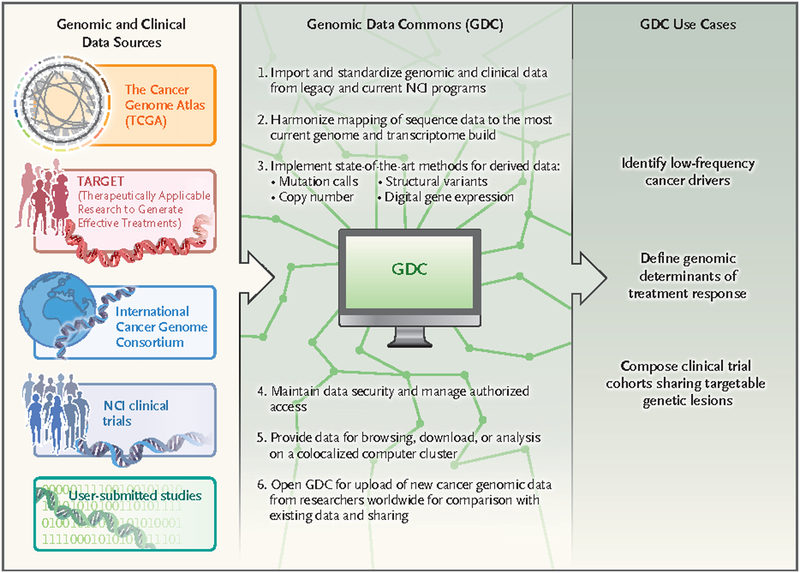
For the past 2 years, the National Cancer Institute (NCI), the University of Chicago, the Ontario Institute for Cancer Research, and Leidos Biomedical Research have been developing an information system called the NCI Genomic Data Commons (GDC) (see figure). The GDC will initially contain raw genomic data as well as diagnostic, histologic, and clinical outcome data from NCI-funded projects such as the Cancer Genome Atlas (TCGA) and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) program. Unlike previous versions of these data sets, the genomic data will be “harmonized” using uniform analytic pipelines to align the raw sequencing data to the genome and identify mutations, copy-number alterations, and gene-expression changes. The research community can access the GDC through an interactive portal (https://gdc-portal.nci.nih.gov), computer systems can interact through the GDC Application Programming Interface, and developers can suggest new features based on GDC open-source code.
Figure 1. Functionality and Utility of the National Cancer Institute Genomic Data Commons (GDC).
The GDC will accept cancer genomic and clinical data from a number of different sources, harmonize the data using consistent bioinformatic pipelines, and allow users to make discoveries regarding the genetic basis of cancer and its impact in the clinic and potentially to identify patients whose tumor profiles make them eligible for particular clinical trials.
An unusual and powerful feature of the GDC is that all researchers will be welcome to submit their cancer genomics data and use the system’s computational pipelines, as long as they agree to share their data broadly. The GDC will add value to the researcher’s own project by providing access to state-of-the-art bioinformatics tools, and the researcher’s data will incrementally increase the interpretive power of the GDC. The system enables researchers to meet the data-access standards for publication in scientific journals and the requirements of the National Institutes of Health (NIH) Genomic Data Sharing policy, and it uses the database of Genotypes and Pheno-types (dbGaP) system to ensure proper data use as specified in informed-consent documents.
Clearly, data sharing will have a pivotal role in precision oncology. A worthy goal set forth by the Institute of Medicine is to develop a new taxonomy of disease based on molecular pathogenesis. By facilitating the sharing of cancer genomic and clinical data, the GDC will gather the information needed by the research and medical community to build a new molecular taxonomy of cancer that has clinical utility. Analysis of current cancer genomic data sets suggests that we are still far from uncovering all the genetic alterations that promote malignant phenotypes, which are known as cancer “drivers.” These calculations suggest that in order to discover genes that acquire driver mutations in 2% or more of patients with cancer, more than 100,000 cancers need to be analyzed.1 The prevalence of cancer drivers follows a long-tail distribution, meaning that the driver events causing disease in many patients with cancer are not prevalent alleles, such as BRAF V600E in melanoma, but are, rather, rare alleles, many of which have not yet been described.
The need to identify such rare drivers of cancer is clear: for any given patient with cancer, detection of a rare genetic driver may be the key to successful therapy. For example, translocation and overexpression of the ROS1 gene occurs in roughly 1% of lung adenocarcinomas, and small-molecule ROS1 inhibitors can induce complete or partial responses in many affected patients.2 The cooccurrence of mutations in rare subgroups of cancers can limit the effectiveness of single targeted drugs such as vemurafenib, but in some cases the problem may be overcome by combinations of drugs.3 The clinical heterogeneity of human cancers is also driven by epigenetic diversity. For example, the response of diffuse large B-cell lymphomas to targeted agents can be predicted by gene-expression profiles.4 Hence, multiple genomic methods are required to provide a molecular description of cancer that has maximum clinical import.
