Abstract
Human hepatic cell lines are widely used as an in vitro model for the study of drug metabolism and liver toxicity. However, the validity of this model is still a subject of debate because the expressions of various proteins in the cell lines, including drug-metabolizing enzymes (DMEs), can differ significantly from those in human livers. In the present study, we first conducted an untargeted proteomics analysis of the microsomes of the cell lines HepG2, Hep3B, and Huh7, and compared them to human livers using a sequential window acquisition of all theoretical mass spectra (SWATH) method. Furthermore, high-resolution multiple reaction monitoring (MRM-HR), a targeted proteomic approach, was utilized to compare the expressions of preselected DMEs between human livers and the cell lines. In general, the SWATH quantifications were in good agreement with the MRM-HR analysis. Over 3,000 protein groups were quantified in the cells and human livers, and the proteome profiles of human livers significantly differed from the cell lines. Among the 101 DMEs quantified with MRM-HR, most were expressed at substantially lower levels in the cell lines. Thus, appropriate caution must be exercised when using these cell lines for the study of hepatic drug metabolism and toxicity.
Keywords: Drug-metabolizing Enzymes, Hep3B, HepG2, Huh7, Human Livers, MRM-HR, SWATH
1. Introduction
The liver is the principal organ for drug metabolism, between having high expression levels of most drug-metabolizing enzymes (DMEs) and being involved in first-pass metabolism. Hepatic drug metabolism and liver toxicity have been an active research area for several decades, and are important for drug development as well. Various models have been developed for the study of drug metabolism and toxicity in the liver, such as animal models, immortal cell lines, human primary hepatocytes, human liver tissues, and recombinant enzymes [1]. Each model has its own advantages and limitations. Primary human hepatocytes are generally considered as the ‘gold standard’ for in vitro drug metabolism and liver toxicity studies since the primary cells retain many hepatocyte functions, especially the activity of major DMEs [2]. However, significant batch-to-batch variability, short life spans, limited availability, and high cost have hindered their applications. To address these limitations, several cell lines have been derived from human hepatoma cells, such as HepG2, Hep3B, and Huh7. These cell lines retain some morphological features and functions of hepatocytes, and have robust reproducibility, great availability, relatively low cost and high-throughput capacity. Consequently, these cell lines have been widely used as in vitro models for various studies, such as drug metabolism and toxicity [3–5], lipid metabolism and obesity [6, 7], carcinogenesis and anticancer research [8–10], and virology [11, 12]. For example, a study of resveratrol metabolism was successfully conducted in HepG2 cells, and the phase II DMEs UGT1A1, UGT2B7, and ST1E1 were significantly induced by resveratrol in the cells [3]. Furthermore, Hep3B cells were utilized as an in vitro model to study the mechanisms of acetaminophen-induced hepatotoxicity [5]. However, many DMEs were found to be expressed at lower levels in hepatoma cell lines in comparison with primary human hepatocytes and human liver tissues, which challenges the usefulness of these cells in the study of drug metabolism and drug-induced liver injury [13–16].
Over recent decades, LC-MS/MS-based proteomics has emerged as a powerful technology for protein identification and quantification. Data-dependent acquisition (DDA) has been the main stay proteomics method for untargeted analysis. However, several drawbacks have been recognized for the DDA approach, such as its inherent bias to abundant peptides and the lack of reproducibility for low-abundance proteins. Recently, data independent acquisition (DIA), also termed sequential window acquisition of all theoretical mass spectra (SWATH) when analysis is performed on an AB Sciex TripleTOF-type instrument, has proven a powerful approach for both untargeted and targeted protein quantification. DIA/SWATH is capable of quantifying protein expression with high reproducibility and accuracy via overcoming the biased sampling problem seen with DDA [17]. In addition, a new targeted proteomics assay named parallel reaction monitoring (PRM) or high resolution multiple reaction monitoring (MRM-HR) has shown several advantages over conventional targeted proteomics (e.g., MRM or SRM), such as greater selectivity and robustness [18, 19]. To date, these new proteomics approaches have not been utilized to determine the proteome profiles of hepatoma cell lines, much less compare them with those of human livers. The purposes of this study were to 1) compare proteome profiles between human livers and the three commonly used hepatic cell lines Hep3B, HepG2, and Huh7 using SWATH; 2) quantify selected DMEs in human livers and the cell lines using an MRM-HR strategy for evaluation of differences in expressions; 3) compare quantification results from SWATH and MRM-HR. A better understanding of the differences of protein expression between hepatic cell lines and human livers, especially differences in DMEs abundance, will lead to a more appropriate use of these cell lines for in vitro studies of drug metabolism and liver toxicity.
2. Materials and Methods
2.1. Chemicals and Reagents
Human liver microsomes (pooled from 200 donors with mixed-gender) were purchased form Sekisui XenoTech (Kansas City, KS). Dulbecco’s Modified Eagle Medium (DMEM), Fetal bovine serum (FBS), and 100× antibiotics mixture containing 100 IU/mL penicillin and 100 μg/mL streptomycin (P/S) were products of Invitrogen (Carlsbad, CA, USA). HyClone™ RPMI-1640 medium was purchased from Thermo Scientific (Waltham, MA). Urea and dithiothreitol were purchased from Fisher Scientific Co. (Pittsburgh, PA). Trifluoroacetic acid, formic acid, and acetonitrile were from Sigma-Aldrich (St. Louis, MO). Iodoacetamide and ammonium bicarbonate were the products of Acros Organics (Morris Plains, NJ). TPCK-treated trypsin was obtained from Worthington Biochemical Corporation (Freehold, NJ). Water Oasis HLB columns were from Waters Corporation (Milford, MA). Bovine serum albumin (BSA) standard was purchased from Thermo Fisher Scientific (Waltham, MA).
2.2. Cell Culture
HepG2 cells (HB-8065™) were purchased from ATCC (Manassas, VA, USA). Hep3B and Huh7 cell lines were kindly provided by Drs. Theodore H. Welling III and Lei Yin (University of Michigan), respectively. Cells were cultured in DMEM supplemented with FBS, 1% P/S and 2 mM glutamine. The concentrations of FBS were 10% for HepG2 and Hep3B cells, and 5% for Huh7 cells. Cell passages were performed with 0.25% trypsin-EDTA (Gibco, Life Technologies) when cells reached 80~90% confluence.
