Abstract
As a contribution to the celebration of the year 2014, declared by the United Nations to be “The International Year of Crystallography”, the FEBS Journal is dedicating this issue to papers showcasing the intimate union between macromolecular crystallography and structural biology, both in historical perspective and in current research. Instead of a formal editorial piece, by way of introduction, this review discusses the most important, often iconic, achievements of crystallographers that led to major advances in our understanding of the structure and function of biological macromolecules. We identified at least 42 scientists who received Nobel Prizes in Physics, Chemistry, or Medicine for their contributions that included the use of X-rays or neutrons and crystallography, including 24 who made seminal discoveries in macromolecular sciences. Our spotlight is mostly, but not only, on the recipients of this most prestigious scientific honor - presented in roughly chronological order. As a summary of the review, we attempt to construct a genealogy tree of the principal lineages of protein crystallography, leading from the founding members to the present generation.
Early days of crystallography
Humans have been fascinated by crystals for millennia, but the understanding of their nature and utilization of their properties for endeavors other than creating expensive jewelry had to wait until the 20th century. Two dates have to be particularly kept in mind. Although Wilhelm Conrad Röntgen (1845–1923) discovered X-rays in 1895 in Germany (published for the English-speaking audience a year later [1,2]), another 17 years had to pass before Max von Laue (1879–1960), suspecting that the wavelength of X-rays might be comparable with the interatomic distances, shone them, with the help of two assistants, on a blue crystal of copper sulfate pentahydrate (CuSO4·5H2O) [3]. While Laue was able to provide a physical explanation of the observed diffraction images, the work of the father-and-son team of Sir William Henry Bragg (1862–1942) and Sir William Lawrence Bragg (1890–1971) in England was crucial to the introduction of diffraction as a tool for crystal structure investigation. It was the younger Bragg who soon developed an elegant mathematical explanation of the images generated by Laue, in the form of the famous Bragg’s Law, nλ = 2dsinθ, describing the relation between the angles of diffraction θ, wavelength of the X-rays λ, and interplanar spacings d in the crystal lattice [4]. The early papers of the Braggs have withstood the test of time and their interpretation is still used more than a century later [5–8]. W. H. Bragg went on to construct the first X-ray spectrometer [6] and, of course, one of the first crystal structures determined by the Braggs (next to rock salt) was that of diamond, the perennial favorite crystal of the wealthier part of the human race [9]. The monumental importance of the discoveries of Laue and the Braggs was immediately recognized, leading to the award of the Nobel Prize in Physics to Laue in 1914, and to both Braggs in 1915. Incidentally, W. L. Bragg was, at the age of 25, the youngest ever recipient of the Nobel Prize, a feat that is unlikely to be overshadowed any time soon.
The Nobel Prizes awarded to Laue and the Braggs open a long list of this (Table 1) and other major honors given to crystallographers during the last hundred years. In this review we will primarily concentrate on the achievements of the Nobel Prize winners, with less emphasis on other important accomplishments, especially the more recent ones. It is clear that many more results of macromolecular crystallographers deserve mention, but this could not be done in a brief review. The subject of the history of crystallography, including macromolecular crystallography, has been covered in a recent book by Authier [10] which we strongly recommend to those interested in learning more details of this fascinating field.
Table 1.
Nobel Prizes related to crystallography with prize motivations as provided by the Nobel Committee. The recipients of prizes related to macromolecular crystallography are shown in bold. Nationalities are listed as shown on the Nobel Foundation web page, indicating the country where the award-winning work was primarily done.
| Wilhelm Conrad Röntgen | 1901 | Physics | Germany | In recognition of the extraordinary services he has rendered by the discovery of the remarkable rays subsequently named after him |
| Max von Laue | 1914 | Physics | Germany | For discovery of the diffraction of X-rays by crystals |
| William Henry Bragg | 1915 | Physics | UK | For their services in the analysis of crystal structure by means of X-rays |
| William Lawrence Bragg | 1915 | Physics | UK | |
| Peter Debye | 1936 | Chemistry | Germany | For his contributions to our knowledge of molecular structure through his investigations on dipole moments and on the diffraction of X-rays and electrons in gases |
| Clinton Joseph Davisson George Paget Thomson |
1937 | Physics | USA | For their experimental discovery of the diffraction of electrons by crystals |
| UK | ||||
| James Batcheller Sumner | 1946 | Chemistry | USA | For his discovery that enzymes can be crystallized |
| John Howard Northrop | 1946 | Chemistry | USA | For their preparation of enzymes and virus proteins in a pure form |
| Wendell Meredith Stanley | 1946 | Chemistry | USA | |
| Linus Pauling | 1954 | Chemistry | USA | For his research into the nature of the chemical bond and its application to the elucidation of the structure of complex substances |
| John Kendrew | 1962 | Chemistry | USA | For their studies of the structures of globular proteins |
| Max Perutz | 1962 | Chemistry | UK | |
| Francis Crick | 1962 | Medicine | UK | For their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material |
| James Watson | 1962 | Medicine | UK | |
| Maurice Wilkins | 1962 | Medicine | UK | |
| Dorothy Hodgkin | 1964 | Chemistry | UK | For her determinations by X-ray techniques of the structures of important biochemical substances |
| William Lipscomb | 1976 | Chemistry | USA | For his studies on the structure of boranes illuminating problems of chemical bonding |
| Aaron Klug | 1982 | Chemistry | UK | For his development of crystallographic electron microscopy and his structural elucidation of biologically important nucleic acid-protein complexes |
| Herbert Hauptman | 1985 | Chemistry | USA | For their outstanding achievements in the development of direct methods for the determination of crystal structures |
| Jerome Karle | 1985 | Chemistry | USA | |
| Johann Deisenhofer | 1988 | Chemistry | Germany | For the determination of the three-dimensional structure of a photosynthetic reaction centre |
| Robert Huber | 1988 | Chemistry | Germany | |
| Hartmut Michel | 1988 | Chemistry | Germany | |
| Pierre-Gilles de Gennes | 1991 | Physics | France | For discovering that methods developed for studying order phenomena in simple systems can be generalized to more complex forms of matter, in particular to liquid crystals and polymers |
| Georges Charpak | 1992 | Physics | France | For his invention and development of particle detectors, in particular the multiwire proportional chamber |
| Bertam Brockhouse | 1994 | Physics | Canada | For the development of neutron spectroscopy |
| Clifford Shull | 1994 | Physics | USA | For the development of the neutron diffraction technique |
| John Walker | 1997 | Chemistry | UK | For the elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP) |
| Roderick MacKinnon | 2003 | Chemistry | USA | For structural and mechanistic studies of ion channels |
| Roger Kornberg | 2006 | Chemistry | USA | For his studies of the molecular basis of eukaryotic transcription |
| Venkatraman Ramakrishnan | 2009 | Chemistry | UK | For studies of the structure and function of the ribosome |
| Thomas Steitz | 2009 | Chemistry | USA | |
| Ada Yonath | 2009 | Chemistry | Israel | |
| Andre Geim | 2010 | Physics | UK | For groundbreaking experiments regarding the two-dimensional material graphene |
| Konstantin Novoselov | 2010 | Physics | UK | |
| Dan Shechtman | 2011 | Chemistry | Israel | For the discovery of quasicrystals |
| Robert Lefkowitz | 2012 | Chemistry | USA | For studies of G-protein-coupled receptors |
| Brian Kobilka | 2012 | Chemistry | USA | |
| Martin Karplus | 2013 | Chemistry | USA | For the development of multiscale models for complex chemical systems |
| Michael Levitt | 2013 | Chemistry | USA | |
| Arieh Warshel | 2013 | Chemistry | USA |
Crystallization of macromolecules
The subject of crystallization of proteins has been very recently discussed in a review in this journal [11], thus it will be covered here only very briefly. It is not really possible to trace the first mention of crystals of macromolecules such as proteins, but the description of the serendipitously obtained “blood crystals” of earthworm hemoglobin can be found in a book published as early as 1840 [12]. Hemoglobin from various sources continued to be the favorite protein for crystallization, and a volume containing 600 microscopic photographs of hemoglobin crystals from about 200 organisms was published by Reichert and Brown in 1909 [13]. However, it took another 50 years of titanic effort before the first three-dimensional structure of the hemoglobin molecule could finally meet the human eye [14]. Crystallization of the first enzyme (urease) was reported by James Sumner (1887–1955) in 1926 [15]. This breakthrough was the basis for the award of the 1946 Nobel Prize in Chemistry that went to Sumner, as well as John Northrop (1891–1987) and Wendell Stanley (1904–1971). That prize was awarded essentially for the crystallization of pure proteins and viruses, the achievements proving that “living molecules” could be crystallized or purified and that they did not require any special “élan vital”.
Recording X-ray diffraction images of macromolecular crystals turned out to be quite challenging, since crystals mounted on glass fibers and exposed to air, as is customary for crystals of small molecules, would very quickly deteriorate, losing their crystallinity and diffraction. The first such pictures of protein crystals taken by J. Desmond Bernal (1901–1971; affectionately called “Sage”) were indeed of poor quality but, together with his student Dorothy Crowfoot (later Hodgkin; 1910–1994), they soon realized that crystals of biological macromolecules must be highly hydrated and that sealing them in capillaries with a drop of their mother liquor would efficiently protect them from desiccation. The first reported diffraction was from a crystals of pepsin [16], grown in the laboratory of Theodor Svedberg in Uppsala by John Philpot, who delivered them to Bernal in Cambridge. Those hexagonal crystals had the unit cell lengths reported as a = 67 Å and c = 154 Å (with an expected error of 5%), the latter one being too long for accurate measurements with the equipment available at that time. Thus the structure of this particular form of pepsin was not determined until 1990 (incidentally, by Hodgkin’s former student, Sir Tom Blundell [17]), long after the structure of the protein in the simpler monoclinic crystal form had been published [18]. It turned out that the real length of the c axis was 290.1 Å, about twice as long as originally reported, making the determination of this structure even more challenging. Despite all the problems, Bernal noted [19] that: “…the [X-ray] pictures yielded by protein crystals were of exceptional perfection. They showed large unit cells with great wealth of reflections […] found even at comparatively high angles corresponding to such low spacings as 2 Å. This indicated that not only were the molecules of the proteins substantially identical in shape and size, but also that they had identical and regular internal structures right down to atomic dimensions.” And his quick mind (The Sage!) immediately worked farther: “…the behaviour of the hydrophobe groups of the protein must be such as to hold it together […] the protein molecule in solution must have its hydrophobe groups out of contact with water, that is, in contact with each other […] In this way a force of association is provided which is not so much that of attraction between hydrophobe groups, which is always weak, but that of repulsion of the groups out of the water medium.” His intuitive understanding of the hydrophobic effect can be contrasted with the unfortunate term “hydrophobic force” that is used even today.
The work on urease has an even longer and quite interesting history. When Sumner undertook its crystallization in 1919, he intended to demonstrate by this method that enzymes were proteins. For six years his efforts failed, yielding crystals of concanavalin B, which is an evolutionarily deactivated enzyme (chitinase), a fact that was not known to Sumner. He finally succeeded in crystallizing jack bean urease in 1925 and published the results a year later [15]. The complications with structure determination of jack bean urease were even worse. That particular goal was achieved 85 years later, and the structure of this large (840 residues) metalloenzyme was only published in 2010 [20].
Dorothy Hodgkin, who put bio and crystallography together
Although Dorothy Crowfoot Hodgkin was not the first one to determine the crystal structure of a protein, her contribution to the field of macromolecular crystallography was truly monumental. She initially studied chemistry and then became a coworker of Bernal in Cambridge. Very quickly she acquired excellent mastery of crystallography, buttressed by first class knowledge of chemistry. She worked with Bernal on recording the first protein diffraction images of pepsin crystals [16], and independently, already in Oxford, on obtaining diffraction photographs of insulin [21]. Insulin became her life-long interest, crowned eventually, after almost 35 years of effort, by solving the structure of this important protein hormone [22]. Although the molecule of insulin is not particularly large, solving the structure was complicated by the presence of two molecules in the asymmetric unit in space group R3. This space group lacks centric reflections, which were critical for solving the first crystal structures of hemoglobin and myoglobin (see below). Hodgkin continued structural studies of insulin until the end of her active scientific career, publishing what is most likely the longest paper in the history of protein crystallography, taking up a whole issue of Phil. Trans. R. Soc. Lond. B [23]. The co-authors of this monumental work, listed in alphabetical order and all trained by Hodgkin, include such well-known structural biologists as Ted Baker, Tom Blundell, Eleanor and Guy Dodson, and Mamannamana Vijayan, among others. Guy Dodson (1937–2012), in particular, continued the studies of insulin for many more years, participating in the work that culminated in a key paper describing the structure of its complex with the insulin receptor [24].
Even before her success with insulin, Dorothy Hodgkin was practically a biomacromolecular crystallographer, since the structures successfully solved by her were not only difficult and large for that period (1930–1960), but were also extremely important from the chemical and biological points of view. Chemists at that time were not sure at all about the correct structure of sterols and several possible formulas were around with four aliphatic rings connected in various ways. The crystal structure of an iodo derivative of cholesterol unambiguously established the correct structure of sterols [25]. The crystal structure of penicillin, determined in the early 1940s, had a similarly enormous impact, surprising some chemists with the unexpected four-membered β-lactam ring. This achievement opened the route for making semisynthetic versions of this antibiotic, but was not published until 1949 because of its military use at the end of World War II [26]. The crown jewel of Dorothy Hodgkin’s work, for which she was awarded the 1964 Nobel Prize in Chemistry, was the structure of vitamin B12, the largest crystal structure solved at that time. It again revealed several unexpected features, such as the corrin ring structure and the covalent bond between cobalt and carbon atoms, making vitamin B12 the first identified organometallic compound. This work involved a pioneering application of the early electronic computers in a long-distance collaboration with Ken Trueblood (1920–1998) in California [27].
