Abstract
Background
Polymerase Chain Reaction (PCR) has become an important diagnostic and research tool of modern molecular biology globally. Real-time PCR allows for rapid and reliable quantification of mRNA transcription. Reference genes are used as internal reaction control to normalise mRNA levels between different samples in order to allow for an exact comparison of mRNA transcription level.
Methods
In this study, twelve commonly used human reference genes were investigated in Human Embryonic Kidney Cell Lines (HEK293) using real-time qPCR with SYBR green. The genes included beta-2-microglobulin (B2M), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), succinate dehydrogenase complex subunit A (SDHA), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ). The stability of these reference genes was investigated using the geNorm application.
Results
The range of expression stability in the genes analysed was (from the most stable to the least stable): UBC, TOP1, ATP5B, CYC1, GAPDH, SDHA, YWHAZ, CTB, 18S, EIFA-2, B2M and RPL13A. The optimal number of reference targets in the experiment was calculated to be 2 (geNorm V<0.15) when comparing a normalization factor based on the 2 or 3 most stable targets).
Conclusion
The expression stability varied greatly between the 12 candidate reference genes. UBC, TOP1, ATP5B, CYC1 and GAPDH respectively showed the highest stability in HEK293 cells based on both expression stability and expression level. Overall, our data suggest that UBC and TOP1show the least variation and the highest expression stability. This report validates the need for rational selection of reference genes for data normalization to ensure accuracy of quantitative PCR assays.
Keywords: Reference genes, Nrmalization, PCR, Human cell lines, Gene expression
Introduction
The polymerase chain reaction (PCR) is a molecular biology based method used for amplifying DNA; for RNA-based PCR the RNA sample is first reverse-transcribed to complementary DNA (cDNA) using the reverse transcriptase enzyme (1). Real-time polymerase chain reaction (RT-qPCR) is a polymerase chain reaction that monitors the amplification of a targeted DNA molecule through monitoring of the fluorescence of dyes or probes introduced into the reaction, which is proportional to the amount of product formed during the cycling phase of the PCR (1). Gene expression is regulated by cells in all organisms through the turnover of gene and PCR allows for robust detection and quantification of gene expression from small amounts of RNA (2).
Accurate gene expression in RT-qPCR requires data normalisation as a further step in gene quantification analysis. The inclusion of endogenous controls in the assay serves to correct for sample to sample variations. Additionally, it improves the reliability of relative RT-qPCR experiments (3).
A reference gene is important for relative quantification of gene expression, and it is important to choose a suitable reference gene for each experiment (4). An ideal reference gene for RT-qPCR should not be affected by the experimental conditions and the expression level of the reference gene should also be approximately similar to the target gene under study (4). Recent studies have also shown that no single reference gene is ideal for all experiments hence the determination of the appropriate reference gene(s) is very important for the interpretation of RT-qPCR data.
Common genes for relative quantification include; β-actin, β-2-microglobulin, GAPDH mRNAs, and also 18S rRNA (5,6). Both β-Actin, a cytoskeletal protein, and GAPDH, an enzyme of glycolysis, are the two most commonly used reference genes for relative quantification in gene expression assay (7). The β-actin mRNA was one of the earliest RNAs to be used as a reference target because it is ubiquitously expressed; however, its limitation includes the variability in its transcription levels and the presence of pseudogenes that results in the erroneous detection of genomic DNA during real-time qPCR, thus leading to inaccuracies in quantification (8). GAPDH is also a commonly used reference gene for quantification assays. However, it is also limited as a result of its variation from one individual to the other during different stages of the cell cycle. Additionally, it is unsuitable in some systems, because its abundance also varies following treatment with different drugs (8). Furthermore, all genes used for normalization have different limitations hence reference genes should be validated for one's condition of interest (9–11).
Previous studies have demonstrated that reference gene choice for qPCR data analysis has a significant impact on the study outcome. Hence, it is necessary to choose a suitable reference for reliable expression data (3). The geNorm system allows for selection of the best candidate reference gene for a given experimental assay by measuring the expression of 6 or 12 reference genes in a representative set of their own samples. The reference (house-keeping) genes are ranked in the order of stability of expression. The aim of this research was to select and evaluate the stability of 12 reference genes for the purpose of normalization in studying gene expression in a specific human cell line.
Materials and Methods
Study site: The experiment detailed below was conducted in the Department of Microbial Sciences at the University of Surrey, United Kingdom.