2.3. Microsomes Preparation from HepG2, Hep3B, and Huh7 Cells
Cells pellets were collected following typsin-EDTA digestion and centrifugation, and were homogenized in phosphate buffered saline (PBS, pH 7.4) on ice using an ultrasonic probe (10 s × 4 times). S9 fractions were obtained after the homogenates were centrifuged at 4 °C at 10,000 g for 30 min. The supernatants (S9 fractions) were transferred to a Beckman ultracentrifuge tube and centrifuged at 300,000 g (80,000 rpm) for 20 min. Microsomes were obtained by resuspending the pellets in PBS using a tissue grinder. Protein concentrations of the microsomes were determined using a Pierce™ BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA). The microsome samples were stored at −80°C until use.
2.3. Proteomics Sample Preparation
Protein digestion was conducted according to a previously reported Lys-C/Trypsin combinatorial digestion protocol with some modifications [20]. An aliquot of 100 μg protein of microsomes was mixed with the internal standard 0.5 μg BSA in an Eppendorf Protein LoBind tube. A 10-fold volume of pre-cooled acetone was added. The mixture was briefly vortexed and incubated at −20 °C for at least 2 hours, followed by centrifugation at 17,000 g for 15 min at 4 °C. The supernatants were discarded, and the precipitated proteins were added with 200 μL ice-cold 80% ethanol for a washing step. The mixture was centrifuged again at 17,000 g for 15 min at 4 °C. The supernatants were removed and the precipitated proteins were air-dried at room temperature. The dried proteins were resuspended in 100 μL of freshly prepared 4 mM dithiothreitol in 8 M urea solution containing 100 mM ammonium bicarbonate. Samples were briefly vortexed and sonicated, then incubated at 37 °C for 45 min. After samples were cooled down to room temperature, 100 μL of 20 mM iodoacetamide freshly prepared in 8 M urea/100 mM ammonium bicarbonate solution was added. The mixture was incubated at room temperature for 30 min in the dark for alkylation. Following the incubation, urea concentration was adjusted to 6 M by adding 64.6 μL of 50 mM ammonium bicarbonate, and lysyl endopeptidase (Wako Chemicals, Richmond, VA) was added for the first digestion step (protein to lysyl endopeptidase ratio = 100:1) at 37 °C for 6 h. Samples were then diluted with 50 mM ammonium bicarbonate to decrease urea concentration to 1.6 M urea, followed by the second step of digestion with trypsin at a protein to trypsin ratio of 50:1 for an overnight incubation at 37 °C. Digestion was terminated by the addition of 1 μL trifluoroacetic acid. Digested peptides were extracted and purified using Waters Oasis HLB columns according to the manufacturer’s instruction. Peptides eluted from the columns were dried in a SpeedVac SPD1010 concentrator (Thermo Scientific, Hudson, NH), and reconstituted in 80 μL of 3% acetonitrile solution with 0.1% formic acid. The peptide samples were centrifuged at 17,000 g for 10 min at 4 °C, and half of the supernatant was collected and supplemented with 1 μL of the synthetic iRT standards solution from Biognosys AG (Cambridge, MA) prior to LC-MS/MS analysis.
2.4. LC-MS/MS-Based Protein Quantification
Proteomic analysis was carried out on a TripleTOF 5600+ mass spectrometer (AB Sciex, Framingham, MA) coupled with an Eksigent 2D plus LC system (Eksigent Technologies, Dublin, CA). LC separation was performed via a trap-elute configuration, which includes a trapping column (ChromXP C18-CL, 120 Å, 5 μm, 0.3 mm cartridge, Eksigent Technologies, Dublin, CA) and an analytical column (ChromXP C18-CL, 120 Å, 150 × 0.3 mm, 5 μm, Eksigent Technologies, Dublin, CA). The mobile phase consisted of water with 0.1% formic acid (phase A) and acetonitrile containing 0.1% formic acid (phase B, Avantor, Center Valley, PA). An amount of 6 μg protein was injected for analysis. Peptides were trapped and cleaned on the trapping column with the mobile phase A delivered at a flow rate of 10 μL/min for 3 min before being separated on the analytical column with a gradient elution at a flow rate of 5 μL/min. The gradient time program was set as follows for the phase B: 0 to 68 min: 3% to 30%, 68 to 73 min: 30% to 40%, 73 to 75 min: 40% to 80%, 75 to 78 min: 80%, 78 to 79 min: 80% to 3%, and finally 79 to 90 min at 3% for column equilibration. Each sample was followed by a blank injection to prevent sample carryover. The mass spectrometer was operated in a positive ion mode with an ion spray voltage floating at 5500 v, ion source gas one at 28 psi, ion source gas two at 16 psi, curtain gas at 25 psi and source temperature at 280 °C.
2.4.1. Information-Dependent Acquisition (IDA) and Data Analysis
To generate the reference spectral library for SWATH and MRM-HR data analysis, IDA was performed for the microsomes of human livers and the three hepatic cell lines with three replicates.
The IDA method was set up with a 250 ms TOF-MS scan from 400 to 1250 Da, followed by an MS/MS scan in a high sensitivity mode from 100 to 1500 Da of the top 30 precursor ions from the TOF-MS scan (50 ms accumulation time, 10 ppm mass tolerance, charge state from +2 to +5, rolling collision energy, and dynamic accumulation). Former target ions were excluded from MS/MS scan for 15 s.
All 12 IDA data files were searched using MaxQuant software (version 1.5.3.30, Max Planck Institute of Biochemistry, Germany) with default settings. The human proteome fasta file with 20,237 protein entries downloaded from Uniport on 3/2/2017 was used as the reference sequences for the search. Trypsin/P was used as the protease. Peptide lengths were between 7 and 25 residues with up to two missed cleavage sites allowed. Carbamidomethyl was set as a fixed modification. Variable modifications included acetyl (protein N-term) and oxidation (M). Up to 5 modifications per peptide were allowed. Mass tolerance for precursor was 0.07 for the first search and 0.006 Da for the main search, while the tolerance for fragment ions was 40 ppm. A false discovery rate (FDR) of 0.01 was used as the cutoff for both peptide and protein identification.
2.4.2. SWATH Acquisition and Data Analysis
Microsome samples were analyzed using a SWATH method, which is comprised of a 250 ms TOF-MS scan from 400 to 1250 Da, followed by MS/MS scans from 100 to 1500 Da performed on all precursors in a cyclic manner. A 100-variable isolation window scheme was used in this study (Supplemental Table 1). The accumulation time was 25 ms per isolation window resulting in a total cycle time of 2.8 s.
For untargeted analysis, the Spectronaut software (version 11.0.15038.8, Biognosys AG, Schlieren, Switzerland) was used to process the SWATH data with the reference spectral library generated from the IDA searches [21]. Default settings were used except for the major protein grouping which was set to ‘by Gene id’. The MS2 peak areas of top 3 signature peptides of each protein were used for quantification. The data were normalized using the ‘Local Normalization’ strategy.