Max Perutz, John Kendrew, and the structures of hemoglobin and myoglobin
When young Max Perutz (1914–2002) moved in 1936 from Austria to England, he was encouraged by Bernal to study the structure of proteins by X-ray crystallography. Perutz later wrote: “The story opens in 1936 when I left my hometown, Vienna, for Cambridge, Eng., to seek the Great Sage. […] I asked the Great Sage: “How can I solve the secret of life?” He replied: “The secret of life lies in the structure of proteins, and there is only one way of solving it and that is by X-ray crystallography.”” [28]. A year later Perutz chose determination of the crystal structure of horse hemoglobin as his Ph.D. project and completion of this task took him 22 years. Near the finish line he was outrun by his colleague, Sir John Kendrew (1917–1997), who first determined the structure of the related, but four times smaller, myoglobin [29,30]. These extraordinary achievements brought a great joy to their boss, W. L. Bragg, whose steadfast support and encouragement were crucial for the final success.
However, it was Perutz who pioneered the methodology of protein crystallography, especially the method of isomorphous replacement (see below) for the solution of the phase problem [31]. The structure of hemoglobin followed myoglobin very closely, although it was initially published at low resolution [14]. The crystal structures of many variants and chemical states allowed Perutz, among other things, to understand the allosteric effect of hemoglobin, thanks to which, after the first molecule of oxygen has bound, the additional molecules bind with increased affinity. The structure of hemoglobin immediately explained the molecular basis of sickle cell anemia, a disease resulting from a single residue mutation leading to the formation of fibrous polymers.
In his later recollections, Kendrew said “When my 6 Å model of myoglobin came out one of the first people to see it was Desmond Bernal, one of the gurus of molecular biology and a man so wise that everyone called him Sage; when he saw it he said ‘I always knew proteins would look like that’. What did he mean and how did he know?” [32]. On the other hand, the first view of the three-dimensional structure of proteins was a bit of a shock to the discoverers. Perutz recalls [33] his first impression of the clay model of myoglobin: “Could the search for ultimate truth really have revealed so hideous and visceral-looking an object? Was the nugget of gold a lump of lead? Fortunately, like many other things in nature, myoglobin gains in beauty the closer you look at it.” That to appreciate the beauty, and logic, of macromolecules requires intense looking at (i.e. thinking of) them, could not be more true also today.
Perutz was fascinated and worked with hemoglobin for the rest of his life. Kendrew continued a distinguished career as an animator and organizer of science in Britain and Europe, and was the founding father of the European Molecular Biology Organization (EMBO) and Laboratory (EMBL). He was also the founding editor of the Journal of Molecular Biology at a time when the terms “molecular biology” and “structural biology” were almost synonymous. For their work on the crystal structures of proteins, Perutz and Kendrew shared the Nobel Prize in Chemistry in 1962.
The role of Linus Pauling and the beginnings on the American continent
Linus Pauling (1901–1994), one of the most influential chemists of all time, prided himself on being a crystallographer [34]. He started his scientific career by determining the structure of molybdenite MoS2 for his Ph.D. at CalTech in 1923 [35]. Although he never solved a macromolecular crystal structure, he is quite appropriately credited with the discovery of the fundamental structural motifs of proteins [36], several years before they were found in actual protein crystal structures. He deduced the structure of the α-helix from the geometry of the chemical bonds (his favorite subject, that won him the 1954 Nobel Prize in Chemistry), including the planarity of the peptide group [37], from his intuitive faith in the role of hydrogen bonds, and from the logical assumption that regular structure should arise from repetition of stable motifs. Pauling published the structure of the α-helix together with Robert Corey in 1951 [38] even though the model was in slight disagreement with diffraction data on α-keratin (which is a fibrous, rather than globular protein). Those X-ray diffraction data, taken by William Astbury (1898–1961) in Leeds, showed a prominent meridional reflection at 5.1 Å [39], while Pauling’s model predicted - quite correctly - that the pitch of undeformed α-helix (so named after α-keratin) would be 5.4 Å [40]. Astbury’s skillful X-ray diffraction experiments showed that on stretching (e.g. undear steam), α-keratin (e.g. from wool) would change its conformation (and the diffraction pattern accordingly) to a new form, which he called β-keratin. In the same year of 1951, Pauling and Corey also proposed the β-sheet structure, composed of extended protein chains [41]. Pauling was less lucky with DNA; with the backbone inside and the bases out, his model was a salient failure [42]. Although sometimes controversial, Pauling imprinted a great mark not only on science, but also on other subjects. He should be always remembered as an untiring advocate of peace and the recipient of the 1962 Nobel Peace Prize.
Several students of Pauling have become famous scientists in their own right. Although William Lipscomb (1919–2011), always referred to as “Colonel” (of the Kentucky branch) received his 1976 Nobel Prize in Chemistry for his work on boranes, he was also very active in macromolecular crystallography. The first protein structure solved in his laboratory was that of carboxypeptidase A, initially reported at a rather low resolution of 6 Å [43], soon thereafter extended to 2.0 Å [44]. The structure of aspartate carbamoyltransferase, a very large enzyme consisting of twelve molecules, was a tour-de-force for its times [45]. One of Lipscomb’s students working on these structures was Tom Steitz, who later won his own Nobel Prize (see below).
Martin Karplus, another student of Pauling and a winner of the 2013 Nobel Prize in Chemistry, is not an experimental crystallographer, yet his introduction of the computational methods combining molecular mechanics with quantum chemistry provided a major tool for the interpretation of macromolecular structures. Interestingly, one of his early publications provided a structure-function analysis of hemoglobin [46]. His postdoctoral associate, Arieh Warshel, was co-recipient of the Nobel Prize together with his former mentor.
David Harker (1906–1991) was a student of Pauling who not only became one of the premier developers of crystallographic methodology, but who also established (as early as in 1950) one of the first groups working in the USA on protein crystallography. His efforts to determine the structure of RNase A took a decade and a half to succeed [47].
Development of methods for macromolecular crystallography
Solving the first protein crystal structures was possible only in close connection with development of macromolecular crystallographic methodology. Of course, nothing would be achieved without diffracting crystals, thus the importance of ways of obtaining them, explored by Sumner and Northrop, and of keeping them wet, introduced by Bernal and Hodgkin (see above). The methods of collecting diffraction data were at first the same as for small structures and used photographic films, which required many crystals and enormous amounts of patience. The introduction of the screenless rotation method and oscillation camera [48], developed by Uli Arndt (1924–2006) and Alan Wonacott especially for macromolecular crystallography, was a welcome improvement, but data collection still required extensive effort and time spent with photographic films in darkrooms, and then measuring the reflection intensities with optical scanners. The real breakthrough was the introduction of computer-controlled automatic devices, first based on a wire chamber detector (Georges Charpak (1924–2010), the developer of one such device used in protein crystallography, received the 1992 Nobel Prize in Physics), to be followed by image plates, charge-coupled devices (CCDs), and today - by active pixel detectors.
The progress with detectors was even surpassed by the enormous advances in the technology of X-ray generation. The early sealed tubes were superseded by rotating anode generators, but the true leap was the introduction of synchrotron radiation [49]. From the modest start in the 1970s [50–52], when crystallographers were treated as nuisance parasites by the physicists - the owners of the machines, to storage rings and to X-ray free electron lasers (FEL) [53] dedicated to the production of radiation, the intensity and quality of the X-ray beams provided by synchrotron facilities has increased by many orders of magnitude. Collecting a full data set used to take months on rotating anode generators, hours at early synchrotron beam lines, but currently may take a few seconds at third-generation synchrotrons.
The powerful X-ray beams generated by modern sources are capable of severely damaging the crystals during data collection. This process can be slowed down if the crystals are kept at very low temperature, usually by cooling them in a stream of cold nitrogen (~100 K). Such croyogenic methods were popularized in late-1980s, mainly by Håkon Hope [54], and are now routinely used in almost all experiments. On the other hand, nothing can prevent immediate destruction of crystals exposed to the FEL sources, but these microcrystals still provide useful diffraction data during the last femtoseconds of their existence [55].
The technology of diffraction data acquisition has evolved a lot, but the methods of structure solution and refinement have also improved dramatically. At first it was not clear at all how to attack this problem, since even very small crystal structures were solved by a trial-and-error approach, which is unthinkable e.g. for hemoglobin with ~5000 non-hydrogen atoms in the tetrameric molecule. The possibility opened up with the introduction of vector space interpretation by Arthur Lindo Patterson (1902–1966) [56] and the heavy-atom isomorphous replacement method, first applied to alums [57] and phthalocyanins [58]. Whereas the use of isomorphous replacement for proteins had been postulated by J. Monteath Robertson and Bernal as early as 1939 [59], the lack of proper understanding of the effect slowed down the progress of protein crystallography for a number of years. Indeed, Perutz wondered later “Why then did I wait until 1953 before trying isomorphous replacement on haemoglobin? Robertson’s and Bernal’s suggestions were just hunches which I did not take seriously, because it seemed unlikely to me that the scattering contribution from one mercury atom could alter measurably the combined contributions from 2500 atoms of carbon, nitrogen and oxygen in the asymmetric unit of haemoglobin.” [59]. Fortunately, Perutz being an experimentalist ultimately tested the method even if he did not fully believe in it, and realized that a few tens of additional electrons in heavy atoms such as mercury, platinum, or gold can have a measurable effect on reflection intensities and therefore may lead to macromolecular structure determination. Thus he was ultimately able to find phases for many centric reflections of hemoglobin crystals [31], and later for acentric reflections as well. This major conceptual breakthrough allowed Kendrew to solve the structure of myoglobin [29] and Perutz to complete his investigation of hemoglobin [14] (see above).
The heavy-atom methods evolved significantly since the times of Perutz. The advancement of data collection technology allows very accurate measurement of reflection intensities and nowadays not only the isomorphous signal but also the much weaker anomalous signal of not necessarily very heavy atoms (such as selenium or even sulfur) is used for phasing novel macromolecular crystal structures. The heavy-atom method evolved into several variants, referred to by various acronyms, such as Multiple- or Single-Isomorphous Replacement (MIR/SIR) [60] with additional use of Anomalous Scattering (MIRAS and SIRAS) and, if only the anomalous signal is utilized, Multi- or Single-wavelength Anomalous Diffraction (MAD or SAD). Although the usefulness of the anomalous signal for phasing was noted quite early [61–63], the practical application of anomalous scattering as the sole source of phase information in macromolecular crystallography was largely due to the efforts of Wayne Hendrickson. The first successful application of the SAD approach to proteins (based on the minute anomalous signal of sulfur) led him to the solution of the structure of crambin [64]. Two other groups of investigators – Roger Fourme (1942–2012) and his colleagues [65], as well as Mitchell Guss and his collaborators [66] solved protein crystal structures using the MAD approach. This technique was further refined and popularized by Hendrickson [67]. Of particular importance was showing the effectiveness of replacing methionine by selenomethionine, introduced to proteins by genetic engineering [68]. Recently Hendrickson and colleagues introduced a single-wavelength multi-crystal approach [69].
At first, the heavy atoms had to be located by interpretation of Patterson maps. However, in 1940s and 1950s, there was a growing awareness that the phase problem could also be attacked in a direct way. As a logical argument one could consider that completely random sets of phases would most likely produce an absurd “electron-density” map. Conversely, a sensible electron-density map should be everywhere non-negative and in fact should be concentrated around atomic cores. Realization of these basic truths led David Sayre (1924–2012) to the derivation of a relation between reflection phases [70]. Developed as a mathematical theory, the so-called Direct Methods [71] earned Jerome Karle (1918–2013) and Herbert Hauptman (1917–2011) the Nobel Prize in Chemistry in 1985. Part of it is the tangent formula [72], which allows estimation of unknown phases from those that are already known. The theory of Direct Methods is based on structure factor probability distributions, which are inversely related to the number of atoms. This is why Direct Methods are very effective for small-molecule structures but fail with large macromolecules. However, they can be still successfully applied to smaller (~1000 non-H atoms) macromolecular structures, provided atomic resolution (1.2 Å) data are available [73], or indeed even at lower resolution when looking for only a subset of a few special (e.g. heavy) atoms, as implemented, for example, in George Sheldrick’s SHELX system of programs for structure solution and refinement serving both small-molecule and macromolecular crystallography [74]. Direct Methods can be applied to such problems in their classic form, or more frequently using the so-called dual-space recycling, also known as Shake-and-Bake [75].
Practically all novel crystal structures, that is those not expected to be similar to any known atomic model, must be solved by some variant of the “special atom” method. However, currently the PDB contains close to 100,000 models of macromolecules, thus often a similar structural analogue is available and can be used as a search model in the method of Molecular Replacement (MR). The “Faltmolekülmethode” suggested early on by Walter Hoppe (1917–1986) did not acquire popularity, perhaps because it was published too early for its time and only as an abstract in German language [76]. The practical use of MR was pioneered by Michael Rossmann and David Blow (1931–2004) [77–79]. This approach has also evolved significantly and presently a majority of protein crystal structures are solved by several powerful MR programs. A recent algorithm, implemented in Rosetta [80], is also capable of optimizing the search probe by modeling. Several programs are able to automatically screen the PDB contents for the most plausible models and try a large number of them in succession.
