Abstract
The centromere is a specialized locus that directs the formation of the kinetochore protein complex for correct chromosome segregation. The specific centromere histone H3 variant CENP-A has been de-scribed as the epigenetic mark of this chromatin region. Several laboratories have explored its properties, its partners, and its role in centromere formation. Specifically, two types of CENP-A nucleosomes have been described, suggesting there may be more complexity involved in centromere structure than previously thought. Recent work adds to this paradox by questioning the role of CENP-A as a unique centromeric mark and highlighting the assembly of a functional kinetochore in the absence of CENP-A. In this review, we discuss recent literature on the CENP-A nucleosomes and the debate on its role in kinetochore formation and centromere identity.
Keywords: CENP-A, nucleosome structure, chromatin dynamic, HJURP
Introduction
The centromere is the constriction site of chromosomes where the architectural protein complex called the kinetochore is formed. This network of 40 plus proteins plays a role at the interface between the chromosome and the microtubule spindles and ensures correct chromosome segregation. The role of centromeres is conserved in eukaryotes, but centromeres are not associated with a conserved DNA sequence across the species, and neocentromere can form at non-centromeric sequences (Blower et al. 2002; Saffery et al. 2000). The establishment of centromeres and kinetochores is dependent on the centromeric histone H3 variant (CENH3 or CENP-A), which presents properties different from that of canonical histone H3, and generates a unique chromatin organization (Sullivan and Karpen 2004). An ongoing controversy within the centromere field is the molecular structure of the centromeric nucleosome. Data from various laboratories have shown two possible modes of organization: an octamer and a tetramer. In addition, others have questioned the essential role of CENP-A nucleosomes as the sole epigenetic mark of centromeres, instead illustrating the importance of other kinetochore partners such as CENP-C. In this review, we present recent literature on CENP-A properties and its participation in different complexes. We also highlight how heterogeneity of structure may contribute to centromere function over the cell cycle. In addition, we discuss provocative new findings suggesting that CENP-A might be dispensable for kinetochore formation. Finally, we summarize the impact of recent results on models of centromere inheritance, and discuss new avenues of research investigating centromere structure and function.
CENP-A and H3: complementary leaders for the establishment of a unique chromatin domain
The smallest unit of chromatin is an octamer of histone proteins called a nucleosome, wrapped by 147 bp of DNA in a left-handed fashion (Luger et al. 1997). In its canonical version, the nucleosome is composed of two H3–H4 heterodimers associated together as a tetramer and surrounded on each side by an H2A–H2B heterodimer. The repetition of nucleosome units constitutes an 11-nm fiber, or “beads on a string” structure, which is further organized into a higher level of condensation by the linker histone H1 to form a 30-nm fiber (Robinson and Rhodes 2006). Chromatin-binding proteins and posttranslational modifications of histones participate in chromatin organization to finally form a panel of structures, ranging from open and accessible to tightly condensed, and resulting in different levels of permissiveness to transcription (Li and Reinberg 2011; van Steensel 2011). Furthermore, the inclusion of histone variants within the fiber increases the number of chromatin configurations that are possible. To date, variants of histones H3, H2A, and H2B have been described, each with specific characteristics such as altered hydrophobicity, capacity for posttranslational modifications, association with protein partners, and sub-chromatin localization, all of which contribute to genome plasticity (Yuan and Zhu 2012).
CENP-A is a histone H3 variant specifically enriched at centromeres. In contrast to other histones, CENP-A has evolved rapidly and is poorly conserved in most species, particularly within its N-terminal tail, which varies in size and sequence (from 20 to 200 amino acids; Malik and Henikoff 2003; Talbert et al. 2002, 2004; Schueler et al. 2010; Hirsch et al. 2009). The highest homology with canonical histone H3.1 (50 %) is observed within the carboxy-terminal histone fold domain, excluding a 42-amino acid region (loop1 and α2-helix) identified as the CENP-A-targeting domain (CATD) (Black et al. 2004; Shelby et al. 1997; Vermaak et al. 2002). Although the CENP-A protein sequence differs amongst species, CENP-A remains the mark of centromeres in all eukaryotes and is thought to be essential for their structure and function.