The SWATH data were also analyzed using the Skyline-daily program (version 3.7.1.11271, University of Washington, Seattle, WA) for targeted DMEs quantifications [22]. The isolation scheme was set as “SWATH (VW 100)”. The MS1 and MS/MS filtering were both set as “TOF mass analyzer” with the resolving power of 30,000 and 15,000, respectively. Retention time prediction was based on the auto-calculate regression implemented in the iRT calculator. Proper peak selections were inspected manually with the automated assistance of Skyline-daily. The surrogate peptides used for the DMEs quantification were provided in Supplemental Table 2. The peak areas of top 3 to 5 fragment ions of all surrogate peptides were summed up and normalized to the internal standard BSA. The BSA-normalized summed peak areas were used to determine the relative abundance of DMEs across the different microsome samples.
2.4.3. MRM-HR Acquisition and Data Analysis
A scheduled MRM-HR strategy was also performed for the quantification of selected DMEs based on a previous report with some modifications [18]. For this method, a shorter LC gradient program was set as follows (phase B): 0 to 22 min: 3% to 30%, 22 to 25 min: 30% to 40%, 25 to 26 min: 40% to 80%, 26 to 27 min: 80%, 27 to 28 min: 80% to 3%, and 28 to 32 min at 3% for column equilibration.
The MRM-HR acquisition consisted of one 200 ms TOF-MS scan from 400 to 1250 Da, followed by MS/MS scans from 100 to 1500 Da (50 ms accumulation time, 50 mDa mass tolerance, +2 to +5 charge states with intensity above 2,000,000 cps, 30 maximum candidate ions to monitor per cycle, rolling collision energy) of the inclusion precursors with the scheduled retention times determined from pilot non-scheduled MRM-HR runs. The intensity threshold of the targeted precursors in the inclusion list was set to 0 and the scheduling window was 270 s. The targeted peptides/precursors were the same as that for the targeted SWATH analysis (Supplemental Table 2).
After MRM-HR acquisition, the data were analyzed by Skyline-daily which automatically detects and matches the MS/MS chromatographic peaks against the spectral library generated from the IDA searches. All peak selections were checked manually after the automated matches. The MS1 and MS/MS filtering were both set as “TOF mass analyzer” with the resolution power of 30,000 and 15,000, respectively, while the “Targeted” acquisition method was defined in the MS/MS filtering. The relative abundance of DMEs was also calculated using the BSA-normalized summed peak areas.
2.5. Statistical and Bioinformatic Analysis
Student’s t-test with FDR correction was used to identify differentially expressed proteins between human livers and each of the three hepatic cell lines. Benjamini-Hochberg procedure was applied to control FDR at 0.05. Proteins with an adjusted p-value (q-value) ≤ 0.05 and a Log2 fold change of expression ≥ 0.58 (i.e. 1.5-fold) were considered significantly different. Correlation and linear regression analysis were conducted using GraphPad Prism (version 6.02, La Jolla, CA). Venn diagram was drawn by an online tool available at http://bioinformatics.psb.ugent.be/webtools/Venn/. Proteome heat map was analyzed by Spectronaut and DMEs heat map was generated using Microsoft Excel 2013. PCA was conducted by SIMCA (version 14.1, MKS Umetrics).
Pathway analysis was implemented for all differentially expressed proteins between the three hepatic cell lines and human livers using the software iPathwayGuide (Advaita Corporation, Plymouth, MI). A q-value less than 0.05 was considered statistically significant for the pathway analysis. Pathway database was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Release 81.0+/01–20, Jan 2017).
2.6. Data Availability
All LC-MS/MS data have been deposited to the ProteomeXchange Consortium via the PRIDE [23] partner repository with the dataset identifier PXD008190.
3. Results
3.1. Proteome profile comparisons among the microsomes of human livers and the Hep3B, HepG2 and Huh7 cell lines
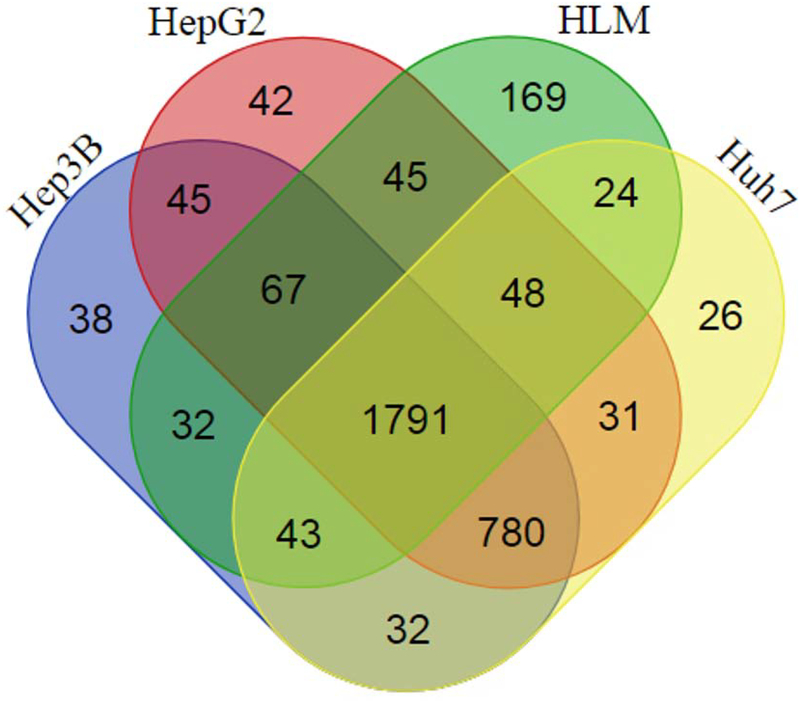
As illustrated in the overall workflow of the present study (Supplemental Figure 1), IDA data were firstly generated for the microsomes samples of human livers and the three hepatic cell lines (n = 3 for each group) on a TripleTOF mass spectrometer. As shown in Table 1, a total of 32,495 peptides, 5,396 proteins, and 3,391 protein groups were identified at a FDR of 0.01. The search results were used to build spectral libraries in both Spectronaut and Skyline. All 12 microsomes samples were then analyzed using a SWATH method, and data analysis with Spectronaut led to the identification of 2219, 2828, 2849, and 2775 protein groups in all three replicates of the microsomes of human livers, Hep3B, HepG2, and Huh7 cells, respectively, at a q-value ≤ 0.01 (Table 1). The total number of protein groups identified among all samples was 3213, in which 1791 protein groups (55.7%) were shared by human livers and the cell lines, 780 protein groups (24.3%) were only shared by the three cell lines, 169 protein groups (5.3%) were unique to human livers, while all remaining protein groups (473, 14.7%) were found in one or two cell lines (Figure 1).