Cell line: Human embryonic kidney (HEK293) cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% heat-treated (56°C, 1 h) fetal bovine serum (FBS; Gibco) and penicillin-streptomycin (50 µg/ml).
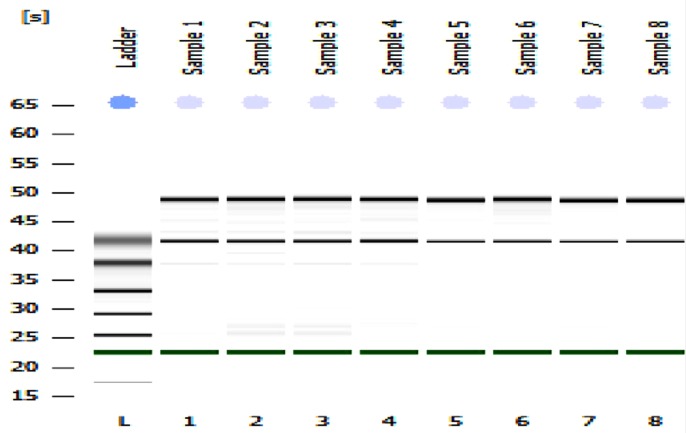
RNA extraction: RNA from HEK293 grown in 6-well plates, was extracted using the ZR MiniPrep TM kit (ZymoResearch, USA) according to the manufacturer's instruction. The RNA was DNAse treated using the Ambion TURBO DNA-free™ Kit (Life Technologies). The concentration and purity of the RNA was measured on a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE) and also analysed on an Agilent 2100 Bioanalyzer (Agilent Technologies) to check the RNA integrity (Fig 1).
Fig 1.
The results of RNA analysis by Agilent bioanalyzer (A) Lane 1 of A shows the molecular marker while lanes 2–9 contain RNA from HEK293 cells.
Quantitative RT-PCR: One µg of total RNA was reverse transcribed to cDNA using the Transcriptor First strand cDNA synthesis kit (Roche) according to the manufacturer's instruction. A cDNA library was created from 10 samples and primers for the 12 reference genes were obtained from PrimerDesign, UK. The reference genes were selected from commonly used reference genes (9,12–14) and the gene symbols, their full names and functions are listed in Table 1.
Table 1.
Reference genes selected with their mean Ct Values and biological functions
| Gene symbol |
Gene Name | CT Values |
Function |
| ATP5B | ATP synthase subunit beta, mitochondrial | 20.12 | ATP synthesis, Hydrogen ion transport, Ion transport, Transport |
| β2M | Beta-2-microglobulin | 20.87 | cytoskeletal protein involved in cell locomotion |
| CTB | Cytochrome B | 21.02 | Electron transport, Respiratory chain, Transport |
| CYC1 | Cytochrome C1 | 22.50 | Accepts electrons from Rieske protein and transfers electrons to cytochrome c in the mitochondrial respiratory chain. |
| EIF4A-2 | Eukaryotic initiation factor 4A-II | 22.89 | Required for mRNA binding to ribosome |
| 18S | 18S ribosomal RNA | 23.09 | RNA component of the 40S ribosome |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 23.78 | carbohydrate metabolism |
| RPL13A | 60S ribosomal protein L13a | 24.62 | Mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes |
| UBC | Polyubiquitin-C | 24.97 | Ubiquitn conjugation |
| SDHA | Succinate dehydrogenase complex, subunit A | 25.86 | Tricarboxylic acid cycle |
| TOP1 | DNA topoisomerase 1 | 28.56 DNA binding | |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein, zeta polypeptide |
30.22 | Protein domain specific binding |
Genes with different functions were chosen in order to avoid genes belonging to the same biological pathways that may be co-regulated. Sequences of reference gene primers (UBC, TOP1, ATP5B, CYC1, CTB, 18S, EIFA-2 and PPC13A, β2M, GAPDH, SDHA, and YWHAZ) are proprietary property of PrimerDesign (15). All qPCR assays were performed on the Quantstudio 7 FLEX Real-Time PCR system (Life Technologies) in 20µL reaction volume containing 10µL of 2x PrecisionPLUS Mastermix (PrimerDesign), 5µL of diluted cDNA (25 ng) and 1µL (300 nmol) each of a gene-specific forward and reverse primer and 3µL of RNAse/DNAse free water. The following standard PCR reaction conditions were used for all transcripts: 95 °C 2 min; 40 cycles of 95 °C 10 s, 60 °C 1 min; 1 cycle of 95 °C 15 s, 60 °C 15 s, 95 °C 15 s. The last cycle provided the Post PCR run melt curve, for assessment of the specificity of amplification. Each of the 10 sample tests and each PCR reaction had three replicates. The experiment was repeated three times to ensure reproducibility.