At the beginning, the protein models could not be refined at all, since in the early 1960s there were no computers capable of such a task. The first, rather simple “refinement” method introduced by Robert Diamond optimized the fitting of a protein model to an electron-density map and was tested on lysozyme [81,82]. The first automatic least-squares refinement of a protein was performed on the structure of rubredoxin by the group of Lyle Jensen (1915–2008) [83,84]. Because of the size of macromolecular structures the refinement had to use approximations (e.g. diagonal matrix) and was interspersed with regularization of the model geometry. A significant improvement was achieved by the introduction of constrained and restrained refinement [85,86]. The contemporary refinement programs support additional features, such as treatment of rigid-body motion or crystal twinning [87,88]. Currently, most of the phasing and refinement algorithms utilize sophisticated probabilistic approaches based on maximum likelihood, advocated and pioneered by Gerard Bricogne [89].
An important part in building, refinement, and validation of macromolecular structures is played by the possibility to display and compare the atomic model with electron-density maps. The maps were initially drawn by hand on glass sheets and stacked at calculated distances apart, making it difficult to build three-dimensional models. The early approach to model-building was to use the so-called Richards box (optical comparator), where maps plotted on plexiglass sheets and stick-wire models were viewed through a semi-transparent mirror [90]. Later the maps were plotted on acetate sheets by computer plotters, framed, stacked, and inspected by eye. A huge breakthrough was the introduction of interactive computer graphics display programs, in particular the Frodo/O programs of Alwyn Jones [91], which later evolved into highly sophisticated systems for displaying, validating, and correcting the atomic models of macromolecules [92].
The accumulation of crystal structures of proteins and nucleic acids led to the development of powerful computational methods for the interpretation of the wealth of structural data generated by crystallography. The pioneers of this approach were Martin Karplus, Michael Levitt, and Arieh Warshel, who combined Newton’s classical physics with the fundamentally different quantum physics into algorithms that allow, for example, to simulate the interactions of drugs with their protein targets. The computer simulations have become very realistic and are today capable of predicting the outcome of traditional experiments. Michael Levitt, in particular, has been directly contributing to crystallographic methodology, first by introducing refinement with energy minimization more than 40 years ago [81] and, more recently, by determining the crystal structure of the eukaryotic chaperonin CCT after analyzing more than 2.5 million possible models [93]. Karplus, Levitt, and Warshel shared the 2013 Nobel Prize in Chemistry.
The advancement of crystallographic algorithms and the ever increasing speed and power of computers have created the possibility to solve macromolecular crystal structures automatically by researchers who need the structural information but do not always have the required knowledge of crystallography. This is a great success of our science, but sometimes may lead to misinterpretation, overinterpretation, or errors [94].
Early structures of enzymes and other important proteins
After Max Perutz opened the way to solving crystal structures of macromolecules, several groups started working on biologically important proteins, including a number of enzymes. The first structure of an enzyme was that of hen egg white lysozyme, solved by the group of Sir David Phillips (later Baron Phillips of Ellesmere; 1924–1999), first at 6 Å resolution [95], later extended to 2 Å [96] and accompanied by complexes with inhibitors [97]. For the first time it was possible to show that enzymes hold their substrates in specific stereochemistry as in a vise and provide appropriate tools for the chemical reaction to proceed with a minimum expense of energy. The presence of two catalytic carboxylates in the active site of lysozyme is characteristic of many glycohydrolases.
A number of protein structures were solved towards the end of 1960s, in several laboratories in England, USA, Germany, the Netherlands, and Sweden. David Blow and his colleagues worked at the MRC Laboratory of Molecular Biology in Cambridge on the structure of chymotrypsin, a serine protease [98]. This was the first enzyme in which the canonical catalytic triad consisting of a serine, a histidine and an aspartate was revealed structurally, and the structure illustrated how the protease hydrolyzes peptide bonds in target proteins [99]. The structure of carboxypeptidase A, already mentioned above, was determined by the Lipscomb’s group at approximately the same time [43,44].
The crystallographic work on bovine pancreatic ribonuclease (RNase A), an enzyme hydrolyzing the 5’-phosphoester bond in RNA, was conducted independently in three laboratories, with the first results published by all of them in 1967. The Buffalo group of David Harker presented their model at 2 Å resolution [47], the Birkbeck group of Harry Carlisle at 5.5 Å [100], and the Yale group of Harold Wyckoff (1926–2003) and Frederick Richards (1925–2009) solved the 3.5 Å structure of RNase S, in which a single peptide bond was cut [101]. These structures, together with other biochemical data, allowed formulation of the two-step enzymatic mechanism of RNases involving the formation of 2’,3’-cyclic phosphate, consecutively hydrolyzed to a terminal 3’-phosphate.
The crystal structure of human erythrocyte carbonic anhydrase C was the subject of investigations by the group of Bror Strandberg in Uppsala [102]. This zinc-containing enzyme catalyzes the conversion of carbon dioxide into carbonate with an extremely fast turnover of 600,000 molecules of CO2 per 1 molecule of enzyme per second.
The crystal structures of two Bacillus proteases were pursued independently by the group of Joseph Kraut in San Diego for subtilisin BPN’ [103] and in the laboratory of Jan Drenth in Groningen for subtilisin Novo [104]. These investigations confirmed that these two enzymes were identical. Papain, a potent cysteine protease from the juice of papaya fruit, was the subject of another crystallographic investigation at Groningen [105]. Its active site contains the Cys-His-Asn triad, similar to the Ser-His-Asp triad of chymotrypsin, confirming an analogous enzymatic mechanism. The structure of bovine pancreatic trypsin inhibitor (BPTI) was investigated in Munich in the laboratory of Robert Huber, initially at the resolution of 2.5 Å [106]. The structure of this small protein, the first one to be later deposited in the PDB at the truly atomic resolution of 1 Å [107], became important in the development of macromolecular NMR and computational methodologies.
Various dehydrogenases were studied in several laboratories. At Purdue, Michael Rossmann and colleagues solved the structure of lactate dehydrogenase [108]. The structures of malate dehydrogenase [109] and horse liver alcohol dehydrogenase [110], solved a little later, confirmed that all these enzymes include nucleotide-binding domains with a characteristic arrangement of α-helices around an open β-sheet, known as the “Rossmann fold”.
Glycogen phosphorylase was, in the 1970s, the largest protein for which detailed structural data became available. The group of Dame Louise Johnson (1940–2012) in Oxford studied the b form of this enzyme [111,112], whereas Robert Fletterick in San Francisco studied the a form [113,114]. Combination of the efforts of both laboratories resulted in a full explanation of the enzymatic activity of this important protein.
At the creation of the Protein Data Bank (PDB) in 1971 [115], there were merely seven protein crystal structures known. In the initial announcement of the operational status of the PDB in 1973 [116], nine sets of atomic coordinates of the following crystal structures were listed: lamprey methemoglobin, cytochrome b5, bovine pancreatic trypsin inhibitor, subtilisin BPN’, chymotrypsin, carboxypeptidase A, lactate dehydrogenase, myoglobin and rubredoxin. Since then the PDB has grown enormously [117], currently containing almost 100,000 structures of macromolecules. However, keeping in mind that only a few of the early structures could be mentioned here, we shall remember the pioneers who paved the way to this success of macromolecular crystallography.
The structure of DNA
Within about one decade, nucleic acids emerged from obscurity and nearly complete ignorance into a prominent structural target. In the early 1950s, several crystallography groups, including at least two in England, were struggling for the Holy Grail. In a Cavendish Laboratory Unit (Cambridge), headed by W. L. Bragg, Francis Crick (1916–2004) and James Watson were working together to build a plausible model of DNA, without much experimental data. In King’s College, London, in a laboratory headed by John T. Randall, Maurice Wilkins (1916–2004) and Rosalind Franklin (1920–1958) were working separately on X-ray diffraction photographs of DNA fibers [118]. The real “queen” of DNA fiber diffraction was Franklin. She could, for example, using very primitive equipment to control the humidity, force the DNA molecules to change conformation from B to A, with a concomitant shrinking of the fiber by ~24%. After her untimely death, Bernal wrote in an obituary that “her photographs are among the most beautiful X-ray photographs of any substance ever taken”. In particular, the iconic “Photograph 51” of sodium salt of B-DNA contained, as we know now, the telltale signature of the DNA structure: (i) a diffraction pattern in the form of a cross, revealing a helical molecule with a diameter (20 Å) related to the angle between the arms; (ii) layers of reflections with a separation indicating that the helical pitch is 34 Å; (iii) a very prominent meridional reflection at layer 10, indicating 10 repeated, largely planar units (modeled as base-paired nucleotides) per turn, with a step of 3.4 Å; (iv) a totally missing layer 4, as a result of two (anti)parallel helical structures with an axial shift creating two gaps (grooves) with 3:5 width ratio. Difficult personalities and mishandling of the situation by the management were the sources of flaming conflicts between Franklin and Wilkins. Wilkins collaborated with Crick and Watson, whereas Franklin worked alone. In contrast to the Cambridge group, she was trying to solve (i.e. calculate) the DNA structure, using inter alia Patterson techniques, and was methodically advancing on her goal [119]. Without her consent, Franklin’s X-ray photographs became known to the competition. For Crick, who was an exquisite crystallographer and had only a year earlier published a paper on Fourier-Transform analysis of helical objects [120], the features of the diffraction pattern immediately set the correct train of thought. When, after a hint from Jerry Donohue (1920–1985), the Cavendish team also corrected their misconception about the chemical structure of the nucleobases (i.e. used the correct keto rather than the incorrect enol tautomers) - all the pieces fell into place and the structure of the DNA double helix was discovered! It was announced in a paper in Nature in 1953 [121], accompanied by papers by Wilkins et al. [122] and Franklin & Gosling [119]. Franklin was not among the Nobel Laureates in Medicine in 1962 (Crick, Watson, and Wilkins), having died four years earlier of cancer, at the age of only 37.
It would be difficult to find a more pointed example illustrating how a molecular structure explains function. Admiring the elegant double-helical DNA with a constant sugar-phosphate backbone and a variable sequence of uniquely paired A-T and G-C bases, even a layman almost intuitively feels how such a molecule can pass its sequence to daughter molecules. The Watson-Crick base pairing between the DNA strands also explained the mystery of Chargaff’s observation [123] that in any DNA the amounts of A and T are always the same, as are the amounts of G and C, without any other correlations. The discovery of the structure of the double helix of DNA is among the greats achievements of mankind, comparable to the discovery of evolution by Darwin or of relativity by Einstein.
The time interval between the proposal of the structure of DNA and its verification at atomic detail was quite long, leading Richard Dickerson to comment that “DNA is probably the most discussed and least observed of all biological macromolecules.” [124]. However, in the late 1970s [125] the structures of the right-handed double helices of B- and A-DNA were confirmed with much more precise data derived from single-crystal diffraction. The champion of those studies was Dickerson, who first published the crystal structure of B-DNA [124] and A-DNA [126], the latter one close in time with a report by Olga Kennard [127]. The base-pairing geometry is the same in all forms of DNA double helices and, in fact, all the possible Watson-Crick base pairs (A-T, T-A, G-C, C-G) have exactly the same connection with the sugar-phosphate backbone: 10.8 Å distance between the C1′ points of attachment and the same angle (51.5°) of any glycosidic N-C1′ bond with the C1′---C1′ line. This allows any nucleotide sequence whatsoever to be inserted in the standard framework of the sugar-phosphate backbone. The difference in the geometry of B- and A-DNA lies in sugar pucker (2′-endo and 3′-endo, respectively), the width of the double helix and its grooves and the tilt of bases, whereas the base pairs are alike..
Also in the late 1970s, the structure of an entirely different, left-handed DNA was discovered in the laboratories of Alexander Rich [128,129], dubbed Z-DNA for the uneven zigzag trace of the alternating purine-pyrimidine polynucleotide chain. In variance with the right-handed forms, Z-DNA exists at high ionic strength [130] and is usually observed for alternating purine-pyrimidine tracts, typically dCdG. The self-complementary d(CGCGCG)2 hexanucleotide Z-DNA duplex is famous for yielding excellent crystals that diffract X-rays to ultimate resolution (0.55 Å), allowing extremely accurate structural studies [131].
The structures of RNA
In variance with the very elegant double-helical form of DNA, biological RNA exists in apparently less regular forms and assumes double-helical conformation more as an exception rather than as a rule. When it does, it can only be A-RNA (with 3′-endo sugar pucker) because of the steric hindrance introduced by the ribose 2′-OH hydroxyl group. The first crystal structures of very short diribonucleotide stumps in A-RNA conformation were reported by Alexander Rich and his colleagues [132,133], but the visualization of a complete A-RNA turn required a longer sequence [134]. It is, therefore, very interesting to note that the first crystallographic studies of a functional polyribonucleotide, the tRNA, were reported several years before the crystal structure determination of synthetic oligoribonucleotides. The tRNA molecule is comprised of seventy-odd nucleotides, many with unusual chemical modifications. The highly structured tRNA molecule indeed contains two stems in double-helical A form. The crystal structure of tRNA was discovered independently and published within a short interval of time by several groups, led by Muttaiya Sundaralingam (1931–2004) [135,136], Alexander Rich [137–139], Sir Aaron Klug [140] and Sung-Hou Kim [141]. The crystal structures revealed a very graceful L-shaped molecule, with the two key sites, the anticodon loop (where a given amino acid is genetically encoded) and the 3′ acceptor arm (where the corresponding amino acid is attached by an ester bond) located as far from each other as is only possible. This discovery produced a bewildering puzzle of how the alphabet of the genetic code is translated into a structural genetic code, where (anti)codon sequences are physically (or rather chemically) coupled with the corresponding amino acids. This riddle was solved about a decade later when the crystal structures of several classes of aminoacyl-tRNA synthetases were determined, revealing how these highly specialized enzymes precisely charge tRNA molecules with the correct amino acids.