Table 1.
Summary of SWATH analysis of protein expression in the Hep3B, HepG2, and Huh7 cell lines and human livers.
| Sample groups | HLM | Hep3B | HepG2 | Huh7 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | #2 | #3 | #1 | #2 | #3 | #1 | #2 | #3 | #1 | #2 | #3 | |
| Numbers of peptides in DDA library | 32495 | |||||||||||
| Numbers of proteins in DDA library | 5396 | |||||||||||
| Numbers of protein groups in DDA library | 3391 | |||||||||||
| Numbers of identified peptides in SWATH | 18911 | 18902 | 19259 | 25336 | 25689 | 25835 | 26290 | 26361 | 26715 | 25215 | 25574 | 25308 |
| Numbers of identified proteins in SWATH | 3785 | 3670 | 3721 | 4663 | 4763 | 4749 | 4668 | 4728 | 4773 | 4589 | 4641 | 4624 |
| Numbers of identified protein groups in SWATH | 2487 | 2436 | 2458 | 2942 | 2990 | 3001 | 2965 | 3002 | 3022 | 2922 | 2944 | 2942 |
| Numbers of quantitative protein groups identified in all replicates (Q-value<0.01) | 2219 | 2828 | 2849 | 2775 | ||||||||
| Numbers and percentages of quantitative protein groups with CV < 25% among replicates | 1884 (84.9%) | 2553 (90.3%) | 2580 (90.6%) | 2491 (89.8%) | ||||||||
Figure 1.
Venn diagram representing the overlaps among the protein groups identified in human livers and the Hep3B, HepG2, and Huh7 cell lines by SWATH. All protein groups were identified in all three biological replicates for each group with a q-value ≤ 0.01. The diagram was produced using a web tool available at http://bioinformatics.psb.ugent.be/webtools/C/.
The quantification of protein groups was based on the average MS2 peak area of top 3 signature peptides with a local normalization strategy by default in Spectronaut. The median CV among three replicates were low (5.3–8.4%) for both human livers and the hepatic cell lines (Supplemental Figure 2). The numbers of the protein groups with CV less than 25% among three replicates were 1884 (84.9%), 2553 (90.3%), 2580 (90.6%) and 2491 (89.8%) for human livers, Hep3B, HepG2 and Huh7, respectively, demonstrating the high throughput and reproducibility of the SWATH method (Table 1). The relative expression levels of all proteins quantified from the SWATH study were summarized in Supplemental Table 3.
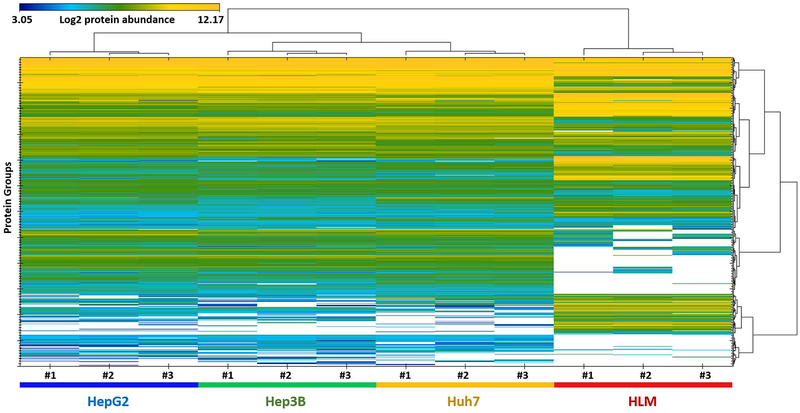
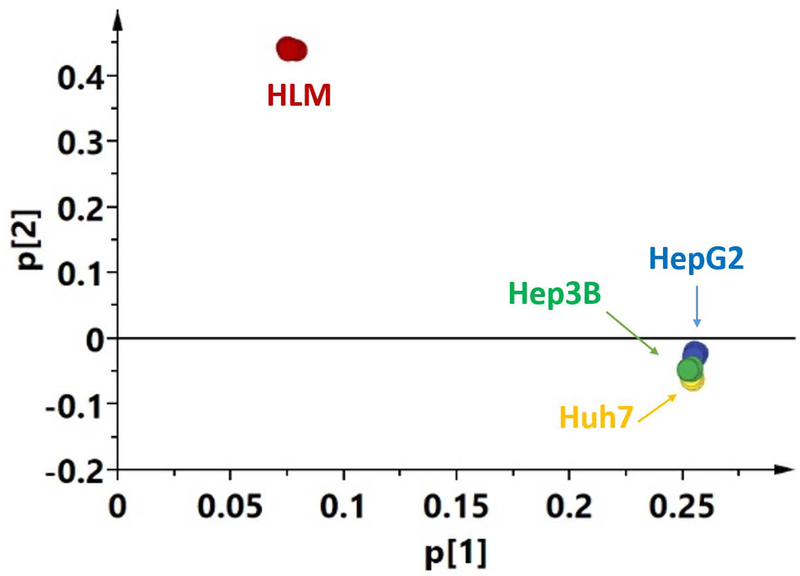
In order to provide a general picture of the proteome profiles of the tested microsome samples, a heat map and PCA results were presented in Figure 2 and 3, respectively. Dramatic differences were observed between human livers and the three cell lines, while the differences among the cell lines were much smaller. Moreover, correlation analysis of the proteomes demonstrated the same trends (Supplemental Figure 3). The correlations between human livers and the cell lines were much lower (R2 = 0.069 – 0.076) than those among the cell lines (R2 = 0.65).
Figure 2.
Heat map analysis of proteins differentially expressed in human livers and the Hep3B, HepG2, and Huh7 cell lines (n= 3 for each group). The MS signal intensity was presented as a Log2 value. White spots indicate zero intensity.
Figure 3.
Principal component analysis of proteome expression profile of human livers and the Hep3B, HepG2, and Huh7 cell lines generated from SWATH data.