Centromeric chromatin is not solely composed of CENP-A nucleosomes, but rather displays a pattern of alternating domains containing both H3.1 and H3.3 nucleosomes (Fig. 1a; Blower et al. 2002; Dunleavy et al. 2011; Sullivan and Karpen 2004). Consistent with condensed chromatin folding, H3-containing nucleosomes within centromeres are hypoacetylated (Sullivan and Karpen 2004). Paradoxically, dimethylation on lysine 4 (H3K4me2), a mark mainly associated with gene expression, is also found at centromeres (Sullivan and Karpen 2004). Indeed, active transcription has been described to be essential for the maintenance of the centromere, demonstrating the permissive nature of this locus and belying its cytological appearance (Topp et al. 2004). Moreover, the loss of permissive posttranslational modifications of H3 affects CENP-A recruitment and, in turn, kinetochore structure and function (Bergmann et al. 2011). Thus, centromeres appear to contain multivalent H3 domains.
Fig. 1.
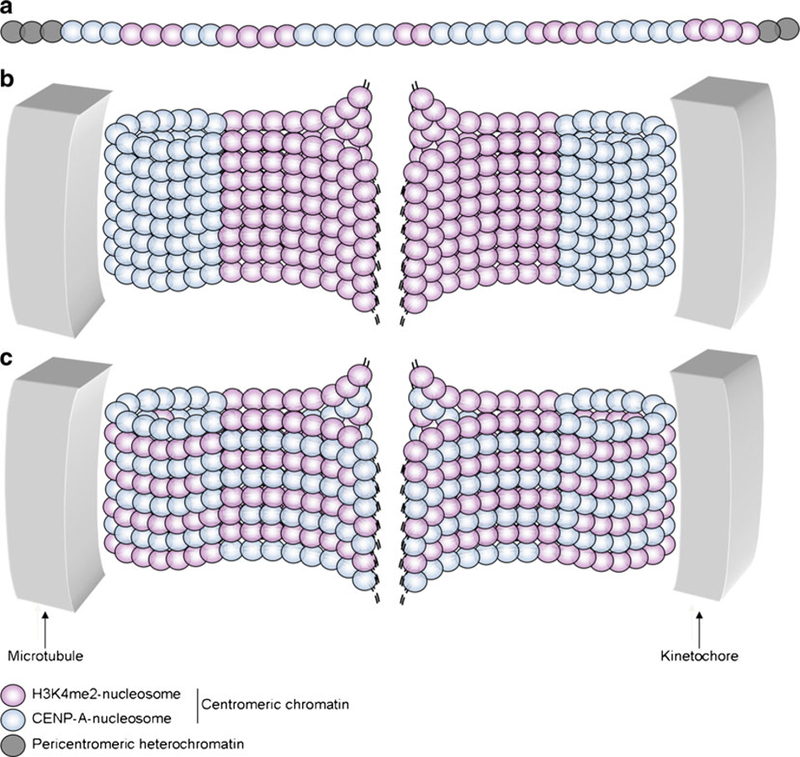
Models of spatial organization of centromeres during chromosome segregation. a Schematic of the CENP-A and H3 (dimethylated on lysine 4—H3K4me2) alternating domains at centromeres. b Model of CENP-A domain exposure at the surface of the mitotic centromere. c Model of boustrophedon organization of the mitotic centromere
During the cell cycle, H3 and CENP-A domains within the centromere recruit centromeric proteins in a specific order, to form the kinetochore and to assure correct chromosome segregation. For example, CENP-C and CENP-N have been described as direct partners of CENP-A nucleosomes, whereas CENP-T and CENP-W show a preference for H3 nucleosomes (Carroll et al. 2009, 2010; Hori et al. 2008). Despite the fact that the identity of most centromeric proteins is known, the organizing principle driving their orderly association has not yet been elucidated. A recent advance comes from three-dimensional views of mitotic chromosomes obtained by electron microscopy (EM), which revealed a precise arrangement of centromeric fibers, so that CENP-A domains are extruded to the surface of the constriction, whereas H3 domains are buried deeper inside (Fig. 1b; Blower et al. 2002; Marshall et al. 2008). However, recent data derived from super-resolution microscopy and field emission scanning EM have challenged this model (Fig. 1c; Ribeiro et al. 2010; Schroeder-Reiter et al. 2012). According to the alternative model, chromatin folding leads to planar sinusoidal layers of centromeric fibers, forming a boustrophedon, which results in the exposure of both, CENP-A and H3 nucleosomes, at the surface of the constriction. This model highlights the fact that both histones are involved in the recruitment of kinetochore proteins (Carroll et al. 2009, 2010; Hori et al. 2008). Thus, chromatin organization at the centromere is elaborate, and structural differences between H3 and CENP-A nucleosomes, and their respective chromatin fibers, may contribute to the basal level of such complexity.