The work on RNA structure gained significant momentum with the subsequent crystallographic studies of catalytic RNA molecules, the best known of which is perhaps the “hammerhead” ribozyme [142,143]. Later, with the discovery of the ribosome structure, a massive amount of rRNA structural data became available (see below).
Viruses
Crystallization of viruses was achieved quite early and was, in fact, rewarded with a Nobel Prize in 1946, given to Wendell Stanley, who crystallized the helical tobacco mosaic virus (TMV) in 1935. An icosahedral virus, tomato bushy stunt virus (TBSV) was first crystallized by Bawden and Pirie in 1937 [144]. The first X-ray diffraction patterns of TBSV and TMV crystals were recorded in 1941 [145]. Helical and icosahedral viruses differ in the architecture of the viral capsid (assembled from many copies of protein subunits) that encapsulates the viral nucleic acid (RNA or DNA genome). In the former ones, the subunits are assembled helically into a rod, in the latter ones they form a sphere-like capsule. The first crystal structure of a helical virus (TMV) was determined at 2.8 Å by Aaron Klug [146], corroborating the structure determined at 4.0 Å using fiber diffraction by Kenneth Holmes et al. [147]. Klug was “infected” with the idea of crystallographic studies of TMV by Rosalind Franklin, who was his mentor. X-Ray crystallographic studies of icosahedral viruses were initiated about the same time by Stephen Harrison who solved the structure of TBSV [148], Michael Rossmann (southern-bean mosaic virus [149] and human common cold virus [150]), Lars Liljas (satellite tobacco necrosis virus [151]), James Hogle (polio virus [152]), and David Stuart (foot-and-mouth disease virus [153]), to name only the early structures, all determined at highly respectable resolution (2.5–2.9 Å). Today the PDB stores several hundred virus structures.
However, long before the crystal structures of icosahedral viruses were solved, a theory of icosahedral triangulation of a sphere was proposed by Donald Caspar and Aaron Klug [154]. The icosahedron is the most complex Platonic solid (tetrahedron-cube-octahedron-dodecahedron-icosahedron) with 20 equilateral triangular facets. Caspar and Klug realized that the principle of icosahedral architecture, in which 60 copies (3 on each face) of the same construction element are repeated with exactly the same environment (exact equivalence) can be extended to larger assemblies with icosahedral symmetry if the principle of equivalence is relaxed to quasi-equivalence, where individual subunit have similar but not necessarily identical environment. Unlike the tetrahedron-cube-octahedron trio, the icosahedron has fivefold (5) symmetry that is incompatible with classical crystallographic symmetry (limited to 1, 2, 3, 4 and 6 axes). (However, the aperiodic quasicrystals can accommodate fivefold symmetry; their discovery by Dan Shechtman was awarded in 2011 with Nobel Prize in Chemistry). To cover (tile) the icosahedral surface with asymmetric objects, one needs in general 60T copies, where T is the triangulation number. In their derivation of T, Caspar and Klug utilized the ideas of the famous architect, Buckminster (Bucky) Fuller, who was able to design sphere-like buildings, called geodesic domes. (Fuller is immortalized in the name of fullerene, given to spherical C60 molecules of carbon, the discovery of which was rewarded with Nobel Prize in Chemistry in 1996.) In view of the enormous importance and scientific challenge connected with the atomic structure of viruses, it is quite surprising that the Nobel Committee did not find these achievements deserving of a separate Nobel Prize. Indirectly, the virus work was rewarded in the 1982 Nobel Prize in Chemistry to Aaron Klug, but the citation stressed crystallographic electron microscopy and general nucleic-acid protein complexes.
The Munich group’s work on integral membrane proteins
At the beginning of the 1980s the crystal structures of more than 50 proteins were already available, but they all shared one common property – the proteins were water soluble. However, since about 1/3 of all proteins in a given organism are located in the cell membranes, crystallization and structure determination of such proteins became the next big challenge. The first report of successful growth of three-dimensional crystals of an integral transmembrane protein, Halobacterium halobium rhodopsin, was published by Hartmut Michel and Dieter Oesterhelt in 1980 [155]. The authors established a protocol that has been successfully used for crystallization of many membrane proteins. They utilized detergents, such as octyl glucoside, to solubilize proteins that have a hydrophobic surface (unlike soluble proteins which are covered by hydrophilic moieties on the surface). However, the complete crystal structure of bacteriorhodopsin was a long time coming, and, in the meantime, another transmembrane protein, namely the photosynthetic reaction center (PSRC) from Rhodopseudomonas viridis, was crystallized, also by Michel [156].
The PSRC is a complex molecule containing four subunits that include a four-heme cytochrome, plus a collection of cofactors (such as bacteriochlorophyll and quinone derivatives). PSRC is responsible for the primary charge separation during photosynthesis. With a total molecular weight of ~125 kDa and no internal symmetry, the PSRC structure presented a true challenge. Nevertheless, an electron-density map at 3 Å resolution, with phases obtained through multiple isomorphous replacement using five heavy atom derivatives, was calculated and interpreted within only two years [157]. A fully interpreted structure was published a year later [158] and its resolution was ultimately extended to 2.3 Å [159]. Amazingly, the protein chains were initially fitted even in the absence of complete amino acid sequence information, and the numerous prosthetic groups were modeled as well. The importance of this structure was two-fold – it provided new and important information regarding the mode of action of the photosynthetic reaction centers, but maybe even more importantly, it has shown that it is possible to determine the structures of integral membrane proteins by crystallographic methods. It is thus not surprising that the 1988 Nobel Prize in Chemistry went to Johann Deisenhofer, Hartmut Michel and Robert Huber.
ATP synthesis and energy metabolism
Adenosine triphosphate (ATP) is the most important energy carrier in the cells, thus the knowledge of the mechanism of its production through harnessing the energy of a transmembrane proton gradient is crucial. ATP synthase (also known as ATPase), an enzyme that accomplishes that task, is a membrane-associated protein consisting of both transmembrane and soluble domains. It was the soluble multi-subunit F1 ATPase domain (α3β3γδε) that was the subject of the initial crystallographic studies. Crystal structure of the bovine heart mitochondrial F1-ATPase was determined in 1994 by Jan-Pieter Abrahams, Andrew Leslie, Sir John Walker, and their colleagues at 2.8 Å resolution and was, at that time, the largest asymmetric structure solved at medium-to-high resolution [160]. The molecule of F1-ATPase consists of alternating α and β subunits arranged around the central γ subunit, but its overall structure is highly asymmetric, due to different interactions of the outer subunits with the γ subunit that influence their nucleotide affinities. This asymmetry confirmed the earlier the proposal of a rotational motion of the F1 domain, which behaves as a molecular motor analogous to the bacterial flagellum. The structure of the holoenzyme from Saccharomyces cerevisiae mitochondria, solved already after Walker received the 1997 Nobel Prize in Chemistry, validated the initial hypothesis and provided a very detailed view of this fascinating protein [161].
Roderick MacKinnon and the membrane channels
Transport of ions, such as potassium, across the cell membrane is crucial to maintaining homeostasis, as well as to diverse phenomena, such as electrical signaling in the nervous system. Such transport is accomplished by dedicated integral membrane proteins that are capable of distinguishing, for example, the K+ ion with its ionic radius of 1.33 Å from that of Na+ (0.95 Å), while maintaining a throughput rate of up to 108 ions per second. However, the potassium channel is quite permeable for ions such as Rb+, and these electron-rich ions were used to visualize the ion-binding sites within the channel molecule. The channel consists of four symmetrically-arranged subunits, each containing two transmembrane helices, with a gated pore spanning the membrane in the middle of the complex and a selectivity filter, lined with carbonyl oxygen atoms, present at the wider end of the opening [162]. Although the size of the protein molecules is not large (fewer than 100 amino acids are visible in each of the engineered subunits of the tetrameric channel), determination of the structure was by no means easy. However, once discovered, this elegant model provided explanation of the mechanistic aspects of a crucial cellular process. Structures of other channels, such as the calcium-gated potassium channel [163], voltage-dependent potassium channel [164], and aquaporin [165] followed in quick succession. Roderick MacKinnon was the recipient of the 2003 Nobel Prize in Chemistry for structural and mechanistic studies of ion channels.
Roger Kornberg’s studies of the transcription machinery
During his stay in the Laboratory of Molecular Biology in Cambridge in the 1970s, Roger Kornberg became involved, under the guidance of Aaron Klug, in the development of methods for the preparation of two-dimensional crystals suitable for structural investigation using electron microscopy. He later used such approaches to study the structure of DNA-dependent RNA polymerase, a crucial enzyme responsible for the transcription of the information encoded by genomic DNA into mRNA. The RNA polymerase is a complicated, multi-subunit enzyme, much simpler in prokaryotes than in eukaryotes (although its molecular weight is still ~450,000 Daltons). The initial breakthrough in structural terms involved electron microscopy studies of the E. coli enzyme that consists of only five subunits, α2ββ’ω, with the β and β’ subunits highly homologous to their eukaryotic counterparts. Two-dimensional crystals were grown on layers of positively charged lipids and, when stained with uranyl acetate, they yielded very low resolution (27 Å) maps which were sufficient, however, to indicate visible similarity of the enzyme’s subunits to DNA polymerase I, and to pinpoint the location of the active site [166]. In their further work, Kornberg and his colleagues switched to RNA polymerase II from yeast, a more tractable enzyme than its mammalian counterpart. Nevertheless, this is still a very complicated enzyme consisting of 12 distinct polypeptides with a molecular weight exceeding 500,000 Daltons. Its structure determination at 16 Å resolution was a true tour-de-force, although finer structural details could not yet be resolved [167]. It took another decade until the crystal structure of a 10-subunit variant of the yeast enzyme was determined in two crystal forms, at the resolution of 2.8–3.1 Å [168]. At such resolution it was possible not only to trace the polypeptide chains quite accurately, but also to locate the divalent metal cations in the active site of the enzyme. The structure enabled a better understanding of the multiple steps of the transcription mechanism. In appreciation of these achievements the 2006 Nobel Prize in Chemistry was awarded to Roger Kornberg.
Ribosome, the translation machine
The determination of the structure of the ribosome resulted from a long and exceedingly difficult project that ultimately led to the award of the 2009 Nobel Prize in Chemistry to Ada Yonath, Tom Steitz, and Venkatraman (Venki) Ramakrishnan. The ribosome is a versatile molecular machine that can translate any genetic message in all living cells (and even in a cell-free context) into a protein when provided with the code in the form of mRNA and the necessary chemical ingredients. All ribosomes are assembled from a small and a large subunit. A bacterial ribosome contains more than 50 proteins but its main component is RNA (66% of the total mass of over 2.5 million Daltons) comprised of one and two chains in the small and large subunits, respectively.
Based on the observation that ribosomes can spontaneously crystallize in the cells, microcrystals of ribosomes from hypothermic chick embryos were isolated as early as 1970 [169]. However, it took another decade before single crystals of much better behaving ribosomes from E. coli were reported [170]. The crystals of complete ribosomes were not amenable to crystallographic studies at that time and, therefore, the individual subunits of E. coli and Bacillus stearothermophilus ribosomes were crystallized separately [171]. The introduction of cryocrystallography became very important in enabling data collection [172]. Since the ribosomal particles from eubacteria are not stable in the presence of salt, a crucial step towards the elucidation of the structure of ribosomes was the crystallization of the subunits of an archeal ribosome from Haloarcula marismortui, an organism that lives at the saturated salt concentration environment of the Dead Sea [173]. However, only after appropriate heavy-atom compounds were used to derivatize the ribosome crystals (including complex ions such as (P2W18O62)6-) and the methods of electron-density modification were sufficiently improved, was it possible to determine medium-to-high resolution structures of the ribosomal subunits [174–176].
The most remarkable result of these studies was the realization that the ribosome is in fact a ribozyme (i.e. an RNA enzyme), in which the creation of the peptide bonds (at the rate of even ~20 per second) is catalyzed solely by the RNA component, and not by the ribosomal proteins. Almost concurrently with the Nobel-winning discoveries by Yonath, Steitz and Ramakrishnan, the structure of the intact bacterial ribosome, complete with the tRNA molecules and a piece of mRNA, was determined in the laboratory of Harry Noller, but at a rather low (5.5 Å) resolution [177]. This feat was later repeated at an improved resolution (up to 3.2 Å) for the E. coli ribosome by Jamie Cate [178] and by Ramakrishnan at the spectacular resolution, for the size of this structure, of 2.8 Å [179]. Although the ribosome is a highly conserved universal machine, responsible for the creation of all proteins on Earth for billions of years, the present eukaryotic ribosome is visibly more complex than its bacterial counterpart. For instance, its molecular mass is about 4 million Daltons. Ultimately, it was possible to determine relatively high-resolution structures (up to 3.0 Å) of the eukaryotic ribosome from Saccharomyces cerevisiae [180].
Membrane receptors (GPCR)
A groundbreaking work to elucidate the structural features and function of the cellular membrane-bound G-protein-coupled receptors (GPCRs) was rewarded in 2012 with the Nobel Prize in Chemistry for Robert Lefkowitz and Brian Kobilka. Kobilka has been working since the mid 1980s on the β2-adrenergic receptor (β2AR), sensitive to hormones such as adrenaline, first with Lefkowitz, and later with his own team.