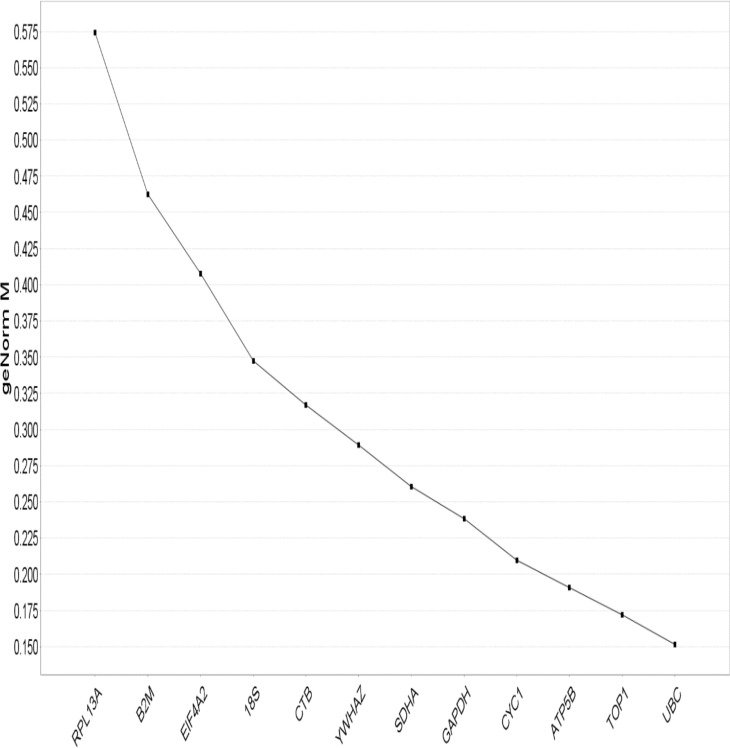
geNorm analysis: The gene expression stability (M) of the 12 reference genes was derived using the software geNorm (15,16). The geNorm program is a Visual Basic application tool for Microsoft Excel and is based on the principle of keeping the expression ratio of two perfect reference genes in a constant state throughout the different experimental conditions (17). The average pair-wise variation of a certain gene with all other tested reference genes is referred to as the M value, whereas the variation of this certain reference gene to another is determined as the standard deviation of the log2-transformed expression level ratios. The gene expression stability M values of the 12 reference genes are shown in Figure 2. The gene with the lowest M value is considered as the most stable expression, while the highest M value has the least stable expression. Prior to the entry of Ct values from the quantitative real-time PCR into geNorm, all Ct values were transformed into relative quantification data, by subtraction of the highest Ct value from all other Ct values for each gene measured. The levels of variation in average reference gene stability with the sequential addition of each reference gene to the equation was also assessed (Figure 2).
Fig. 2.
Gene expression stability of candidate reference genes in HEK293 analyzed by the geNorm program.
Threshold for eliminating a gene as unstable was M ≥ 1.5. The chart generated indicates the average expression stability value M of reference genes at each step during stepwise exclusion of the least stable expressed reference gene. Starting from the least stable gene at the left, the genes are ranked according to increasing expression stability, ending with the two most stable genes on the right. In this example TOP1 and UBC are the two most stable genes.
Results
Analysis of the raw expression levels across all samples identified some variation among reference genes. Quantification cycle (Ct) values for the 12 genes studied ranged from 15 to 30.9, while the majority of these values were between 18.6 and 24.6.
The gene encoding UBC was highly expressed compared to the protein coding genes, reaching threshold fluorescence after only 8.2 amplification cycles, whereas the Ct average of all reference genes within the datasets was approximately 20.7 cycles. The range of expression stability in the genes analysed was (from the most stable to the least stable): UBC, TOP1, ATP5B, CYC1, CTB, 18S, EIFA-2 and RPL13A, β2M, GAPDH, SDHA, and YWHAZ. The optimal number of reference targets in the experiment was calculated to be 2 (geNorm V<0.15) when comparing a normalization factor based on the 2 or 3 most stable targets).