CENP-A: a histone variant with a panel of potential nucleosome structures
The conservative view of the CENP-A nucleosome: the octamer
The smallest unit of canonical H3 chromatin is an octamer. Consequently, this structure has also been proposed as a model for the CENP-A nucleosome, and has been confirmed in vitro using purified proteins from several species in the presence or the absence of histone chaperones, such as nucleosome assembly protein 1 (NAP1) and HJURP/Scm3 (Holliday junction recognition protein; Black et al. 2004; Camahort et al. 2009; Dechassa et al. 2011; Kingston et al. 2011; Sekulic et al. 2010; Tachiwana et al. 2011; Yoda et al. 2000). These studies also revealed specific characteristics of the CENP-A nucleosome (Conde e Silva et al. 2007; Kingston et al. 2011; Yoda et al. 2000). First, the hypothesis of a hybrid octamer, i.e., containing one copy of H3 and one copy of CENP-A, has been excluded. Mixing both histones in chromatin reconstitution assays in vitro leads to the formation of either CENP-A- or H3-containing nucleosomes, but not a mixture containing both (Kingston et al. 2011). The basis for this selectivity is unclear, and in vivo, the consequences of such exclusivity have not yet been explored. Second, CENP-A complexes migrate slower than canonical nucleosomes in sucrose or glycerol gradient and electromobility shift assays, suggesting a larger volume (Dechassa et al. 2011; Yoda et al. 2000). In addition, small-angle X-ray scattering suggests an elongated CENP-A nucleosome structure (Dechassa et al. 2011). However, DNA digestion stud-ies seemingly contradict this finding. Whereas the micrococcal nuclease (MNase)-protected DNA fragment of the H3 nucleosome is 147 bp, the CENP-A nucleosome protects only 120–125 bp (Dechassa et al. 2011; Kingston et al. 2011). Reduced interaction of DNA with the CENP-A octamer, and reduced binding at the entry and exit of the centromeric nucleosome, have both been observed by EM. These data suggest that the slower migration does not result from an increased octameric volume, but rather from unbound DNA protruding off the edges of the CENP-A nucleosome (Tachiwana et al. 2011). Third, an unanticipated instability of the CENP-A nucleoprotein complex has been revealed. Within in vitro-reconstituted CENP-A octamers, the dissociation of one H2A-H2B dimer in the presence of the chaperone NAP1 is faster than in the H3 octamer, and the (CENP-A-H4)2 tetramers are more easily destabilized than (H3-H4)2 tetramers (Conde e Silva et al. 2007). More intriguingly, DNA binding to the (CENP-A-H4)2 tetramer is unfavorable compared to the corresponding (H3-H4)2 tetramer, leading to a low efficiency of (CENP-A-H4)2 tetramer reconstitution (only 0.05 % of the total input protein; Camahort et al. 2009; Conde e Silva et al. 2007). Yet, octamer reconstitution using purified histones from yeast is more efficient for CENP-A than for H3 (Camahort et al. 2009). Adding to the confusion, analysis of reconstituted CENP-A chromatin after prolonged storage highlights the lack of stability of octameric CENP-A nucleosomes in vitro. Whereas H3 and CENP-A particles are equally stable when reconstituted on generic DNA (601 sequence or fragment of pBR322 plasmid), CENP-A nucleosomes (but not H3 nucleosomes) on centromeric DNA are completely destabilized (Dechassa et al. 2011). The discrepancy between the efficiency of CENP-A nucleosome reconstitution and its subsequent stability suggests that even though the CENP-A octamer can be formed, this structure is inherently unstable on centromeric DNA. This implies that, in vivo, CENP-A octamers either exist only transiently or require non-histone factors for stability.