The GPCRs are eukaryotic seven-helix transmembrane proteins that span the cell membrane, with an extracellular fragment that senses specific molecules outside the cell, and an intracellular part that forms a complex with a trimeric G-protein composed of subunits αβγ. In its inactive (and intact) state, the G-protein contains a bound GDP molecule and the C terminus of its subunit α is docked in a cavity created by a mobile cytoplasmic part of the receptor. When a ligand is bound to the extracellular sensory part of a GPCR molecule, it causes a conformational change of the receptor that is transmitted to the cytosolic part and effectively converts the receptor to a guanine-nucleotide exchange factor (GEF). The GEF function of the GPCR then activates the associated G-protein by exchanging its bound GDP to GTP. The G-protein’s subunit α, together with the bound GTP, can then dissociate from the β and γ subunits initiating an intracellular signaling cascade, whose outcome depends on the type of the α subunit.
Crystallization of the GPCRs, which are integral membrane proteins and contain flexible extra- and intra-cellular parts, was a formidable task, and Kobilka (together with Ray Stevens) achieved this goal using ingenious protein tinkering tricks, such as complexation with a nanobody or fusion with a lysozyme domain [181–183]. An even more daunting challenge was the crystal structure determination of a complex, with both the signaling molecule and then also with the G-protein partner. Working patiently and methodically, Kobilka was able to achieve both goals and in fact was able to capture the GPCR complex at the crucial moment of relaying the signal to the G-protein partner [184,185].
The structure of the light-sensing rhodopsin from the eye’s retina, which is another GPCR protein, was solved even earlier, in 2000, by Palczewski et al. [186]. Thanks to all these efforts we now know that the GPCR proteins are very versatile receptors, working like cassettes with different sensing elements and different G-protein partners for different signaling pathways. There are nearly 1000 different GPCR receptors in the human body. Some GPCRs are highly specific, others are multifunctional, i.e. can recognize several signals. Among the ligands that activate GPCR pathways are light-sensitive molecules, odor molecules and pheromones, hormones and neurotransmitters. The GPCR proteins are ideal targets for rational design of drug molecules. Indeed, almost half of all modern drugs in use today target the GPCR receptors.
Macromolecular crystallography and drug design
Even in the early days, after the first crystal structures of proteins had been solved, it was quite clear that their availability might play a very important role in understanding human health and disease at the molecular level. The crystal structure of hemoglobin, for example, established the molecular basis for the hereditary disease sickle cell anemia and led to efforts of developing therapeutically useful agents that would reverse the sickling process [187]. Another direction was an attempt to design compounds that would stabilize deoxyhemoglobin, thus promoting oxygen liberation [188]. A different early target was insulin, which was engineered for more long lasting retention and thus for improved treatment of diabetes [189]. However, although these early efforts relied very much on the availability of structural information, they were not dependent on using the structures directly.
The field truly blossomed in the 1980s, when a number of large pharmaceutical companies became interested, and new startup companies were created with the specific purpose of using structural data (mostly crystallographic, but later also obtained by NMR spectroscopy) for designing drugs that would specifically affect selected protein or nucleic acid targets. One of the first such small companies was Agouron Pharmaceuticals, established in 1984 in La Jolla, California. The story of Agouron Pharmaceuticals, and the associated Agouron Institute, was described in interesting detail by one of its founders, John Abelson [190]. Their first target was thymidylate synthase, selected in an effort to find a better drug than the quite toxic 5-fluorouracil, then a first-line cancer drug. Although this program did result in the discovery of a number of very potent inhibitors of human thymidylate synthase, it did not directly lead to the creation of new drugs, but rather served as a platform for learning how best to use this new methodology. Another company created around the same time was Molecular Discovery Ltd, where Peter Goodford was developing the GRID software [191] for fitting ligands to their targets. That computer program helped in the development of zanamivir, an influenza virus neuraminidase inhibitor originally discovered by Peter Colman, who also established in 1985 a company, Biota Holdings. Some other software tools developed within academia, such as DOCK [192], have also played an important role in promoting progress in this area.
Some of the earliest efforts in drug design had to rely on substitute targets, as structures of the relevant human proteins were not known at that time. Thus, for example, the development of antihypertensive drugs, such as captopril, functioning as inhibitors of the angiotensin-converting enzyme, had to initially rely on models based on the available coordinates of carboxypeptidase A [193]. Similarly, work on another antihypertensive target, human renin, had to rely on the then known structures of fungal aspartic proteases and on computer models of human renin derived from them [194]. The work on renin inhibitors resulted in the creation of a large number of compounds in many pharmaceutical companies, but, until recently, of no drugs. However, the lessons learned were applied directly to the later development of inhibitors of HIV protease, and, finally, to the approval in 2007 by the Food and Drug Administration (FDA) of the renin inhibitor aliskiren for essential (primary) hypertension.
A poster child of the rational drug design in the early 1990s was the aspartic protease encoded by HIV, with almost every major pharmaceutical company, as well as some of the startups such as Agouron or Vertex, and academic institutions, designing inhibitors. These efforts were, to a large extent, based on the unrestricted availability of the structure of this enzyme and its complexes with inhibitors [195–197], and even earlier of a related RSV protease-based model [198,199]. Work on the inhibitors of HIV protease resulted in the FDA approval, by 1997, of four very successful drugs (Roche Pharmaceuticals’ saquinavir, Abbot’s ritonavir, Merck’s indinavir, and Agouron’s nelfinavir). The availability of these drugs, as well as additional protease inhibitors developed later, together with drugs targeting other HIV proteins, allowed the introduction of combination therapy which changed an irrevocably mortal disease to a manageable infection.
Later efforts, such as the introduction of fragment-based drug discovery, primarily championed by another startup company, Astex Therapeutics (now part of Otsuka Pharmaceutical Ltd.), made even more direct use of the crystal structures of the macromolecular targets. In this approach, cocktails of small molecule ligands are soaked into macromolecular crystals and the structures of two or more ligands binding in adjacent sites are the starting point for creating much more potent inhibitors. Structure-based drug design has not yet been much in evidence for drugs that target GPCR receptors, but that may change in the future due to the recent availability of the structures of a variety of GPCRs.
This is, of course, only a very incomplete and selective summary of the early efforts in structure-based drug design. Many reviews cover this area in considerable detail, starting with the early description of the process coming from the Blundell’s laboratory [200], through later papers by Navia [201] or Colman [202], to list just a few.
Crystallography in the era of structural genomics
Whereas progress in the development of macromolecular crystallography in the first three decades following the early protein crystal structure determinations was mainly driven by accomplishments of individual scientists, the situation started changing later. Already in the 1990s, there was evidence of steep progress in several methodological aspects of structural biology, from genome sequencing and annotating, genetic engineering, recombinant protein production in diverse variants, to crystallization and diffraction data acquisition. Rapid progress was also visible in methods development for structure solution by X-ray crystallography and NMR, as well as for theoretical modeling. Automation was being introduced to many stages of these processes. Combined with constant increase of computer power, these advances made it possible to determine large numbers of protein structures on a genome-wide scale, and this possibility has led to the creation of a number of structural genomics (SG) initiatives in America, Europe, and Asia [203–208]. These activities were modeled in part on the Human Genome Project, an earlier large-scale biological initiative that had been expected to benefit in an unprecedented way our ability to combat various human diseases [209,210]. The sequencing of the human genome and of the genomes of many other organisms has created favorable conditions for rational selection of targets for structural genomics.
One of the goals of these initiatives is to identify novel protein folds, to ensure that the structural databases contain representatives of all possible folds as a basis for subsequent homology modeling, functional studies, and identification of targets relevant to the development of new medicines and therapies. Some of the SG centers are more focused on specific aims, concentrating, for example, on targets from infectious microbes, the flora of the human intestine, or membrane proteins. An important aspect of SG is to develop efficient high-throughput methodology for rapid evaluation of protein structures, and this activity has led to very significant advances, considerably benefitting the entire community of structural biologists, not just the SG centers. Practically all existing SG centers are supported by public funds and, therefore, the results they generate, as well as the developed methodologies are freely available to all.
Currently, out of close to 100,000 macromolecular structures in the PDB, nearly 10% (9355 until the end of 2013) resulted from X-ray crystallographic and NMR studies carried out at various SG centers (http://biosync.sbkb.org/stats.do?stats_sec=SG&stats_focus_lvl=GLBL). It may be anticipated that future research based on the protein structures produced by the SG consortia will, in the longer term, bring the expected results in the form of new medical treatments. As of now, the most significant benefits to the community from the SG activities are seen in innovative methodologies for structural biology, in various novel, highly efficient and effective, automated procedures, and in the creation of advanced, new-age research infrastructures.
Outlook: XFELs, diffraction before destruction, femtosecond crystallography at nanoscale, single-particle imaging, and more…
Over the last hundred years, beginning with the discovery of X-ray diffraction by Laue, crystallography has undergone a tremendous advancement, fueling progress in such disciplines as physics, chemistry, materials science and biology. The leap is particularly visible in structural biology, which, starting boldly with merely two similar protein crystal structures at the beginning of the 1960s, has now accumulated almost 100,000 structures in the PDB, ~90% of which were determined by crystallography-related methods. The boom coincided in the 1990s with the rapid development of methods for recombinant protein production and of computing technology, but was mostly sparked by widespread use of very powerful synchrotron X-ray sources. The progress has not stalled there, and in fact continues to be driven by accelerator physics, now offering astronomically bright radiation from X-ray Free Electron Lasers (XFELs). The power of those beams destroys any sample within a fraction of a picosecond. However, pioneering work of Janos Hajdu, John Spence, Henry Chapman and their colleagues demonstrated that in a flash no longer than 100 femtoseconds, a constructive diffraction image can be captured before destruction [211]. As proof of principle, the first protein crystal structures have been already determined that way [53,55,212]. Because of the enormous flux of the X-ray beam, the size of the crystals can be accordingly smaller, and a few tens of a nanometer is enough. This femtosecond nanoscale crystallography can be extended even beyond the constraints of a crystal: simulated experiments have shown that it will be possible to study X-ray scattering from single molecules [213] and X-ray imaging of microscopic biological objects (such as intact cells) has been already demonstrated in practice [214,215]. The future of macromolecular crystallography looks therefore very bright, in both figurative and literal terms. An encouraging perspective for the International Year of Crystallography!
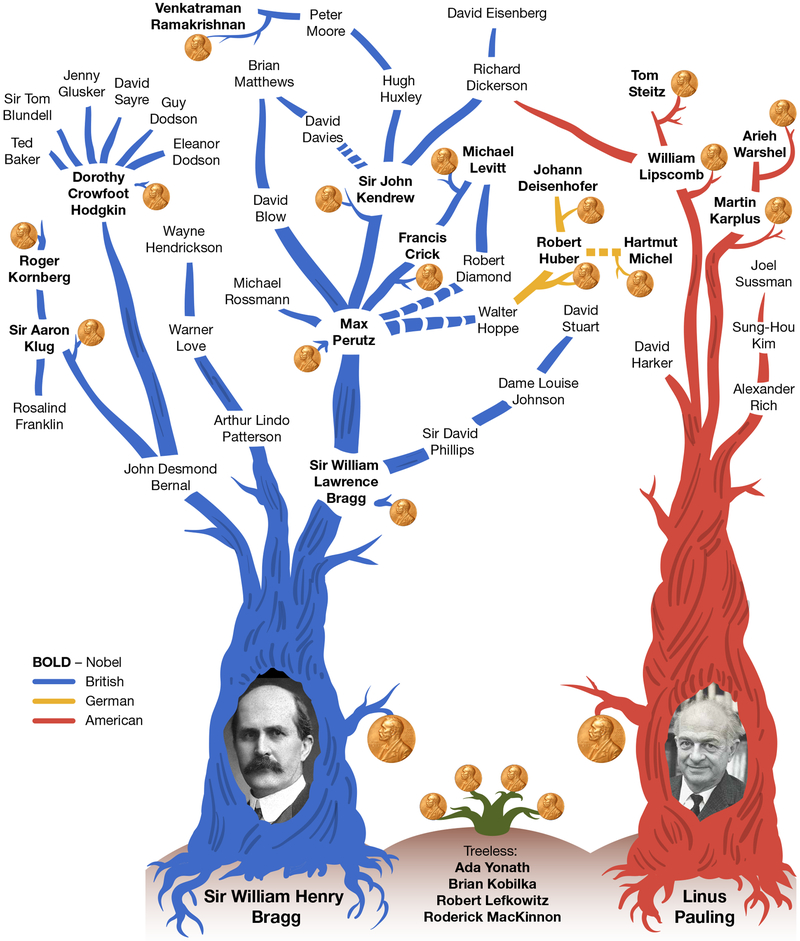
A family tree of macromolecular crystallography
At the beginning of the 20th century, crystallography in Germany was very strongly connected with physics, while it was chemistry-oriented in England. This explains why Max von Laue did not educate any structural crystallographers, and why macromolecular crystallography in Germany was later seeded from the British soil. It also confirms that the discipline of chemistry is naturally more productive in fertilizing structure-oriented innovations, such as those propagated by the pioneers of crystallography. The task and honor of establishing a dynasty fell on Sir William Henry Bragg, even if he himself, being a physicist, had very little connection to biology. Many crystallographers of the previous generation (or even the ones active today, including the authors of this review) can trace their roots more or less directly to him. Thus we present here, with certain caveats, a family tree of macromolecular crystallographers (Fig. 1). The caveats are that the tree, with its two principal trunks, is very much the result of our own bias and the space limitation of a single page. We were not able, therefore, to include all the crystallographers that certainly deserve a place in this genealogy. Also, we consider mentoring in a very broad sense – we mark scientists as descendants if they worked with their mentors in any capacity, not necessarily as students or postdocs only.
Figure 1.