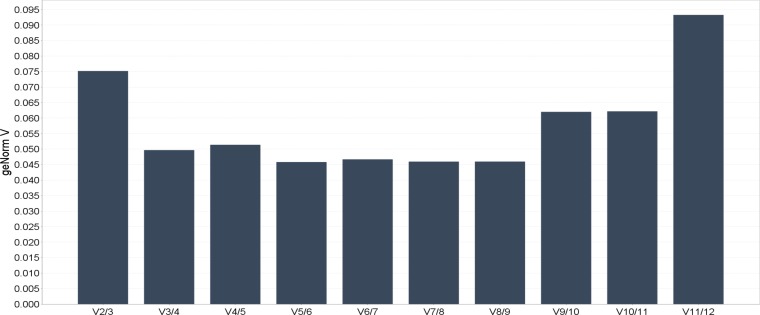
Based on both the expression stability and expression level, our data suggested that UBC and TOP1 can be used as a reference gene for high abundance gene transcripts, CTB and 18S for medium abundance transcripts, and YWHAZ for low abundance transcripts in gene expression studies (Figure 3).
Fig. 3.
The relative expression level of the reference candidate genes in HEK293 cell lines. The chart illustrates the levels of variation in average reference gene stability with the sequential addition of each reference gene to the equation (for calculation of the normalization factor). Starting with the two most stably expressed genes on the left, with the inclusion of a 3rd, 4th 5th gene etc. moving to the right. This measure is known as the “pairwise variation V”. A V score of below 0.15 is recommended as the ideal for this test system. In the example above the two most stable genes from Figure 2, would give high quality data.
Discussion
Quantitative PCR is one of the most sensitive and flexible quantification methods, and it provides simultaneous measurement of gene expression in many different samples for a number of genes. A major limitation of real-time PCR, particularly for relative quantification assays, is that several factors, including the selection of ideal reference genes, may significantly affect the results generated. The use of reference gene helps to control for differences in the amount of starting material, efficiency of amplification, and differences in expression from cells as well as overall level of transcription (5). For accurate gene expression measurements, it is essential to normalise results from qPCR experiments to a fixed reference gene. The ideal reference gene should be unaffected by the experimental treatment and should be expressed at a constant level among different tissues of an organism. Expectedly, no one single gene is expressed at such a constant level in all situations (4,14).
Although Ribonucleic acid (RNA) is a thermodynamically stable molecule, it is highly susceptible to rapid digestion in the presence of RNase enzymes. Hence, a number of electrophoretic methods have been applied to evaluate the integrity of RNA in samples through separation according to the size of the comprised molecule (18,19). RNA quantity and integrity have also been shown to be critical for successful gene expression analysis as misleading qPCR results are obtained following the use of degraded or inaccurately quantified RNA. In this study, the total RNA was extracted from lysates from HEK293 cell lines and careful RNA analysis was performed using an Agilent 2100 Bioanalyzer (Agilent Technologies) prior to the gene expression study. The results suggested that all our RNA samples were of good quality (Fig 1).
The geNorm software provides a useful algorithm for determining the most stable reference genes from a set of tested candidate reference genes in a given sample panel. Other software for ranking of reference genes include the BestKeeper and NormFinder, all three programs are based on different algorithms and analytical procedures. In the current study, only the geNorm software was used for determining overall gene stability, as previous comparative studies have shown that all three software packages, geNorm, BestKeeper, and NormFinder generated similar results when used to analyse data from the same tissue (20).
The geNorm software is based on the prediction that none of the genes being analysed are co-regulated as this would lead to an erroneous choice of optimum normaliser pair, hence the use of a wide array of genes with diverse regulation (Table 1). The optimal number of reference targets in the experiment was calculated to be 2 (geNorm V<0.15) when comparing a normalization factor based on the 2 or 3 most stable targets). Both UBC and TOP1 were considered as useful reference genes for high abundance gene transcripts based on their observed expression stability and expression level. Additionally, our data suggested that both genes CTB and 18S were useful for medium abundance transcripts, while YWHAZ could be considered for low abundance transcripts in gene expression studies.
Although a number of studies have reported that only one single gene is required as an internal control for normalization, it has also been widely suggested that the use of two or more reference genes for RT-qPCR studies might generate more reliable results (9–11). Based on the cut-off value of 0.15 proposed by geNorm program, below which the inclusion of an additional reference gene is not required, the two most stable reference genes of each series subset would be sufficient for accurate normalization. The optimal number of reference targets in this experimental situation is 2 (geNorm V<0.15 when comparing a normalization factor based on the 2 or 3 most stable targets). As such, the optimal normalization factor can be calculated as the geometric mean of reference targets TOP1 and UBC (Figure 3).