To better understand the differences between CENP-A and H3, several groups have analyzed the atomic structure of CENP-A complexes. Confirming previous results (Black et al. 2004), the crystal structure of the DNA-free (CENP-A-H4)2 tetramer, compared to computational modeling of the (H3-H4)2 tetramer, shows a more compact and rigid complex (Fig. 2a; Sekulic et al. 2010). Compaction of the tetramer results from a rotational difference between CENP-A/H4 dimer pairs, leading to a stronger contact between residues at their interface. A key difference appears to be a positively charged bulge in the N-terminal part of loop1, where aspartate residues of H3 are replaced by lysine and arginine in CENP-A (at positions 77 and 80, respectively). Other areas present differences in hydrophobicity, particularly at the extremities of the interface between CENP-A and H4, potentially contributing to the loss of flexibility. In contrast, the recently solved crystal structure of the entire CENP-A octamer did not confirm either the subtle rotation at the CENP-A/CENP-A interface or the rigidity of the (CENP-A-H4)2 tetramer (Tachiwana et al. 2011). Surprisingly, the CENP-A nucleosome was super-imposable with the H3 octamer (Fig. 2b). Still, the αN helix, which contributes to DNA stability around the octamer, is shorter in CENP-A than in H3, resulting in an unfixed conformation of the 13 bp at both entry and exit of the nucleosome. Consequently, only 121 bp of DNA are observed in the crystal structure. Tachiwana et al (2011). extend an attractive hypothesis for the potential usefulness of the exposure of DNA ends in the CENP-A nucleosome. They propose that the binding of the CENP-B protein to its centromeric DNA motif (CENP-B box) is enhanced by the proximity of this motif to the entry or exit of the CENP-A nucleosome (Tanaka et al. 2005). Consequently, the flexible entry/ exit DNA could participate in the spatial organization of centromeric DNA-binding proteins such as CENP-B and CENP-C. In addition, CENP-A loop1 projects outside the nucleosome, thus maintaining its accessibility to centromeric factors, but is less flexible than the corresponding region in the (CENP-A-H4)2 tetramer crystal (Fig. 2a; Sekulic et al. 2010, Tachiwana et al. 2011). The importance of loop1 is illustrated by the fact that two residues in loop1, Arginine80 and Glycine81, are required for stable CENP-A maintenance at centromeres (Tachiwana et al. 2011). Factors which bind loop1 remain unknown. Finally, in the octamer, the C-terminal domain of CENP-A also remains unstructured, suggesting a potential role as a docking site for non-histone proteins.
Fig. 2.
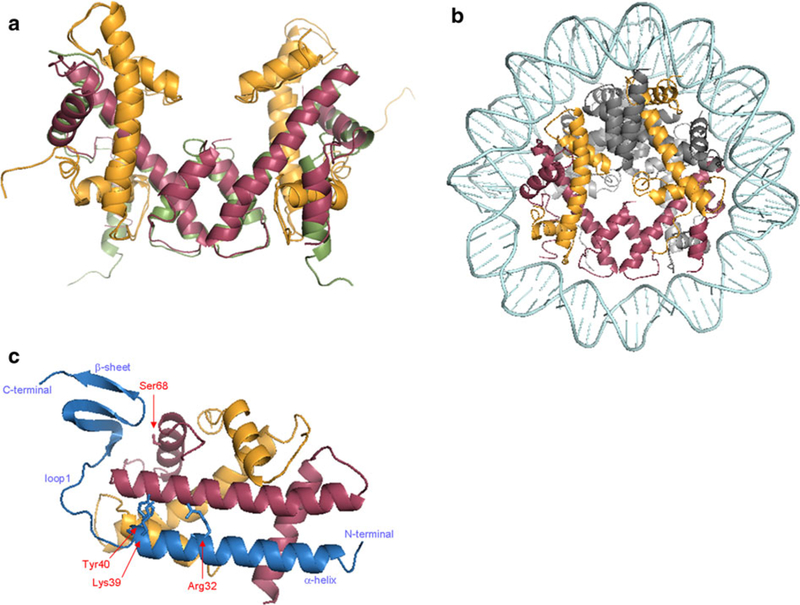
Crystal structure of human CENP-A complexes. a Superimposition of the (CENP-A-H4)2 tetramer (PDB 3NQJ; Sekulic et al. 2010) and H3 (PDB 1AOI; Luger et al. 1997). b View in the axis of the DNA helix of the CENP-A octamer (PDB 3AN2; Tachiwana et al. 2011). c Structure of the HJURP-CENP-A-H4 trimer (PDB 3R45; Hu et al. 2011). Important residues for the interaction between these three proteins are localized: Serine68 (Ser68) on CENP-A, Arginine32 (Arg32), Lysine39 (Lys39), and Tyrosine40 (Tyr40) on HJURP. All ribbon diagrams were generated in Pymol (CENP-A in red, H4 in orange, H3 in green, H2A and H2B in gray, and HJURP in blue)