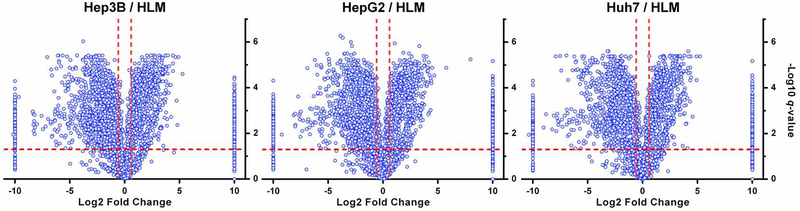
In the present study, 2510, 2529, and 2483 proteins were found to be differentially expressed between human livers and the hepatic cell lines, Hep3B, HepG2, and Huh7, respectively (Figure 4). To further understand the functional role of the differently expressed proteins, we performed a pathway analysis using the iPathwayGuide online software. iPathwayGuide scores pathways utilizing an ‘Impact Analysis’ approach, which uses not only the number of differentially expressed genes (enrichment analysis), but also pathway topological information. As shown in Table 2, 12, 11, and 11 biological pathways were found to be significantly impacted in the Hep3B, HepG2, and Huh7 cells, respectively. Most of the pathways were downregulated in the hepatic cell lines, such as those related to xenobiotics metabolism, non-alcoholic fatty liver disease, Alzheimer’s disease and Parkinson’s disease. Only three pathways were upregulated, including spliceosome, ribosome, and mRNA surveillance pathways.
Figure 4:
Comparison of protein groups expressed in the Hep3B, HepG2, and Huh7 cell lines with human livers. The volcano plot was plotted as Log2 intensity fold changes of the protein groups in Hep3B, HepG2 and Huh7 cells relative to human livers versus −Log10 q-value. For undetected proteins in numerator (cell line) or denominator (HLM), a “−10” or “10” was given as its Log2 abundance fold change value, respectively. The significant thresholds for q-value (≤ 0.05) and intensity fold change (≥ 1.5-fold) were indicated by the red dashed lines.
Table 2.
Significantly altered biological pathways in the Hep3B, HepG2, and Huh7 cell lines compared to human livers.
| Hep3B vs HLM | HepG2 vs HLM | Huh7vs HLM | ||||||
|---|---|---|---|---|---|---|---|---|
| Pathway name | q-value | Trend | Pathway name | q-value | Trend | Pathway name | q-value | Trend |
| Non-alcoholic fatty liver disease | 6.0E−03 | ↓ | Chemical carcinogenesis | 4.5E−04 | ↓ | Ribosome | 5.4E−05 | ↑ |
| Metabolism of xenobiotics by cytochrome P450 | 6.0E−03 | ↓ | Metabolism of xenobiotics by cytochrome P450 | 5.2E−04 | ↓ | Metabolism of xenobiotics by cytochrome P450 | 2.2E−04 | ↓ |
| Steroid hormone biosynthesis | 7.0E−03 | ↓ | Parkinson’s disease | 7.0E−03 | ↓ | Drug metabolism-cytochrome P450 | 2.0E−03 | ↓ |
| Alzheimer’s disease | 7.0E−03 | ↓ | Alzheimer’s disease | 7.0E−03 | ↓ | Alzheimer’s disease | 2.0E−03 | ↓ |
| Spliceosome | 9.0E−03 | ↑ | Drug metabolism-cytochrome P450 | 1.9E−02 | ↓ | Steroid hormone biosynthesis | 3.0E−03 | ↓ |
| Drug metabolism-cytochrome P450 | 1.2E−02 | ↓ | Metabolic pathways | 1.9E−02 | ↓ | Chemical carcinogenesis | 5.0E−03 | ↓ |
| Metabolic pathways | 1.3E−02 | ↓ | Biosynthesis of amino acids | 1.9E−02 | ↓ | Non-alcoholic fatty liver disease | 2.4E−02 | ↓ |
| Peroxisome | 1.5E−02 | ↓ | Non-alcoholic fatty liver disease | 1.9E−02 | ↓ | Drug metabolism - other enzymes | 2.6E−02 | ↓ |
| Chemical carcinogenesis | 2.7E−02 | ↓ | Spliceosome | 2.3E−02 | ↑ | Retinol metabolism | 3.8E−02 | ↓ |
| Parkinson’s disease | 2.9E−02 | ↓ | Drug metabolism - other enzymes | 3.0E−02 | ↓ | Spliceosome | 3.9E−02 | ↑ |
| Oxidative phosphorylation | 3.4E−02 | ↓ | Steroid hormone biosynthesis | 3.0E−02 | ↓ | Parkinson’s disease | 4.5E−02 | ↓ |
| mRNA surveillance pathway | 4.0E−02 | ↑ | ||||||
3.2. MRM-HR analysis of DMEs protein expression in the microsomes of human livers and hepatic cell lines
A comprehensive list of clinically relevant DMEs was generated based on previous publications [13, 24–26] and the human protein atlas (http://www.proteinatlas.org), which consists of more than 200 enzymes. This list was screened against the human liver microsome IDA data generated in our laboratory, resulting in 101 hits which include 78 phase I enzymes and 23 phase II enzymes. MRM-HR, a newly introduced targeted proteomics acquisition method, was used for the quantifications of the selected DMEs since the method provides improved sensitivity, reproducibility, and quantitative dynamic range compared with other quantitative proteomics techniques such as DDA, SRM, and SWATH.
The surrogate peptides for the 101 DMEs were selected based on MS detectability, peptide uniqueness, and no ragged ends. Finally, a total of 277 peptides were included for the quantification of the 101 DMEs and BSA (Internal standard) (Supplemental Table 2). It is noted that 25 enzymes only have one unique peptide for quantification due to the high degree of conservation within the enzyme family. Following the MRM-HR acquisition, data were analyzed using Skyline. The protein expression levels of the 101 DMEs in the three hepatic cell lines relative to human livers were calculated as log2 ratios and plotted as a heat map in Supplemental Figure 4.
A striking finding was that 41 out of 101 (40.6%) DMEs could not be detected in any of the three hepatic cell lines under our experimental conditions, including most CYP450s (19 out of 24) and uridine 5’-diphospho-glucuronosyltransferases (UGTs) (10 out of 11). The numbers of undetectable enzymes in HepG2, Hep3B and Huh7 cells were 44 (43.6%), 54 (53.5%) and 49 (48.5%), respectively. In addition, most enzymes detected in the cell lines were expressed at significantly lower levels than those in human livers. Nevertheless, a few enzymes had comparable or even higher abundance in the cell lines relative to human livers, which include ALDH1A1, ALDH1B1, ALDH4A1, ALDH5A1, ALDH7A1, DHRS2, GSR, HNF4A, and METAP1.