A family tree of macromolecular crystallographers. The tree includes only a subset of the most notable pioneers of macromolecular crystallography, particularly those mentioned in the article. The “British” and “American” trunks are designated only in order to show the roots, but they do not in any way describe the nationality of the scientists (who may be citizens of a variety of countries, or even have as many as four simultaneous citizenships, making it rather difficult to assign them to a particular region of the world). Mentorship (in solid lines) has to be taken with a grain of salt, since it does not necessarily reflect an official relationship. We aimed to show only the simplest connections to the roots – of course, there were many other interactions among these scientists that could not be indicated here, since that would make the chart impossibly complicated. Dashed lines indicate working together, but not necessarily as a mentor and a mentee.
It is very clear that the single most important institution responsible for the development of macromolecular crystallography has been the Laboratory of Molecular Biology (LMB) of the Medical Research Council in Cambridge, UK. That laboratory was established in 1947, upon recommendation of W. L. Bragg, for the specific purpose, as stated on the LMB history web page, “to enable Max Perutz and John Kendrew to develop their work using X-ray diffraction to study proteins.” LMB has been and still is remarkably successful in this task and almost half of the Nobel Prizes related to macromolecular crystallography were awarded to scientists who were either employees or alumni of that institution.
Although W. H. Bragg himself was not directly involved in macromolecular crystallography, three of his associates, his son W. L. Bragg, Desmond Bernal, and Lindo Patterson, were responsible for training many of the most influential scientists who brought the field to its current prominence. Patterson, in particular, although again not a macromolecular crystallographer himself, was one of the first to bring macromolecular crystallography to the United States. Whereas many American crystallographers can trace their roots to the founders of the LMB, a separate trunk of the tree has grown on the American continent. Its founder was Linus Pauling and, by now, it has four Nobel Prizes to its credit (some other American recipients of this honor are not fruits of this particular tree). There is also a connection linking the German branch of the tree to the LMB roots, since Walter Hoppe, the mentor of Robert Huber, spent about two years there working with Max Perutz.
There are, of course, some famous macromolecular crystallographers who learned the trade on their own and are not part of either the UK- or USA-based branches of the tree (or its German offshoot). The names that come to mind are Brian Kobilka, Robert Lefkowitz, Roderick MacKinnon, and Ada Yonath, among others. However, we can be quite sure that the monumental achievements of the founders of the field must have played a role in the development of their brilliant careers, even if indirectly.
Acknowledgements
We would like to thank Hans Deisenhofer, Jenny Glusker, Robert Huber, Michael Levitt, Brian Matthews, Michael Rossmann, and Joel Sussman for their valuable comments and suggestions. We thank Richard Frederickson and Joseph Meyer (Scientific Publications, Graphics & Media, Leidos Biomedical Research, Inc.) for designing Figure 1. Original work in the laboratories of AW and ZD was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
We dedicate this review to David Davies, David Eisenberg, Keith Hodgson, Zofia Kosturkiewicz, Ivar Olovsson, and Michael Woolfson: our teachers and mentors, to whom we owe our own connection with the roots of crystallography.
References
- 1.Röntgen WC (1896) On a new kind of rays. Science 3, 227–231. [DOI] [PubMed] [Google Scholar]
- 2.Röntgen WC (1896) A new form of radiation. Science 3, 726–729. [DOI] [PubMed] [Google Scholar]
- 3.Friedrich W, Knipping P, & Laue M (1912) Interferenz-Erscheinungen bei Röntgenstrahlen. Sitz Bayer Akad Wiss 303–322. [Google Scholar]
- 4.Bragg WL (1913) The diffraction of short electromagnetic waves by a crystal. Proc Cambridge Phil Soc 17, 43–57. [Google Scholar]
- 5.Bragg WH (1912) X-rays and crystals. Nature 90, 219. [Google Scholar]
- 6.Bragg WH & Bragg WL (1913) The reflection of X-rays by crystals. Proc Roy Soc Lond A 88, 428–438. [Google Scholar]
- 7.Bragg WH (1913) The reflection of X-rays by crystals (II). Proc Roy Soc Lond A 89, 246–248. [Google Scholar]
- 8.Bragg WL (1913) The structure of some crystals as indicated by their diffraction of X-rays. Proc R Soc Lond A 89, 248–277. [Google Scholar]
- 9.Bragg WH & Bragg WL (1913) The structure of the diamond. Proc R Soc Lond A 89, 277–291. [Google Scholar]
- 10.Authier A (2013) Early Days of X-ray Crystallography, Oxford University Press, Oxford, UK. [Google Scholar]
- 11.Giege R (2013) A historical perspective on protein crystallization from 1840 to the present day. FEBS J 280, 6456–6497. [DOI] [PubMed] [Google Scholar]
- 12.Hünefeld FL (1840) in Die Chemismus in der tierescher Organisation p. 160, Leipzig. [Google Scholar]
- 13.Reichert ET & Brown AP (1909) The differentiation and specificity of corresponding proteins and their vital substances in relation to biological classification and organic evolution, Carnegie Institution of Washington, Washington DC. [Google Scholar]
- 14.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, & North ACT (1960) Structure of Haemoglobin: A three-dimensional Fourier synthesis at 5.5-Å resolution, obtained by X-ray analysis. Nature 185, 416–421. [DOI] [PubMed] [Google Scholar]
- 15.Sumner JB (1926) The isolation and crystallization of the enzyme urease. J Biol Chem 69, 435–441. [Google Scholar]
- 16.Bernal JD & Crowfoot D (1934) X-ray photographs of crystalline pepsin. Nature 133, 794–795. [Google Scholar]
- 17.Cooper JB, Khan G, Taylor G, Tickle IJ, & Blundell TL (1990) X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 Å resolution. J Mol Biol 214, 199–222. [DOI] [PubMed] [Google Scholar]
- 18.Andreeva NS, Fedorov AA, Gushchina AE, Riskulov RR, Shutskever NE, & Safro MG (1978) [X-ray structural analysis of pepsin. V. Conformation of the main chain of the enzyme]. Mol Biol (Mosk) 12, 922–936. [PubMed] [Google Scholar]
- 19.Bernal JD (1939) Structure of proteins. Nature 143, 663–667. [Google Scholar]
- 20.Balasubramanian A & Ponnuraj K (2010) Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J Mol Biol 400, 274–283. [DOI] [PubMed] [Google Scholar]
- 21.Crowfoot D (1935) X-ray single crystal photographs of insulin. Nature 135, 591–592. [Google Scholar]
- 22.Adams MJ, Blundell TL, Dodson EJ, Dodson GG, Vijayan M, Baker EN, Harding MM, Hodgkin DC, Rimmer B, & Sheat S (1969) Structure of rhombohedral 2 zinc insulin crystals. Nature 224, 491–495. [Google Scholar]
- 23.Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD, et al. (1988) The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Phil Trans R Soc Lond B Biol Sci 319, 369–456. [DOI] [PubMed] [Google Scholar]
- 24.Menting JG, Whittaker J, Margetts MB, Whittaker LJ, Kong GK, Smith BJ, Watson CJ, Zakova L, Kletvikova E, Jiracek J, et al. (2013) How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlisle CH & Crowfoot D (1945) The crystal structure of cholesteryl iodide. Proc R Soc Lond A 184, 64–83. [Google Scholar]
- 26.Crowfoot D, Bun CW, Rogers-Low BW, & Turner-Jones A (1949) in Chemistry of Penicillin pp. 310–367. University Press, Princeton, NJ. [Google Scholar]
- 27.Hodgkin DC, Kamper J, Lindsey J, MacKay M, Pickworth J, Robertson JH, Shoemaker CB, White JG, Prosen RJ, & Trueblood KN (1957) The structure of vitamin B12 I. An outline of the crystallographic investigation of vitamin B12. Proc R Soc Lond A 242, 228–263. [Google Scholar]
- 28.Perutz M (1997) Science is Not a Quiet Life, World Scientific Publishing Co, Singapore. [Google Scholar]
- 29.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, & Phillips DC (1958) A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666. [DOI] [PubMed] [Google Scholar]
- 30.Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, & Shore VC (1960) Structure of myoglobin. A three-dimensional Fourier synthesis at 2 Å resolution. Nature 185, 422–427. [DOI] [PubMed] [Google Scholar]
- 31.Green DW, Ingram VM, & Perutz MF (1954) The structure of haemoglobin. IV. Sign determination by the isomorphous replacement method. Proc R Soc Lond A 225, 287–307. [Google Scholar]
- 32.Kendrew J (1996) Protein crystallography and computing: recollections of the 50’s. Acta Crystallogr A52, C–7. [Google Scholar]
- 33.Perutz MF (1964) The hemoglobin molecule. Sci Am 211, 64–76. [DOI] [PubMed] [Google Scholar]
- 34.Pauling L (1992) in The Chemical Bond Structure and Dynamics pp. 3–16. Academic Press, [Google Scholar]
- 35.Dickinson RG & Pauling L (1923) The crystal structure of molybdenite. J Am Chem Soc 45, 1466–1471. [Google Scholar]
- 36.Pauling L (1993) How my interest in proteins developed. Protein Sci 2, 1060–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edison AS (2001) Linus Pauling and the planar peptide bond. Nature Struct Biol 8, 201–202. [DOI] [PubMed] [Google Scholar]
- 38.Pauling L, Corey RB, & Branson HR (1951) The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci USA 37, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Astbury WT & Street A (1932) X-ray studies of the structure of hair, wool, and related fibres. Phil Trans Roy Soc A 230, 75–101. [Google Scholar]
- 40.Astbury WT & Woods HJ (1934) X-ray studies of the structure of hair, wool, and related fibres. II. The molecular structure and elastic properties of hair keratin. Phil Trans, Roy Soc A 232, 333–394. [Google Scholar]
- 41.Pauling L & Corey RB (1951) The pleated sheet, a new layer configuration of polypeptide chains. Proc Natl Acad Sci U S A 37, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauling L & Corey RB (1953) A proposed structure for the nucleic acids. Proc Natl Acad Sci USA 39, 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipscomb WA, Coppola JC, Hartsuck JA, Ludwig ML, Muirhead H, Searl J, & Steitz TA (1966) The structure of carboxypeptidase A: III. Molecular structure at 6 Å resolution. J Mol Biol 19, 423–441. [Google Scholar]
- 44.Reeke GN, Hartsuck JA, Ludwig ML, Quiocho FA, Steitz TA, & Lipscomb WN (1967) The structure of carboxypeptidase A, VI. Some results at 2.0 Å resolution, and the complex with glycyl-tyrosine at 2.8 Å resolution. Proc Natl Acad Sci USA 58, 2220–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren SG, Edwards BF, Evans DR, Wiley DC, & Lipscomb WN (1973) Aspartate transcarbamoylase from Escherichia coli: electron density at 5.5 A resolution. Proc Natl Acad Sci U S A 70, 1117–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabo A & Karplus M (1972) A mathematical model for structure-function relationships in hemoglobin. Biochem Biophys Res Commun 46, 855–860. [DOI] [PubMed] [Google Scholar]
- 47.Kartha G, Bello J, & Harker D (1967) Tertiary structure of ribonuclease. Nature 213, 862–865. [DOI] [PubMed] [Google Scholar]
- 48.Arndt UW & Wonacott AJ (1977) The rotation method in crystallography: data collection from macromolecular crystals, North Holland Pub. Co., [Google Scholar]
- 49.Dauter Z, Jaskolski M, & Wlodawer A (2010) Impact of synchrotron radiation on macromolecular crystallography: a personal view. J Synchrotron Radiat 17, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbaum G, Holmes KC, & Witz J (1971) Sychrotron radiation as a source for x-ray diffraction. Nature 230, 434–437. [Google Scholar]
- 51.Phillips JC, Wlodawer A, Yevitz MM, & Hodgson KO (1976) Applications of synchrotron radiation to protein crystallography: preliminary results. Proc Natl Acad Sci USA 73, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harmsen A, Leberman R, & Schulz GE (1976) Comparison of protein crystal diffraction patterns and absolute intensities from synchrotron and conventional X-ray sources. J Mol Biol 104, 311–314. [DOI] [PubMed] [Google Scholar]
- 53.Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, et al. (2011) Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hope H (1988) Cryocrystallography of biological macromolecules: a generally applicable method. Acta Crystallogr D44, 22–26. [DOI] [PubMed] [Google Scholar]
- 55.Barends TR, Foucar L, Botha S, Doak RB, Shoeman RL, Nass K, Koglin JE, Williams GJ, Boutet S, Messerschmidt M, et al. (2013) De novo protein crystal structure determination from X-ray free-electron laser data. Nature 505, 244–247. [DOI] [PubMed] [Google Scholar]
- 56.Patterson AL (1934) A Fourier series method for the determination of the components of interatomic distances in crystals. Phys Rev 46, 372–376. [Google Scholar]
- 57.Lipson H & Beevers CA (1935) The crystal structure of the alums. Proc R Soc Lond A 148, 664–680. [Google Scholar]
- 58.Robertson JM (1936) An X-ray study of phtalocyanines. Part II. Quantitative structure determination of the metal-free compound. J Chem Soc 1936, 1195–1209. [Google Scholar]
- 59.Perutz MF (1983) Review of “Structural crystallography in chemistry and biology: benchmark papers in physical chemistry and chemical physics.Vol. 4”. Acta Crystallogr B39, 139–141. [Google Scholar]
- 60.Blow DM & Crick FH (1959) The treatment of errors in the isomorphous replacement method. Acta Crystallogr 12, 794–802. [Google Scholar]