The liberal view of the CENP-A nucleosome: the heterotetramer
An alternative tetrameric structure for CENP-A nucleosomes was proposed based on findings in Drosophila, subsequently leading to an energetic debate in the field (Dalal et al. 2007). In vivo chromatin cross-linking experiments of the CENP-A nucleosome in fly detected a tetrameric complex, whereas in vitro, both CENP-A octamers and tetramers were observed. Further experimental dissection of fly centromeric nucleosomes shed more light on these findings. Biochemical purifications demonstrated one copy of each H2A, H2B, H4, and CENP-A within the nucleosome, nuclease protection assays showed less than 1.5 turns of DNA, and native polyacrylamide gel electrophoresis analysis supported the existence of a smaller or more compact particle than the H3 octamer. Two biophysical approaches, EM and atomic force microscopy (AFM), demonstrated that purified CENP-A nucleosomes were smaller than, and half the height of, H3 octamers (Dalal et al. 2007). Subsequent AFM studies coupled with recognition imaging using a specific antibody against CENP-A demonstrated that single CENP-A epitopes could be detected with the tetrameric structures, confirming previous results (Wang et al. 2008). These results indicate that, in vivo, fly CENP-A nucleosomes can exist in a non-canonical tetramer form, which was also recently found to exist in human cells under varying salt conditions (Dimitriadis et al. 2010). In addition, immuno-EM experiments confirmed the heterotetrameric composition of this nucleosome by detecting H2B and CENP-A within these tetrameric nucleosomes, subsequently called “hemisomes” (Dimitriadis et al. 2010; Tatchell and Van Holde 1979; Lavelle and Prunell 2007).
In yeast, MNase digestion of centromeric DNA generates a ~130 bp product, too short to wrap around an octamer and too long for a tetramer (Bloom and Carbon 1982). A hypothesis proposed by Cole et al. (2011) is that centromeric proteins CBF1 and CBF3 (centromere binding factors 1 and 3, respectively), which interact directly with centromeric DNA and promote the formation of a loop in vitro, might be tightly associated with the tetramer to form a bigger centromeric complex. This model has been supported by recent crystallographic data showing that Ndc10, a subunit of CBF3, binds as a dimer and protects 50 bp of centromeric DNA, leaving only ~80 bp DNA—i.e., a single turn—to wrap around the CENP-A nucleosome (Cho and Harrison 2012).
Finally, in vitro and in vivo studies highlight an alternative topology induced by the presence of yeast CENP-A nucleosome, consistent with right-handed DNA wrapping (Furuyama and Henikoff 2009; Huang et al. 2011). This positive supercoiling was previously described in vitro for the (H3–H4)2 tetramer, which can wrap DNA in either direction (Hamiche et al. 1996). However, the (CENP-A–H4)2 tetramer cannot be positively wrapped by DNA efficiently, suggesting that such wrapping in vivo might occur only around the (CENP-A–H4–H2A–H2B) tetramer or require additional factors (Conde e Silva et al. 2007). If right-handed nucleosomal wrapping were discovered for other species’ CENP-A nucleosomes in vivo, it would change not only the orientation of the nucleosome on chromatin, exposing novel sites for specific centromeric partners, but also the folding of the chromatin fiber.