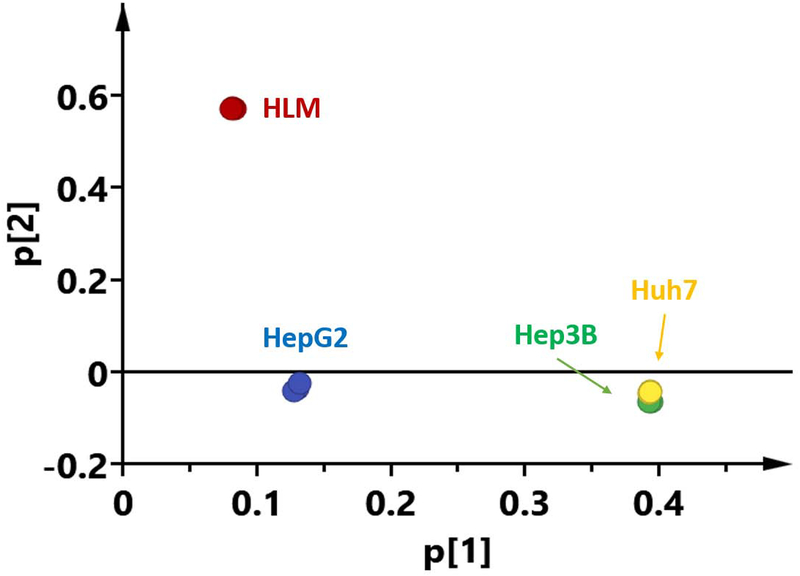
The similarities and discrepancies of DMEs protein expression profiles between human livers and the three hepatic cell lines were further illustrated by PCA. Figure 5 showed a PCA scores plot using the first two principal components (t[1] and t[2]) to demonstrate the expression patterns among the three cell lines and human livers. The replicates of each group were clustered tightly, indicating a good reproducibility of the MRM-HR assay. Three distinct patterns in DMEs expression profiles were found in the scores plot: human livers, HepG2, Hep3B and Huh7. While the significant differences between human livers and the hepatic cell lines were expectedly observed, it’s interesting that the Hep3B and Huh7 were very similar in terms of DMEs expression patterns, and HepG2 exhibited a distinctive expression pattern from the Hep3B and Huh7 cells.
Figure 5.
Principal component analysis of 101 drug-metabolizing enzymes expression profiles generated from the MRM-HR data of human liver and the Hep3B, HepG2, and Huh7 cell lines. The relative contribution of the variance is shown by two major principal components.
Furthermore, correlation analysis of DMEs protein expressions among human livers and the hepatic cell lines was conducted. The results demonstrated that the expressions in human livers did not significantly correlate with that of any of the cell lines, while significant correlations were observed among the three cell lines. Consistent with the PCA results, the correlations between HepG2 and the other two cell lines (R2 = 0.58 (Huh7) and 0.67 (Hep3B)) were smaller than that between Hep3B and Huh7 (R2 = 0.83).
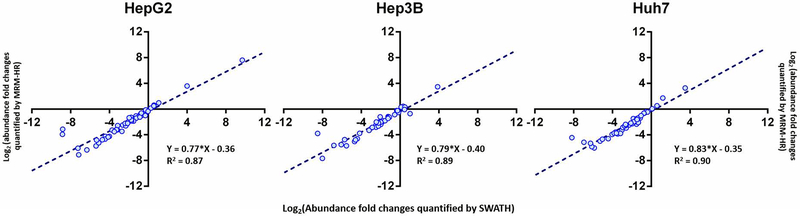
3.3. Correlations of the quantifications of DMEs protein expressions between MRM-HR and SWATH
In order to directly compare the quantification results between MRM-HR and SWATH, SWATH data were also processed by Skyline for the targeted analysis of the DMEs quantified by the MRM-HR method using the same surrogate peptides and transitions. Figure 6 shows the comparison between the MRM-HR and SWATH results regarding the Log2-fold changes of DMEs expressions in the cell lines compared to human livers. A high correlation between the two methods (R2 = 0.87–0.90) was observed for DMEs quantified by both methods. Several DMEs were only detected by the MRM-HR but not the SWATH method, which include DHRSX and HNF4A in HepG2, ADH6, AKR1A1, AKR1D1, CYP2J2, CYP4F12, DHRSX, HNF4A and UGT2B4 in Hep3B, AKR1A1, CYP2J2 and HNF4A in Huh7.
Figure 6.
Comparison of quantification results of drug-metabolizing enzymes between SWATH and MRM-HR. Linear regression analysis of the results from the two methods was performed for the Log2-fold changes of the expressions in the Hep3B, HepG2, and Huh7 cell lines relative to human livers. Several proteins were only detected by the MRM-HR method, and were excluded from the analysis.
4. Discussion
Several mRNA microarray studies have been conducted to compare mRNA expressions among selected hepatic cell lines, primary human hepatocytes, and human liver tissues [14, 27–29]. However, it has been well-recognized that mRNA and protein expression levels are poorly correlated for many genes including those encoding DMEs because protein expression can be influenced by many post-transcriptional factors [30–32]. Since proteins are the functional molecule for most biological processes, it is important to take protein expression profiles into consideration when choosing a hepatic cell line for the in vitro study of drug metabolism or liver toxicity. However, very few studies have been conducted to compare protein expressions between human livers and hepatic cell lines, especially using new quantitative proteomics strategies, such as DIA/SWATH and PRM/MRM-HR. Studies that have been done with DDA methods include: Slany et al. compared the proteome profiles of primary human hepatocytes and the hepatocyte cell lines HepG2 and Hep3B using 2D-PAGE and DDA proteomics [33]; Megger and colleagues applied DDA and tandem mass tag label-based strategies to proteome analysis of HepG2, Hep3B and SK-Hep-1 [34]; and recently, proteomic characteristics of HepG2, Upcyte, and HepaRG were compared to primary human hepatocytes using a DDA strategy [16].
The MS/MS scans in conventional DDA methods are acquired for the top N (e.g., top 30) precursors detected in an MS1 scan with narrow isolation windows [35], and quantifications are based on MS1 scans. Relative to the DIA/SWATH method, DDA is more amenable for searching against a proteome database for the assignment of peptides to spectra; however, it has inherent limitations in reproducibility and selectivity for quantification. In contrast, DIA/SWATH acquired MS/MS scans of all precursors within a selected m/z (400–1250 m/z in this study) [36]. A growing body of research has demonstrated this method’s high coverage, reproducibility and precision for both discovery and quantitative proteomics [37–39].
In the current study, we generated a comprehensive spectral library using the data obtained from DDA runs. A total of 5396 proteins (3391 protein groups) were identified in HLM and the three hepatic cell lines at a FDR of 0.01, comparing to 1578 [34], 1995 [33], and 4696 [16] proteins identified in previous DDA proteomics studies with hepatic cell lines. Moreover, we used SWATH to compare the proteome profiles from microsome preparations of three widely used human hepatoma cell lines, i.e. HepG2, Hep3B and Huh7, and human liver. Furthermore, we utilized a MRM-HR approach for targeted quantitative analysis of pre-selected DMEs, and compared its results with the SWATH data. With the SWATH approach, the total numbers of protein groups identified were 2219, 2828, 2849 and 2775 in all replicates of the microsomes of human livers, Hep3B, HepG2 and Huh7 cells, respectively (q-value ≤ 0.01, Table 1). Low median CV values (5.3–8.4%) among the replicates in all groups demonstrated excellent reproducibility of the assay (Supplemental Figure 2).