In conclusion, the current study has shown that UBC, TOP1 and ATP5B were the most reliable internal controls for accurate normalization when looking at the expression data set as a whole, because these three genes were always classified among the 3 best performing reference genes analyzed by geNorm in all the sample pools. On the other hand, β2M, EIFA2 and RPL13A ranked poorly based on the geNorm software program, indicating that these three genes were not consistently expressed and should be avoided as internal controls when doing gene expression studies in our experimental setup.
This study details the validation of a procedure to select the most stable and the optimal number of control genes for normalization of RT-PCR in a human cell line. Both UBC and TOP1genes can be used for highabundance mRNA, and also allows for more accurate normalization. Our data suggest that UBC and TOP1 may be suitable reference genes in gene expression studies of HEK293 cell lines. Additionally, there is a need to evaluate reference genes in each experimental setting to allow for rational basis for choosing reference genes needed for quantitative expression studies. A number of molecular biology based studies from developing countries are conducted without rational selection of reference target genes. This is because of financial constraints due to sparse funding for research and lack of technical expertise. This study therefore provided a panel of tested reference targets that could be utilized for RTPCR studies in mammalian cell lines.
Acknowledgment
The technical assistance of Dr Alison Davis of PrimerDesign, UK is hereby acknowledged. Laboratory resources were kindly provided by Prof David Blackbourn of the Department of Microbial Sciences, School of Biosciences and Medicine, University of Surrey, UK. This work was partly supported by the PhD Gold student sponsorship package from PrimerDesign, UK to Dr. Adeola Fowotade. The content is solely the responsibility of the author and does not necessarily represent the official views of the funding organization.
References
- 1.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. “The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments”. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 2.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Molecular Aspects of Medicine. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of Suitable Reference Genes for qPCR Analysis of Serum microRNA in Gastric Cancer Patients. Digestive Diseases and Sciences. 2012;57(4):897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 4.Thellin O, Zorzi W, Lakaye B, DeBorman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. Journal of Biotechnology. 1999;75:291. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 5.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochemical Biophysical Research Communication. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 7.Gilliland G, Perrin S, Bunn HF. Competitive PCR for quantitation of mRNA. In: Innis MA, editor. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 60–69. [Google Scholar]
- 8.Glare EM, Divjak M, Bailey MJ, et al. β-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–770. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T, Higgins PJ, DR C. Control selection for RNA quantitation. BioTechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 10.Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphatedehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scandinavian Journal of Immunology. 2004;59(6):566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 11.Yperman J, De Visscher G, Holvoet P, Flameng W. Beta-actin cannot be used as a control for gene expression in ovine interstitial cells derived from heart valves. Journal of Heart Valve Dieases. 2004;13(5):848–853. [PubMed] [Google Scholar]
- 12.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiology Genomics. 2005;21(3):389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 13.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 14.Haberhausen G, Pinsl J, Kuhn CC, Markert-Hahn C. Comparative study of different standardization concepts in quantitative competitive reverse transcription-PCR assays. Journal of Clinical Microbiology. 1998;36:628–633. doi: 10.1128/jcm.36.3.628-633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildyard JC, Wells DJ. PLOS Currents Muscular Dystrophy. 2014. Identification and Validation of Quantitative PCR Reference Genes Suitable for Normalizing Expression in Normal and Dystrophic Cell Culture Models of Myogenesis. PrimerDesign geNorm system Reference Link, 6:ecurrents.md.faafdde4bea8 df4aa7d0cd5553119a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nygard AB, Jørgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Molecular Biology. 2007;8(7):67. doi: 10.1186/1471-2199-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong Z, Gao Z, Wang F, Zhou J, Zhang Z. Selection of reliable reference genes for gene expression studies in peach using realtime PCR. BMC Molecular Biology. 2009;10:71. doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auer H, Lyianarachchi S, Newsome D, Klisovic M, Marcucci G, Kornacker K, Marcucci U. Chipping away at the chip bias: RNA degradation in microarray analysis. Nature Genetics. 2003;35:292–293. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- 19.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Research. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCulloch RS, Ashwell MS, O'Nan AT, Mente PL. Identification of stable normalization genes for quantitative real-time PCR in porcine articular cartilage. Journal of Animal Science and Biotechnology. 2012;3(1):1. doi: 10.1186/2049-1891-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]