The moderate view of the CENP-A nucleosome: the dynamic model
The observation of different structures for CENP-A nucleosomes is controversial and has not yet been resolved. It has been suggested that different techniques in different studies could generate artifacts. However, this hypothesis is not satisfying because studies described herein combine various approaches using CENP-A from more than one species to arrive at their conclusion. The octamer has been mainly observed in vitro, whereas the tetramer has been shown preferentially in vivo. One attractive possibility is that in the cell, the microenvironment (chaperones, protein partners, posttranslational modifications) may regulate CENP-A nucleosome structure and stability, contributing to a potential switch between stable octamers and tetramers. Mimicking the cellular context in vitro is difficult and may favor one configuration over another. Sample fixation method may also contribute to the confusion, as might be the case with two conflicting studies examining CENP-A nucleosome structure and stability in Drosophila cells (Dalal et al. 2007; Zhang et al. 2012). When the classical intranucleosomal cross-linker dimethylsuberimidate (DMS) was used for fixing CENP-A nucleosomes, octameric structures were observed in vitro; both octamers and tetramers were detected in mitotic cells, but only tetramers were observed in asynchronous cells (Dalal et al. 2007). DMS cross-linking relies on the availability of primary amines (lysines), thus trapping cross-wise interactions between H4, H2A, H2B, and H3 or CENP-A within nucleosome (Thomas and Kornberg 1975a, b). In contrast, a recent study inferred the presence of CENP-A octamers in asynchronous fly cells using the cysteine cross-linking agent copper phenanthroline (Zhang et al. 2012). Cysteines are found in H3 (position 110) and CENP-A (position 75), but not in H2A, H2B, or H4 (Zhang et al. 2012). Therefore, this approach cannot directly discriminate between tetrameric (CENP-A–H4–H2A–H2B) or octameric configurations, but can inform on CENP-A/CENP-A interactions. Interestingly, cysteine/cysteine CENP-A interactions were detected in non-physiological salt (50 mM NaCl), but not in physiological salt (150 mM NaCl), leading to the interpretation that the CENP-A octamer is unstable under physiological conditions found in vivo. An important implication of this result is that CENP-A nucleosomes could interconvert between hemisomes and octamers in vivo depending on environmental cues. Two caveats exist to temper this interpretation with caution. The first is that copper phenanthroline has long been used in the chromatin field as a chemical nuclease (Brogaard al. 2012; Jessee et al. 1982), leaving open the possibility that non-nucleosomal/solubilized CENP-A could contribute to the results obtained above. The second is that the stability of human CENP-A octamer was shown to remain unchanged under varying salt conditions in vitro (Sekulic et al. 2010). Thus, additional factors might be necessary to contribute to potential CENP-A dynamics in situ.
The possibility that the CENP-A nucleosome can interconvert between two forms, and the physiological relevance of such bistability, is an exciting new avenue of research that is being actively pursued (Fig. 3; Dalal and Bui 2010; Probst et al. 2009). CENP-A domains are replicated in late S phase in human cells, a time at which no new CENP-A is available, leading to the dilution of this centromeric mark (Shelby et al. 2000). To maintain centromere identity, it has been proposed that disassembled CENP-A nucleosomes may be recycled as heterotetramers after the replication fork has passed (Probst et al. 2009; Dalal and Bui 2010). Consequently, it is feasible that following its synthesis during G2, when new CENP-A is incorporated in the early G1 phase of the next cell cycle (Hemmerich et al. 2008; Jansen et al. 2007), it forms an octamer in advance of the next replication (Dalal and Bui. 2010). If such dynamics do exist, the histone chaperone HJURP likely plays a pivotal role in this process. It has been shown that HJURP accumulates at centromeres at the late telophase–early G1, the time of CENP-A incorporation, and remains for a couple of hours (Dunleavy et al. 2009; Foltz et al. 2009). It is currently unknown whether HJURP could return after the S phase to recycle CENP-A. If correct, such a model reconciles specific properties of CENP-A and could potentially explain why laboratories have obtained different data, while pointing to an entirely novel mechanism involved in epigenetic inheritance.