From the inception of cancer genomics, the value of data sharing has been evident. First, the complexity of genomic data inevitably means that only a fraction of the insights inherent in the data can be reported in any one publication. Second, researchers cannot realistically generate within any one project all the genomic data necessary to draw important conclusions, but they can enrich their study by reusing genomic data from other projects.
However, a major challenge for researchers working with cancer genomic data sets is their sheer size. The TCGA data set alone is over a petabyte in size and consists of more than 575,000 files. Just to download the data using a 10-Gbit-per-second connection would take over 3 weeks. Setting up a secure, compliant infrastructure of sufficient scale to store and analyze the data is not only technically challenging, but also expensive. In 2016, the computing equipment required to analyze the raw TCGA sequencing data costs over $1 million, not including the cost of systems maintenance, security, and compliance that are necessary when working with human genomic and clinical data. The GDC addresses these logistic and economic barriers by democratizing access to cancer genomics data, enabling researchers to bring their hypotheses to the data.
The recent explosion of cancer genome analysis has left in its wake a trail of data ambiguity that must be addressed and rectified. Often, genomic studies are published without the authors’ providing raw sequencing data in a public repository, making it impossible to judge the validity of the reported genetic aberrations. Identifying somatic genetic alterations in cancer samples is challenging because of variable contributions of nonmalignant cells, changes in gene copy number, and the presence of tumor subclones. Similarly, the description of copy-number alterations and the quantification of gene expression have not been standardized, posing a problem for researchers trying to compare data from different studies. The GDC addresses these issues by using a harmonization process in which raw sequencing reads are processed through uniform analytic pipelines, and it provides results from multiple analytic methods when there is no single standard. As the human genome sequence is further refined and annotated, and as better analytic pipelines are developed, the GDC will reharmonize its entire genomic content.
The GDC is the foundation of a multiyear NCI effort to foster a new molecular taxonomy of cancer that provides prognostic information and predicts response or resistance to particular therapies. This project will require the curation of data from clinical trials with embedded genomics and from laboratory experiments that assign biologic phenotypes to particular cancer variants, as well as the development of new methods for integrating multiple clinical and molecular data types. Achievement of this goal will be greatly facilitated by genomic data sharing through the GDC, which will be required for all researchers supported by the NCI and should prove attractive to any investigator whose research would benefit from GDC tools. The genomic and clinical data from NCI-sponsored precision-medicine trials such as the NCI Molecular Analysis for Therapy Choice (MATCH) study will be shared through the GDC, allowing researchers to discern the molecular basis of response and resistance to the many targeted agents being investigated.
The recent explosion of cancer genome analysis has left in its wake a trail of data ambiguity that must be addressed and rectified.
The NCI expects investigators to share clinical trial data as outlined in a recent editorial5 and believes that the GDC will be an appropriate venue for sharing data from NCI-supported clinical trials. As the number of cases in the GDC grows, GDC data could provide evidence of drugs working in cancer subtypes that are too rare to be discerned in smaller clinical trial cohorts.
The GDC could expand rapidly as the acquisition of genomic data becomes routine in the course of cancer care. In time, it may be possible for individual patients to become “cancer information donors” and allow their genomic data to be shared through the GDC. Mechanisms for enabling such donation are being developed under the NIH Precision Medicine Initiative Cohort program. Given appropriate informed-consent systems, the GDC could identify patients with rare molecular subtypes of cancer who could be contacted for potential participation in clinical trials appropriate for their particular cancer.
Clearly, the principles and practice of precision oncology will be accelerated by sharing data from thousands of patients with cancer. We hope that the GDC will be embraced by researchers, clinicians, regulatory agencies, patients, and other interested parties as a means to achieve this goal.
Footnotes
Disclosure forms provided by the authors are available at NEJM.org.
References
- 1.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AT, Ou S-HI, Bang Y-J, et al. Crizotinib in ROS1-rearranged non–small-cell lung cancer. N Engl J Med 2014; 371: 1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larkin J, Ascierto PA, Dréno B, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014; 371: 1867–76. [DOI] [PubMed] [Google Scholar]
- 4.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med 2015; 21: 922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taichman DB, Backus J, Baethge C, et al. Sharing clinical trial data — a proposal from the International Committee of Medical Journal Editors. N Engl J Med 2016; 374: 384–6. [DOI] [PubMed] [Google Scholar]