] [PubMed] [Google Scholar]
- 20.Venner J.M., Hidalgo L.G., Famulski K.S. The molecular landscape of antibody-mediated kidney transplant rejection: evidence for NK involvement through CD16a Fc receptors. Am J Transplant. 2015;15:1336–1348. doi: 10.1111/ajt.13115. [DOI] [PubMed] [Google Scholar]
- 21.Halloran P.F., Famulski K.S., Reeve J. Molecular assessment of disease states in kidney transplant biopsy samples. Nat Rev Nephrol. 2016;12:534–548. doi: 10.1038/nrneph.2016.85. [DOI] [PubMed] [Google Scholar]
- 22.Halloran P.F., de Freitas D.G., Einecke G. The molecular phenotype of kidney transplants. Am J Transplant. 2010;10:2215–2222. doi: 10.1111/j.1600-6143.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- 23.Halloran P.F., Pereira A.B., Chang J. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM) Am J Transplant. 2013;13:2865–2874. doi: 10.1111/ajt.12465. [DOI] [PubMed] [Google Scholar]
- 24.Dominy K.M., Roufosse C., de Kort H. Use of quantitative real time polymerase chain reaction to assess gene transcripts associated with antibody-mediated rejection of kidney transplants. Transplantation. 2015;99:1981–1988. doi: 10.1097/TP.0000000000000621. [DOI] [PubMed] [Google Scholar]
- 25.Kikic Z., Kainz A., Kozakowski N. Capillary C4d and kidney allograft outcome in relation to morphologic lesions suggestive of antibody-mediated rejection. Clin J Am Soc Nephrol. 2015;10:1435–1443. doi: 10.2215/CJN.09901014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauiyyedi S., Crespo M., Collins A.B. Acute humoral rejection in kidney transplantation: II. Morphology, immunopathology, and pathologic classification. J Am Soc Nephrol. 2002;13:779–787. doi: 10.1681/ASN.V133779. [DOI] [PubMed] [Google Scholar]
- 27.Djamali A., Muth B.L., Ellis T.M. Increased C4d in post-reperfusion biopsies and increased donor specific antibodies at one-week post transplant are risk factors for acute rejection in mild to moderately sensitized kidney transplant recipients. Kidney Int. 2013;83:1185–1192. doi: 10.1038/ki.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas M., Ratner L.E., Montgomery R.A. C4d staining of perioperative renal transplant biopsies. Transplantation. 2002;74:711–717. doi: 10.1097/00007890-200209150-00021. [DOI] [PubMed] [Google Scholar]
- 29.Nickeleit V., Zeiler M., Gudat F. Detection of the complement degradation product C4d in renal allografts: diagnostic and therapeutic implications. J Am Soc Nephrol. 2002;13:242–251. doi: 10.1681/ASN.V131242. [DOI] [PubMed] [Google Scholar]
- 30.Dickenmann M., Steiger J., Descoeudres B. The fate of C4d positive kidney allografts lacking histological signs of acute rejection. Clin Nephrol. 2006;65:173–179. doi: 10.5414/cnp65173. [DOI] [PubMed] [Google Scholar]
- 31.Regele H., Böhmig G.A., Habicht A. Capillary deposition of complement split product C4d in renal allografts is associated with basement membrane injury in peritubular and glomerular capillaries: a contribution of humoral immunity to chronic allograft rejection. J Am Soc Nephrol. 2002;13:2371–2380. doi: 10.1097/01.asn.0000025780.03790.0f. [DOI] [PubMed] [Google Scholar]
- 32.Mengel M., Bogers J., Bosmans J.L. Incidence of C4d stain in protocol biopsies from renal allografts: results from a multicenter trial. Am J Transplant. 2005;5:1050–1056. doi: 10.1111/j.1600-6143.2005.00788.x. [DOI] [PubMed] [Google Scholar]
- 33.Koo D.D., Roberts I.S., Quiroga I. C4d deposition in early renal allograft protocol biopsies. Transplantation. 2004;78:398–403. doi: 10.1097/01.tp.0000128328.68106.54. [DOI] [PubMed] [Google Scholar]
- 34.Wasowska B.A. Mechanisms involved in antibody- and complement-mediated allograft rejection. Immunol Res. 2010;47:25–44. doi: 10.1007/s12026-009-8136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ollert M.W., Kadlec J.V., David K. Antibody-mediated complement activation on nucleated cells. A quantitative analysis of the individual reaction steps. J Immunol. 1994;153:2213–2221. [PubMed] [Google Scholar]
- 36.Nishi S., Imai N., Ito Y. Pathological study on the relationship between C4d, CD59 and C5b-9 in acute renal allograft rejection. Clin Transplant. 2004;18(Suppl 11):18–23. doi: 10.1111/j.1399-0012.2004.00242. [DOI] [PubMed] [Google Scholar]
- 37.Sapir-Pichhadze R., Curran S.P., John R. A systematic review of the role of C4d in the diagnosis of acute antibody-mediated rejection. Kidney Int. 2015;87:182–194. doi: 10.1038/ki.2014.166. [DOI] [PubMed] [Google Scholar]
- 38.Brocker V., Pfaffenbach A., Habicht A. Beyond C4d: the ultrastructural appearances of endothelium in ABO-incompatible renal allografts. Nephrol Dial Transplant. 2013;28:3101–3109. doi: 10.1093/ndt/gft373. [DOI] [PubMed] [Google Scholar]
- 39.de Kort H., Moran L., Roufosse C. The role of electron microscopy in renal allograft biopsy evaluation. Curr Opin Organ Transplant. 2015;20:333–342. doi: 10.1097/MOT.0000000000000183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Formalin-fixed paraffin-embedded (FFPE) and RNAlater comparison. A comparison of NanoString counts obtained from samples from the same patient and stored by different methods.
Comparison of geometric mean from ABO-incompatible (ABOi) and ABO-compatible (ABOc) C4d-positive biopsies without evidence of rejection (C4d+ WER) biopsies. There was no difference in geometric mean transcript expression between ABOi and ABOc C4d+ WER biopsies (Mann-Whitney test, P = 0.1532).
Comparison of geometric mean from donor-specific antibody (DSA)-positive and DSA-negative C4d-positive biopsies without evidence of rejection (C4d+ WER). There was no difference in geometric mean transcript expression between DSA-positive and DSA-negative C4d+ WER biopsies (Mann-Whitney test, P = 0.7307).