Fig. 3.
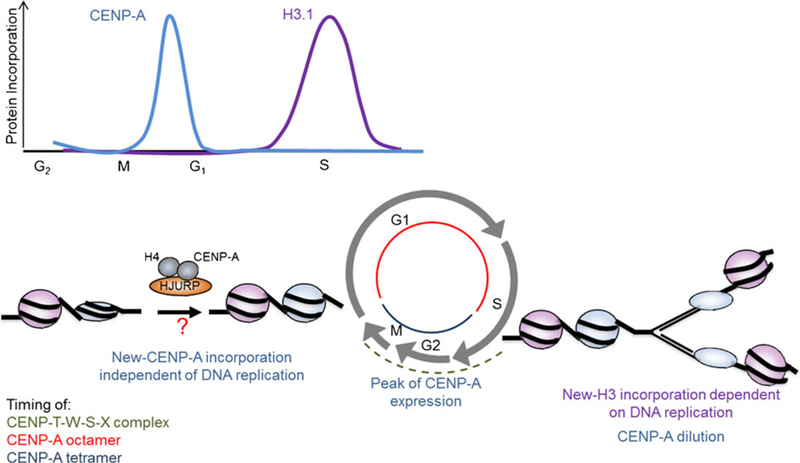
Model of CENP-A nucleosome dynamics over the cell cycle. In the S phase, centromeric domain replication is accompanied by the dilution of CENP-A proteins and potentially its distribution as a heterotetramer, whereas H3 nucleosomes conserve their octameric structure within the centromere. Then, kinetochore proteins are recruited at the centromere, such as the complex composed of CENP-T, CENP-W, CENP-S, and CENP-X, for the formation of the kinetochore and the correct chromosome segregation. At the exit of mitosis–early G1, newly synthesized CENP-A is incorporated into the chromatin to form an octameric nucleosome. This remodeling process requires the histone chaperone HJURP (Holliday junction recognition protein); still, the exact mechanism is not known
Potential physiological roles for a dynamic CENP-A nucleosome
If the CENP-A octamer is unstable in vivo, as has been demonstrated experimentally in vitro, such a structure may be disrupted at mitosis, leading to kinetochore instability, centromere fragility, and segregation defects. Consequently, evolutionary selection for a stable CENP-A tetramer may have occurred to avoid accidental dissociation of the nucleosome. We speculate that the CENP-A tetramer might be flexible to deformation during mitosis. Indeed, evidence pointing to this possibility comes from recent analysis of kinetochore–microtubule interactions, which reveals large distortions of the inner, but not outer, kinetochore when bound to microtubules (Fig. 4; Suzuki et al. 2011).
Fig. 4.
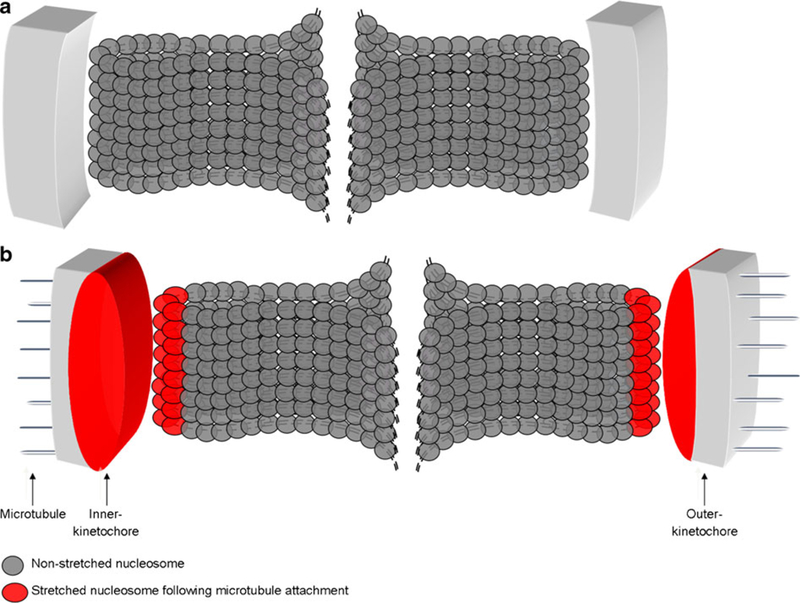
Deformation of the inner kinetochore after microtubule attachment during mitosis. a For the correct chromosome segregation, kinetochore is formed at the centromeric locus. b The correct attachment of microtubules to the outer kinetochore leads to produce a tension, which will trigger structural changes of the inner kinetochore (oval red structure in b), but not the outer kinetochore (gray rectangle in b). The stretch of the inner kinetochore involved constitutive centromere-associated network (CCAN) proteins, composed for example of CENP-A, CENP-C, CENP-N, CENP-T, CENP-W, CENP-S, or CENP-X
A second advantage to an unstable octamer is the prevention of non-centromeric incorporation. Secure CENP-A nucleosomes may require the presence of another centromeric protein whose recruitment depends not only on CENP-A, but also on other centromeric cues. The strongest such candidate is CENP-C, which interacts with H3 and CENP-A nucleosomes at the centromere, binds centromeric DNA, is recruited by centromeric RNA, and whose depletion causes CENP-A loss (Du et al. 2010; Erhardt et al. 2008; Hori et al. 2008; Moree et al. 2011; Orr and Sunkel 2011). Indeed, recent data have shown that CENP-C directly binds the C-terminus of CENP-A (Carroll et al. 2010). Finally, as for other histones, the loading of CENP-A is dependent on its histone chaperone HJURP, which plays an important role in its targeting and retention at the correct location, as discussed below.