- 61.Ramachandran GN & Raman S (1956) A new method for the structure analysis of non-centrosymmetric crystals. Curr Sci 25, 348–351. [Google Scholar]
- 62.Rossmann MG (1961) The position of anomalous scatterers in protein crystals. Acta Crystallogr 14, 383–388. [Google Scholar]
- 63.Karle J (1980) Some developments in anomalous dispersion for the structural investigation of macromolecular systems in biology. Int J Quant Chem S7, 357–367. [Google Scholar]
- 64.Hendrickson WA & Teeter MM (1981) Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulfur. Nature 290, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kahn R, Fourme R, Bosshard R, Chiadmi M, Risler JL, Dideberg O, & Wery JP (1985) Crystal structure study of Opsanus tau parvalbumin by multiwavelength anomalous diffraction. FEBS Lett 179, 133–137. [DOI] [PubMed] [Google Scholar]
- 66.Guss JM, Merritt EA, Phizackerley RP, Hedman B, Murata M, Hodgson KO, & Freeman HC (1988) Phase determination by multiple-wavelength x-ray diffraction: crystal structure of a basic “blue” copper protein from cucumbers. Science 241, 806–811. [DOI] [PubMed] [Google Scholar]
- 67.Yang W, Hendrickson WA, Crouch RJ, & Satow Y (1990) Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science 249, 1398–1405. [DOI] [PubMed] [Google Scholar]
- 68.Hendrickson WA, Horton JR, & LeMaster DM (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three dimensional structure. EMBO J 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Q, Liu Q, & Hendrickson WA (2013) Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr D69, 1314–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sayre D (1952) The squaring method: a new method for phase determination. Acta Cryst 5, 60–65. [Google Scholar]
- 71.Hauptman H & Karle J. (1953) Solution of the phase problem. ACA Monograph No. 3, Polycrystal Book Service, Pittsburgh, PA. [Google Scholar]
- 72.Germain G & Woolfson MM (1971) The application of phase relationships to complex structures III. The optimum use of phase relationships. Acta Crystallogr A27, 368–376. [Google Scholar]
- 73.Sheldrick GM (1990) Phase annealing in SHELX-90: direct methods for larger structures. Acta Crystallogr A46, 467–473. [Google Scholar]
- 74.Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A64, 112–122. [DOI] [PubMed] [Google Scholar]
- 75.Miller R, Gallo SM, Khalak HG, & Weeks CM (1994) SnB: crystal structure determination via Shake-and-Bake. J Appl Crystallogr 27, 613–621. [Google Scholar]
- 76.Hoppe W (1957) Die ‘Faltmolekülmethode’ - eine neue Methode zur Bestimmung der Kristallstruktur bei ganz oder teilweise bekannter Molekülstruktur. Acta Crystallogr 10, 750–751. [Google Scholar]
- 77.Rossmann MG & Blow DM (1962) The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr 15, 24–31. [Google Scholar]
- 78.Rossmann MG & Blow DM (1963) Determination of phases by the conditions of non-crystallographic symmetry. Acta Crystalogr 16, 39–45. [Google Scholar]
- 79.Rossmann MG (1972) The Molecular Replacement Method, a Collection of Papers on the Use of Non-crystallographic Symmetry, Gordon & Breach, NY. [Google Scholar]
- 80.DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, Valkov E, Alon A, Fass D, Axelrod HL, et al. (2011) Improved molecular replacement by density and energy guided protein structure optimization. Nature 473, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diamond R & Levitt M (1971) A refinement of the structure of lysozyme. Biochem J 125, 92P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diamond R (1971) A real-space refinement procedure for proteins. Acta Crystallogr A27, 436–452. [Google Scholar]
- 83.Watenpaugh KD, Sieker LC, Herriott JR, & Jensen LH (1972) The structure of a non-heme iron protein: rubredoxin at 1.5 Angstrom resolution. Cold Spring Harb Symp Quant Biol 36, 359–367. [DOI] [PubMed] [Google Scholar]
- 84.Watenpaugh KD, Sieker LC, Herriott JR, & Jensen LH (1973) Refinement of the model of a protein: Rubredoxin at 1.5 Å resolution. Acta Cryst B29, 943–956. [Google Scholar]
- 85.Sussman J, Holbrook SR, Church GM, & Kim S-H (1977) A structure factor least squares refinement procedure for macromolecular structures using constrained and restrained parameters. Acta Crystallogr A33, 800–804. [Google Scholar]
- 86.Konnert JH & Hendrickson WA (1980) A restrained-parameter thermal-factor refinement procedure. Acta Crystallogr A36, 344–350. [Google Scholar]
- 87.Murshudov GN, Vagin AA, & Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D53, 240–255. [DOI] [PubMed] [Google Scholar]
- 88.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, & Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Fortelle E & Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 90.Richards FM (1985) Optical matching of physical models and electron density maps: early developments. Methods Enzymol 115, 145–154. [DOI] [PubMed] [Google Scholar]
- 91.Jones TA (1978) A graphics model building and refinement system for macromolecules. J Appl Crystallogr 11, 268–272. [Google Scholar]
- 92.Emsley P & Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 93.Kalisman N, Schroder GF, & Levitt M (2013) The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure 21, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dauter Z, Wlodawer A, Minor W, & Jaskólski M (2014) Avoidable errors in deposited macromolecular structures - an impediment to efficient data mining. IUCrJ; in press., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blake CC, Fenn RH, North AC, Phillips DC, & Poljak RJ (1962) Structure of lysozyme. A Fourier map of the electron density at 6 Å resolution obtained by x-ray diffraction. Nature 196, 1173–1176. [DOI] [PubMed] [Google Scholar]
- 96.Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, & Sarma VR (1965) Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Å resolution. Nature 206, 757–761. [DOI] [PubMed] [Google Scholar]
- 97.Johnson LN & Phillips DC (1965) Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Å resolution. Nature 206, 761–763. [DOI] [PubMed] [Google Scholar]
- 98.Matthews BW, Sigler PB, Henderson R, & Blow DM (1967) Three-dimensional structure of tosyl-alpha-chymotrypsin. Nature 214, 652–656. [DOI] [PubMed] [Google Scholar]
- 99.Blow DM (1976) Structure and mechanism of chemotrypsin. Acc Chem Res 9, 145–152. [Google Scholar]
- 100.Avey HP, Boles MO, Carlisle CH, Evans SA, Morris SJ, Palmer RA, Woolhouse BA, & Shall S (1967) Structure of ribonuclease. Nature 213, 557–562. [DOI] [PubMed] [Google Scholar]
- 101.Wyckoff HW, Hardman KD, Allewell NM, Inagami T, Johnson LN, & Richards FM (1967) The structure of ribonuclease-S at 3.5 Å resolution. J Biol Chem 242, 3984–3988. [PubMed] [Google Scholar]
- 102.Fridborg K, Kannan KK, Liljas A, Lundin J, Strandberg B, Strandberg R, Tilander B, & Wiren G (1967) Crystal structure of human erythrocyte carbonic anhydrase C. 3. Molecular structure of the enzyme and of one enzyme-inhibitor complex at 5–5 Å resolution. J Mol Biol 25, 505–516. [DOI] [PubMed] [Google Scholar]
- 103.Wright CS, Alden RA, & Kraut J (1969) Structure of subtilisin BPN’ at 2.5 Å resolution. Nature 221, 235–242. [DOI] [PubMed] [Google Scholar]
- 104.Drenth J, Hol WG, Jansonius JN, & Koekoek R (1972) Subtilisin Novo. The three-dimensional structure and its comparison with subtilisin BPN’. Eur J Biochem 26, 177–181. [DOI] [PubMed] [Google Scholar]
- 105.Drenth J, Jansonius JN, Koekoek R, Swen HM, & Wolthers BG (1968) Structure of papain. Nature 218, 929–932. [DOI] [PubMed] [Google Scholar]
- 106.Huber R, Kukla D, Ruhlmann A, Epp O, & Formanek H (1970) The basic trypsin inhibitor of bovine pancreas. I. Structure analysis and conformation of the polypeptide chain. Naturwissenschaften 57, 389–392. [DOI] [PubMed] [Google Scholar]
- 107.Wlodawer A, Walter J, Huber R, & Sjolin L (1984) Structure of bovine pancreatic trypsin inhibitor. Results of joint neutron and X-ray refinement of crystal form II. J Mol Biol 180, 301–329. [DOI] [PubMed] [Google Scholar]
- 108.Adams MJ, Ford GC, Koekoek R, Lentz PJ, McPherson A Jr., Rossmann MG, Smiley IE, Schevitz RW, & Wonacott AJ (1970) Structure of lactate dehydrogenase at 2.8 Å resolution. Nature 227, 1098–1103. [DOI] [PubMed] [Google Scholar]
- 109.Hill E, Tsernoglou D, Webb L, & Banaszak LJ (1972) Polypeptide conformation of cytoplasmic malate dehydrogenase from an electron density map at 3.0 Å resolution. J Mol Biol 72, 577–589. [DOI] [PubMed] [Google Scholar]
- 110.Branden CI, Eklund H, Nordstrom B, Boiwe T, Soderlund G, Zeppezauer E, Ohlsson I, & Akeson A (1973) Structure of liver alcohol dehydrogenase at 2.9 Å resolution. Proc Natl Acad Sci USA 70, 2439–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson LN, Madsen NB, Mosley J, & Wilson KS (1974) The crystal structure of phosphorylase beta at 6 Å resolution. J Mol Biol 90, 703–717. [DOI] [PubMed] [Google Scholar]
- 112.Weber IT, Johnson LN, Wilson KS, Yeates DG, Wild DL, & Jenkins JA (1978) Crystallographic studies on the activity of glycogen phosphorylase b. Nature 274, 433–437. [DOI] [PubMed] [Google Scholar]
- 113.Fletterick RJ, Sygusch J, Semple H, & Madsen NB (1976) Structure of glycogen phosphorylase a at 3.0 Å resolution and its ligand binding sites at 6 Å. J Biol Chem 251, 6142–6146. [PubMed] [Google Scholar]
- 114.Fletterick RJ, Sygusch J, Murray N, & Madsen NB (1976) Low-resolution structure of the glycogen phosphorylase a monomer and comparison with phosphorylase b. J Mol Biol 103, 1–13. [DOI] [PubMed] [Google Scholar]
- 115.Protein Data Bank (1971) Nature New Biol 233, 223.20480989 [Google Scholar]
- 116.Protein Data Bank (1973) Acta Crystallogr B29, 1746. [Google Scholar]
- 117.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, & Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klug A (2004) The discovery of the DNA double helix. J Mol Biol 335, 3–26. [DOI] [PubMed] [Google Scholar]
- 119.Franklin RE & Gosling RG (1953) Molecular configuration in sodium thymonucleate. Nature 171, 740–741. [DOI] [PubMed] [Google Scholar]
- 120.Cochran W, Crick FH, & Vand V (1952) The structure of synthetic polypeptides. I. The transform of atoms on a helix. Acta Cryst 5, 581–586. [Google Scholar]
- 121.Watson JD & Crick FH (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 122.Wilkins MHF, Stokes AR, & Wilson HR (1953) Molecular structure of deoxypentose nucleic acids. Nature 171, 738–740. [DOI] [PubMed] [Google Scholar]
- 123.Vischer E & Chargaff E (1948) The composition of the pentose nucleic acids of yeast and pancreas. J Biol Chem 176, 715–734. [PubMed] [Google Scholar]
- 124.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, & Dickerson RE (1980) Crystal structure analysis of a complete turn of B-DNA. Nature 287, 755–758. [DOI] [PubMed] [Google Scholar]
- 125.Viswamitra MA, Kennard O, Jones PG, Sheldrick GM, Salisbury S, Favello L, & Shakked Z (1978) DNA double helical fragment at atomic resolution. Nature 273, 687–688. [DOI] [PubMed] [Google Scholar]
- 126.Conner BN, Takano T, Tanaka S, Itakura K, & Dickerson RE (1982) The molecular structure of d(ICpCpGpG), a fragment of right-handed double helical A-DNA. Nature 295, 294–299. [DOI] [PubMed] [Google Scholar]
- 127.Shakked Z, Rabinovich D, Cruse WB, Egert E, Kennard O, Sala G, Salisbury SA, & Viswamitra MA (1981) Crystalline A-DNA: the X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc R Soc Lond B Biol Sci 213, 479–487. [DOI] [PubMed] [Google Scholar]
- 128.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der MG, & Rich A (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686. [DOI] [PubMed] [Google Scholar]
- 129.Drew HR & Dickerson RE (1981) Conformation and dynamics in a Z-DNA tetramer. J Mol Biol 152, 723–736. [DOI] [PubMed] [Google Scholar]
- 130.Drew HR, Dickerson RE, & Itakura K (1978) A salt-induced conformational change in crystals of the synthetic DNA tetramer d(CpGpCpG). J Mol Biol 125, 535–543. [DOI] [PubMed] [Google Scholar]
- 131.Brzezinski K, Brzuszkiewicz A, Dauter M, Kubicki M, Jaskolski M, & Dauter Z (2011) High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 Å. Nucleic Acids Res 39, 6238–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seeman NC, Rosenberg JM, Suddath FL, Kim JJ, & Rich A (1976) RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3’,5’-uridine hexahydrate. J Mol Biol 104, 109–144. [DOI] [PubMed] [Google Scholar]
- 133.Rosenberg JM, Seeman NC, Day RO, & Rich A (1976) RNA double-helical fragments at atomic resolution. II. The crystal structure of sodium guanylyl-3’,5’-cytidine nonahydrate. J Mol Biol 104, 145–167. [DOI] [PubMed] [Google Scholar]
- 134.Dock-Bregeon AC, Chevrier B, Podjarny A, Johnson J, de Bear JS, Gough GR, Gilham PT, & Moras D (1989) Crystallographic structure of an RNA helix: [U(UA)6A]2. J Mol Biol 209, 459–474. [DOI] [PubMed] [Google Scholar]
- 135.Ichikawa T & Sundaralingam M (1972) X-ray diffraction study of a new crystal form of yeast phenylalanine tRNA. Nature New Biol 236, 174–175. [DOI] [PubMed] [Google Scholar]
- 136.Stout CD, Mizuno H, Rubin J, Brennan T, Rao ST, & Sundaralingam M (1976) Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res 3, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, & Rich A (1973) Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science 179, 285–288. [DOI] [PubMed] [Google Scholar]
- 138.Suddath FL, Quigley GJ, McPherson A, Sneden D, Kim JJ, Kim SH, & Rich A (1974) Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0 Å resolution. Nature 248, 20–24. [DOI] [PubMed] [Google Scholar]