Dramatically different proteome profiles were observed between the cell lines and human livers, while the differences among the cell lines were much smaller (Figure 2, 3). Pathway analysis suggests that many metabolism-related biological processes, such as xenobiotics metabolism, were significantly downregulated in the hepatoma cell lines compared with human livers, an observation consistent with previous reports [13, 16, 38]. Spliceosome proteins were significantly upregulated in all three cell lines, which may reflect the cancerous natures of the cells [40, 41].
Unlike SWATH, targeted proteomics approaches including SRM and PRM/MRM-HR detect preselected surrogate peptides in order to quantify proteins of interest. SRM, performed on a triple quadruple-like instrument, only detects pre-selected fragment ions from the selected precursors [42] whereas PRM/MRM-HR, performed on quadrupole-high resolution mass spectrometers such as TripleTOF and Orbitrap, records all fragments from the selected precursors with high-resolution scans [18, 19, 43]. Thus, PRM/MRM-HR requires much less effort for assay development and optimization, and also provides higher selectivity due to the higher resolution of MS/MS scans.
To verify the quantification results from the SWATH analysis and compare the performance of SWATH with targeted proteomic methods, we used MRM-HR to quantify 101 DMEs and BSA utilizing 277 surrogate peptides. In agreement with Nakamura et al’s results [38], our data showed that the SWATH quantification results highly correlated with those from the MRM-HR analysis (Figure 6, R2 = 0.87–0.90). However, several DMEs with low expression levels in the cell lines could only be detected in MRM-HR, and not in SWATH. This is expected because, compared to MRM-HR, the wider isolation windows in SWATH (m/z: 5–10, 100 variable windows in this study) can result in a higher level of background noise, and thus lower signal-to-noise ratios.
With respect to DMEs expression, the three hepatoma cell lines displayed very different patterns compared to human livers. Under our experimental conditions, most CYP450s (19/24) and UGTs (10/11) could not be detected in any of the three cell lines. Therefore, the use of these cells for assessing the biotransformation of xenobiotics mediated by CYPs or UGTs is unlikely to yield pharmacologically or toxicologically relevant data. However, some phase I enzymes had considerable expression in the cell lines. For example, most aldehyde dehydrogenase (ALDH) enzymes, which are responsible for the oxidation of many aldehydes to their corresponding carboxylic acid metabolites, were expressed at significant levels in all three cell lines. Therefore, these cell lines can be a legitimate in vitro model for ALDH-related experiments [44, 45]. Surprisingly, HNF4A, known as a regulator of many DMEs [46], had a much higher expression level in the cell lines than in human livers. In addition, all three cell lines significantly expressed some phase II enzymes, such as glutathione S-transferases, microsomal glutathione S-transferases, and sulfotransferases.
Based on the PCA results presented in Figure 5, it is interesting to note that Hep3B and Huh7 were very similar in terms of DMEs expression patterns, while HepG2 differed significantly from them both. This result might be associated with the different origins of these cells. HepG2 was derived from neonatal liver stem cells obtained from a liver biopsy sample of hepatoblastoma [47], which may have more differential potential, whereas Hep3B and Huh7 were isolated from more differentiated hepatocellular carcinoma cells [48–50].
In conclusion, using the two new quantitative proteomics strategies SWATH and MRM-HR, we compared proteome profiles among the microsomes of human livers and three widely used hepatoma cell lines, i.e. HepG2, Hep3B, and Huh7, with a focus on DMEs. Our results indicate that protein expression profiles, including DMEs, differ significantly between human livers and the hepatic cell lines. Therefore, appropriate caution must be exercised when considering these cell lines as in vitro models for drug metabolism and liver toxicity studies. This study improved our understanding of the similarities and discrepancies between human livers and the commonly used hepatoma cell lines at the protein level. The data can assist in making an appropriate choice of hepatic cell lines for the study of biotransformation and toxicity of drugs metabolized by specific enzymes.
Supplementary Material
Acknowledgements
This work was partially supported by the University of Michigan MCubed program and the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01HL126969, Hao-Jie Zhu].
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
References:
- [1].Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, et al. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J Clin Pharmacol 2008;48:662–70. [DOI] [PubMed] [Google Scholar]
- [2].Lecluyse EL, Alexandre E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol Biol 2010;640:57–82. [DOI] [PubMed] [Google Scholar]
- [3].Lancon A, Hanet N, Jannin B, Delmas D, Heydel JM, Lizard G, et al. Resveratrol in human hepatoma HepG2 cells: metabolism and inducibility of detoxifying enzymes. Drug Metab Dispos 2007;35:699–703. [DOI] [PubMed] [Google Scholar]
- [4].Knasmuller S, Mersch-Sundermann V, Kevekordes S, Darroudi F, Huber WW, Hoelzl C, et al. Use of human-derived liver cell lines for the detection of environmental and dietary genotoxicants; current state of knowledge. Toxicology 2004;198:315–28. [DOI] [PubMed] [Google Scholar]
- [5].Manov I, Hirsh M, Iancu TC. Acetaminophen hepatotoxicity and mechanisms of its protection by N-acetylcysteine: a study of Hep3B cells. Exp Toxicol Pathol 2002;53:489–500. [DOI] [PubMed] [Google Scholar]
- [6].Meex SJ, Andreo U, Sparks JD, Fisher EA. Huh-7 or HepG2 cells: which is the better model for studying human apolipoprotein-B100 assembly and secretion? J Lipid Res 2011;52:152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao H, Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J Biol Chem 2008;283:849–54. [DOI] [PubMed] [Google Scholar]
- [8].Jin CY, Park C, Kim GY, Lee SJ, Kim WJ, Choi YH. Genistein enhances TRAIL-induced apoptosis through inhibition of p38 MAPK signaling in human hepatocellular carcinoma Hep3B cells. Chem Biol Interact 2009;180:143–50. [DOI] [PubMed] [Google Scholar]
- [9].Ji L, Liu T, Liu J, Chen Y, Wang Z. Andrographolide inhibits human hepatoma-derived Hep3B cell growth through the activation of c-Jun N-terminal kinase. Planta Med 2007;73:1397–401. [DOI] [PubMed] [Google Scholar]
- [10].Hsu CM, Hsu YA, Tsai Y, Shieh FK, Huang SH, Wan L, et al. Emodin inhibits the growth of hepatoma cells: finding the common anti-cancer pathway using Huh7, Hep3B, and HepG2 cells. Biochem Biophys Res Commun 2010;392:473–8. [DOI] [PubMed] [Google Scholar]