- 139.Quigley GJ, Suddath FL, McPherson A, Kim JJ, Sneden D, & Rich A (1974) The molecular structure of yeast phenylalanine transfer RNA in monoclinic crystals. Proc Natl Acad Sci USA 71, 2146–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, & Klug A (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250, 546–551. [DOI] [PubMed] [Google Scholar]
- 141.Sussman JL & Kim SH (1976) Idealized atomic coordinates of yeast phenylalanine transfer RNA. Biochem Biophys Res Commun 68, 89–96. [DOI] [PubMed] [Google Scholar]
- 142.Pley HW, Flaherty KM, & McKay DB (1994) Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74. [DOI] [PubMed] [Google Scholar]
- 143.Scott WG, Finch JT, & Klug A (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- 144.Bawden FC & Pirie NW (1937) The isolation and some properties of liquid crystalline substances from solanaceous plants infected with three strains of tobacco mosaic virus. Proc R Soc Lond B 123, 274–320. [Google Scholar]
- 145.Bernal JD & Fankuchen I (1941) X-ray and crystallographic studies of plant virus preparations: I. Introduction and preparation of specimens. II. Modes of aggregation of the virus particles. J Gen Physiol 25, 111–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bloomer AC, Champness JN, Bricogne G, Staden R, & Klug A (1978) Protein disk of tobacco mosaic virus at 2.8 Å resolution showing the interactions within and between subunits. Nature 276, 362–368. [DOI] [PubMed] [Google Scholar]
- 147.Stubbs G, Warren S, & Holmes K (1977) Structure of RNA and RNA binding site in tobacco mosaic virus from 4 Å map calculated from X-ray fibre diagrams. Nature 267, 216–221. [DOI] [PubMed] [Google Scholar]
- 148.Harrison SC, Olson AJ, Schutt CE, Winkler FK, & Bricogne G (1978) Tomato bushy stunt virus at 2.9 Å resolution. Nature 276, 368–373. [DOI] [PubMed] [Google Scholar]
- 149.Abad-Zapatero C, Abdel-Meguid SS, Johnson JE, Leslie AG, Rayment I, Rossmann MG, Suck D, & Tsukihara T (1980) Structure of southern bean mosaic virus at 2.8 Å resolution. Nature 286, 33–39. [DOI] [PubMed] [Google Scholar]
- 150.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, et al. (1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153. [DOI] [PubMed] [Google Scholar]
- 151.Jones TA & Liljas L (1984) Structure of satellite tobacco necrosis virus after crystallographic refinement at 2.5 Å resolution. J Mol Biol 177, 735–767. [DOI] [PubMed] [Google Scholar]
- 152.Hogle JM, Chow M, & Filman DJ (1985) Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229, 1358–1365. [DOI] [PubMed] [Google Scholar]
- 153.Acharya R, Fry E, Stuart D, Fox G, Rowlands D, & Brown F (1989) The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature 337, 709–716. [DOI] [PubMed] [Google Scholar]
- 154.Caspar DL & Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 27, 1–24. [DOI] [PubMed] [Google Scholar]
- 155.Michel H & Oesterhelt D (1980) Three-dimensional crystals of membrane proteins: bacteriorhodopsin. Proc Natl Acad Sci USA 77, 1283–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Michel H (1982) Three-dimensional crystals of a membrane protein complex. The photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol 158, 567–572. [DOI] [PubMed] [Google Scholar]
- 157.Deisenhofer J, Epp O, Miki K, Huber R, & Michel H (1984) X-ray structure analysis of a membrane protein complex. Electron density map at 3 Å resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol 180, 385–398. [DOI] [PubMed] [Google Scholar]
- 158.Deisenhofer J, Epp O, Miki K, Huber R, & Michel H (1985) Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature 318, 618–624. [DOI] [PubMed] [Google Scholar]
- 159.Deisenhofer J, Epp O, Sinning I, & Michel H (1995) Crystallographic refinement at 2.3 Å resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol 246, 429–457. [DOI] [PubMed] [Google Scholar]
- 160.Abrahams JP, Leslie AGW, Lutter R, & Walker JE (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621. [DOI] [PubMed] [Google Scholar]
- 161.Stock D, Leslie AG, & Walker JE (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- 162.Doyle DA, Morais CJ, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, & MacKinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77. [DOI] [PubMed] [Google Scholar]
- 163.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, & MacKinnon R (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522. [DOI] [PubMed] [Google Scholar]
- 164.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, & MacKinnon R (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41. [DOI] [PubMed] [Google Scholar]
- 165.Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WE, Robbins RA, Miercke LJ, & Stroud RM (2009) Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc Natl Acad Sci USA 106, 7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Darst SA, Kubalek EW, & Kornberg RD (1989) Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature 340, 730–732. [DOI] [PubMed] [Google Scholar]
- 167.Darst SA, Edwards AM, Kubalek EW, & Kornberg RD (1991) Three-dimensional structure of yeast RNA polymerase II at 16 Å resolution. Cell 66, 121–128. [DOI] [PubMed] [Google Scholar]
- 168.Cramer P, Bushnell DA, & Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 169.Barbieri M, Pettazzoni P, Bersani F, & Maraldi NM (1970) Isolation of ribosome microcrystals. J Mol Biol 54, 121–124. [DOI] [PubMed] [Google Scholar]
- 170.Wittmann HG, Mussig J, Piefke J, Gewitz HS, Rheinberger HJ, & Yonath A (1982) Crystallization of Escherichia coli ribosomes. FEBS Lett 146, 217–220. [DOI] [PubMed] [Google Scholar]
- 171.Yonath A, Tesche B, Lorenz S, Mussig J, Erdmann VA, & Wittmann HG (1983) Several crystal forms of the Bacillus stearothermophilus 50 S ribosomal particles. FEBS Lett 154, 15–20. [DOI] [PubMed] [Google Scholar]
- 172.Hope H, Frolow F, von Bohlen K, Makowski I, Kratky C, Halfon Y, Danz H, Webster P, Bartels KS, & Wittmann HG (1989) Cryocrystallography of ribosomal particles. Acta Crystallogr B45, 190–199. [DOI] [PubMed] [Google Scholar]
- 173.Makowski I, Frolow F, Saper MA, Shoham M, Wittmann HG, & Yonath A (1987) Single crystals of large ribosomal particles from Halobacterium marismortui diffract to 6 Å. J Mol Biol 193, 819–822. [DOI] [PubMed] [Google Scholar]
- 174.Ban N, Nissen P, Hansen J, Moore PB, & Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 175.Wimberly BT, Brodersen DE, Clemons WM Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, & Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 176.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 177.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, & Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 178.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, & Cate JH (2007) Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nature Struct Mol Biol 14, 727–732. [DOI] [PubMed] [Google Scholar]
- 179.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, & Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942. [DOI] [PubMed] [Google Scholar]
- 180.Ben-Shem A, Garreau de LN, Melnikov S, Jenner L, Yusupova G, & Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529. [DOI] [PubMed] [Google Scholar]
- 181.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. (2007) Crystal structure of the human ß2 adrenergic G-protein-coupled receptor. Nature 450, 383–387. [DOI] [PubMed] [Google Scholar]
- 182.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, et al. (2007) GPCR engineering yields high-resolution structural insights into ß2 adrenergic receptor function. Science 318, 1266–1273. [DOI] [PubMed] [Google Scholar]
- 183.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. (2007) High-resolution crystal structure of an engineered human ß2 adrenergic G protein-coupled receptor. Science 318, 1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. (2011) Structure of a nanobody-stabilized active state of the ß2 adrenoceptor. Nature 469, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011) Crystal structure of the ß2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le T,I, Teller DC, Okada T, Stenkamp RE, et al. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 187.Perutz MF, Rosa J, & Schechter A (1978) Therapeutic agents for sickle cell disease. Nature 275, 369–370. [DOI] [PubMed] [Google Scholar]
- 188.Beddell CR, Goodford PJ, Norrington FE, Wilkinson S, & Wootton R (1976) Compounds designed to fit a site of known structure in human haemoglobin. Br J Pharmacol 57, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pullen RA, Lindsay DG, Wood SP, Tickle IJ, Blundell TL, Wollmer A, Krail G, Brandenburg D, Zahn H, Gliemann J, et al. (1976) Receptor-binding region of insulin. Nature 259, 369–373. [DOI] [PubMed] [Google Scholar]
- 190.Abelson J (2009) From molecular biology to geology: a surprising trajectory. J Biol Chem 284, 35997–36006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Goodford PJ (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28, 849–857. [DOI] [PubMed] [Google Scholar]
- 192.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, & Ferrin TE (1982) A geometric approach to macromolecule-ligand interactions. J Mol Biol 161, 269–288. [DOI] [PubMed] [Google Scholar]
- 193.Ondetti MA, Rubin B, & Cushman DW (1977) Design of specific inhibitors of angiotensin-converting enzyme: new class of orally active antihypertensive agents. Science 196, 441–444. [DOI] [PubMed] [Google Scholar]
- 194.Blundell T, Sibanda BL, & Pearl L (1983) Three-dimensional structure, specificity and catalytic mechanism of renin. Nature 304, 273–275. [DOI] [PubMed] [Google Scholar]
- 195.Wlodawer A, Miller M, Jaskólski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, & Kent SBH (1989) Conserved folding in retroviral proteases: Crystal structure of a synthetic HIV-1 protease. Science 245, 616–621. [DOI] [PubMed] [Google Scholar]
- 196.Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SBH, & Wlodawer A (1989) Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Å resolution. Science 246, 1149–1152. [DOI] [PubMed] [Google Scholar]
- 197.Wlodawer A & Erickson JW (1993) Structure-based inhibitors of HIV-1 protease. Annu Rev Biochem 62, 543–585. [DOI] [PubMed] [Google Scholar]
- 198.Miller M, Jaskólski M, Rao JKM, Leis J, & Wlodawer A (1989) Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature 337, 576–579. [DOI] [PubMed] [Google Scholar]
- 199.Weber IT, Miller M, Jaskólski M, Leis J, Skalka AM, & Wlodawer A (1989) Molecular modeling of the HIV-1 protease and its substrate binding site. Science 243, 928–931. [DOI] [PubMed] [Google Scholar]
- 200.Tickle IJ, Sibanda BL, Pearl LH, Hemmings AM, & Blundell TL (1984) in X-ray Crystallography and Drug Action pp. 427–440. Clarendon Press, Oxford, UK. [Google Scholar]
- 201.Navia MA & Murcko MA (1992) Use of structural information in drug design. Curr Opin Struct Biol 2, 202–210. [Google Scholar]
- 202.Colman PM (1994) Structure-based drug design. Curr Opin Struct Biol 4, 868–874. [DOI] [PubMed] [Google Scholar]
- 203.Gaasterland T (1998) Structural genomics taking shape. Trends Genet 14, 135. [DOI] [PubMed] [Google Scholar]
- 204.Terwilliger TC, Waldo G, Peat TS, Newman JM, Chu K, & Berendzen J (1998) Class-directed structure determination: foundation for a protein structure initiative. Protein Sci 7, 1851–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim SH (1998) Shining a light on structural genomics. Nature Struct Biol 5, 643–645. [DOI] [PubMed] [Google Scholar]
- 206.Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin D, Sali A, Studier FW, & Swaminathan S (1999) Structural genomics: beyond the human genome project. Nature Genet 23, 151–157. [DOI] [PubMed] [Google Scholar]
- 207.Montelione GT & Anderson S (1999) Structural genomics: keystone for a Human Proteome Project. Nat Struct Biol 6, 11–12. [DOI] [PubMed] [Google Scholar]
- 208.Heinemann U, Frevert J, Hofmann K, Illing G, Maurer C, Oschkinat H, & Saenger W (2000) An integrated approach to structural genomics. Prog Biophys Mol Biol 73, 347–362. [DOI] [PubMed] [Google Scholar]
- 209.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 210.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. (2001) The sequence of the human genome. Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 211.Boutet S, Lomb L, Williams GJ, Barends TR, Aquila A, Doak RB, Weierstall U, DePonte DP, Steinbrener J, Shoeman RL, et al. (2012) High-resolution protein structure determination by serial femtosecond crystallography. Science 337, 362–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Redecke L, Nass K, DePonte DP, White TA, Rehders D, Barty A, Stellato F, Liang M, Barends TR, Boutet S, et al. (2013) Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science 339, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Neutze R, Wouts R, van der SD, Weckert E, & Hajdu J (2000) Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757. [DOI] [PubMed] [Google Scholar]
- 214.Bergh M, Huldt G, Timneanu N, Maia FR, & Hajdu J (2008) Feasibility of imaging living cells at subnanometer resolutions by ultrafast X-ray diffraction. Quart Rev Biophys 41, 181–204. [DOI] [PubMed] [Google Scholar]
- 215.Sayre D (2008) Report on a project on three-dimensional imaging of the biological cell by single-particle X-ray diffraction. Acta Crystallogr A64, 33–35. [DOI] [PubMed] [Google Scholar]