- [11].Thongtan T, Panyim S, Smith DR. Apoptosis in dengue virus infected liver cell lines HepG2 and Hep3B. J Med Virol 2004;72:436–44. [DOI] [PubMed] [Google Scholar]
- [12].Bartenschlager R, Pietschmann T. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc Natl Acad Sci U S A 2005;102:9739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo L, Dial S, Shi L, Branham W, Liu J, Fang JL, et al. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 2011;39:528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Olsavsky KM, Page JL, Johnson MC, Zarbl H, Strom SC, Omiecinski CJ. Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol Appl Pharmacol 2007;222:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Westerink WM, Schoonen WG. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro 2007;21:1581–91. [DOI] [PubMed] [Google Scholar]
- [16].Sison-Young RL, Mitsa D, Jenkins RE, Mottram D, Alexandre E, Richert L, et al. Comparative Proteomic Characterization of 4 Human Liver-Derived Single Cell Culture Models Reveals Significant Variation in the Capacity for Drug Disposition, Bioactivation, and Detoxication. Toxicol Sci 2015;147:412–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11:O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schilling B, MacLean B, Held JM, Sahu AK, Rardin MJ, Sorensen DJ, et al. Multiplexed, Scheduled, High-Resolution Parallel Reaction Monitoring on a Full Scan QqTOF Instrument with Integrated Data-Dependent and Targeted Mass Spectrometric Workflows. Anal Chem 2015;87:10222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics 2012;11:1475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Glatter T, Ludwig C, Ahrne E, Aebersold R, Heck AJ, Schmidt A. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J Proteome Res 2012;11:5145–56. [DOI] [PubMed] [Google Scholar]
- [21].Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, Cheng LY, Messner S, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics 2015;14:1400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic acids research 2016;44:D447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sakamoto A, Matsumaru T, Ishiguro N, Schaefer O, Ohtsuki S, Inoue T, et al. Reliability and robustness of simultaneous absolute quantification of drug transporters, cytochrome P450 enzymes, and Udp-glucuronosyltransferases in human liver tissue by multiplexed MRM/selected reaction monitoring mode tandem mass spectrometry with nano-liquid chromatography. Journal of pharmaceutical sciences 2011;100:4037–43. [DOI] [PubMed] [Google Scholar]
- [25].Shawahna R, Uchida Y, Decleves X, Ohtsuki S, Yousif S, Dauchy S, et al. Transcriptomic and quantitative proteomic analysis of transporters and drug metabolizing enzymes in freshly isolated human brain microvessels. Molecular pharmaceutics 2011;8:1332–41. [DOI] [PubMed] [Google Scholar]
- [26].Penner N, Woodward C, Prakash C. Appendix: Drug Metabolizing Enzymes and Biotransformation Reactions. 1st ed. Hoboken, NJ: John Wiley & Sons, Inc; 2012. [Google Scholar]
- [27].Hart SN, Li Y, Nakamoto K, Subileau EA, Steen D, Zhong XB. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos 2010;38:988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, et al. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell biology and toxicology 2012;28:69–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jennen DG, Magkoufopoulou C, Ketelslegers HB, van Herwijnen MH, Kleinjans JC, van Delft JH. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol Sci 2010;115:66–79. [DOI] [PubMed] [Google Scholar]
- [30].Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science 2015;347:664–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature 2011;473:337–42. [DOI] [PubMed] [Google Scholar]
- [32].Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, et al. Genetic basis of proteome variation in yeast. Nat Genet 2007;39:1369–75. [DOI] [PubMed] [Google Scholar]
- [33].Slany A, Haudek VJ, Zwickl H, Gundacker NC, Grusch M, Weiss TS, et al. Cell characterization by proteome profiling applied to primary hepatocytes and hepatocyte cell lines Hep-G2 and Hep-3B. J Proteome Res 2010;9:6–21. [DOI] [PubMed] [Google Scholar]
- [34].Megger DA, Pott LL, Ahrens M, Padden J, Bracht T, Kuhlmann K, et al. Comparison of label-free and label-based strategies for proteome analysis of hepatoma cell lines. Biochim Biophys Acta 2014;1844:967–76. [DOI] [PubMed] [Google Scholar]
- [35].Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 2011;80:273–99. [DOI] [PubMed] [Google Scholar]
- [36].Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11:O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fabre B, Korona D, Groen A, Vowinckel J, Gatto L, Deery MJ, et al. Analysis of Drosophila melanogaster proteome dynamics during embryonic development by a combination of label-free proteomics approaches. Proteomics 2016;16:2068–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nakamura K, Hirayama-Kurogi M, Ito S, Kuno T, Yoneyama T, Obuchi W, et al. Large-scale multiplex absolute protein quantification of drug-metabolizing enzymes and transporters in human intestine, liver, and kidney microsomes by SWATH-MS: Comparison with MRM/SRM and HR-MRM/PRM. Proteomics 2016;16:2106–17. [DOI] [PubMed] [Google Scholar]
- [39].Collins BC, Gillet LC, Rosenberger G, Rost HL, Vichalkovski A, Gstaiger M, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14–3–3 system. Nat Methods 2013;10:1246–53. [DOI] [PubMed] [Google Scholar]
- [40].Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med 2016;22:976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yoshimi A, Abdel-Wahab O. Molecular Pathways: Understanding and Targeting Mutant Spliceosomal Proteins. Clin Cancer Res 2017;23:336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular systems biology 2008;4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics 2012;11:1709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ni R, Leo MA, Zhao J, Lieber CS. Toxicity of beta-carotene and its exacerbation by acetaldehyde in HepG2 cells. Alcohol and alcoholism 2001;36:281–5. [DOI] [PubMed] [Google Scholar]
- [45].Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Molecular cancer research: MCR 2008;6:1146–53. [DOI] [PubMed] [Google Scholar]
- [46].Hwang-Verslues WW, Sladek FM. HNF4alpha--role in drug metabolism and potential drug target? Curr Opin Pharmacol 2010;10:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980;209:497–9. [DOI] [PubMed] [Google Scholar]
- [48].Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 1982;42:3858–63. [PubMed] [Google Scholar]
- [49].Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 1979;282:615–6. [DOI] [PubMed] [Google Scholar]
- [50].Sell S, Leffert HL. Liver cancer stem cells. J Clin Oncol 2008;26:2800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All LC-MS/MS data have been deposited to the ProteomeXchange Consortium via the PRIDE [23] partner repository with the dataset identifier PXD008190.