CENP-A assembly
A soluble CENP-A–H4–HJURP complex for targeted loading
A series of studies compared the crystal structure of the CENP-A nucleosome alone and in complex with its histone chaperone HJURP/Scm3 (Cho and Harrison 2011; Hu et al. 2011; Zhou et al. 2011). These studies help better understand how CENP-A is assembled upon DNA in vivo. Since HJURP/Scm3, CENP-A, and H4 form a stochiometric complex, it was initially suggested that a hexamer composed of two copies of each might be associated with centromeres (Mizuguchi et al. 2007). Indeed, the addition of HJURP replaces histone H2A–H2B dimers from pre-assembled CENP-A octamers in vitro, suggesting the dissociation of the nucleosome into two complexes: HJURP–CENP-A–H4 hexamer/trimer and H2A–H2B dimers (potentially associated with HJURP; Camahort et al. 2009; Mizuguchi et al. 2007). However, in fission yeast and human cells, after chromatin extraction, CENP-A remains associated with H2A, H2B, and H4 in a nucleosomal form without HJURP (Camahort et al. 2009; Pidoux et al. 2009). This observation suggests that HJURP might not be a stable component of centromeric nucleosomes (Xiao et al. 2011).
Crystallographic studies support this interpretation because HJURP and Scm3 binding to their cognate CENP-As is incompatible with octamer formation (Cho and Harrison 2011; Hu et al. 2011; Zhou et al. 2011). HJURP binds to the CENP-A–H4 dimer with its N-terminal α-helix, an irregular loop and a β-sheet domain at the C-terminus (Fig. 2c). Both CENP-A and H4 display several contact points with HJURP, including through its conserved residues within the N-terminal α-helix, such as Arginine32, Lysine39, and Tyrosine40. A corresponding important residue in CENP-A is Serine68 in the α1-helix, which is localized in a hydrophobic pocket and whose mutation results in loss of specific interaction with HJURP (Hu et al. 2011). However, the primary HJURP-binding motif is located within the α2-helix of the CENP-A histone fold domain, which is placed in an anti-parallel fashion to the N-terminal α-helix of HJURP. Crystalographic and mutation analyses confirm the existence of an HJURP–CENP-A–H4 trimer, but disfavor the possibility of a hexameric structure (Cho and Harrison 2011; Hu et al. 2011; Zhou et al. 2011). Indeed, the superimposition of HJURP–CENP-A–H4 and (CENP-A–H4)2 crystals reveals that HJURP would block the CENP-A/CENP-A four-helix bundle interface, involving CENP-A C-terminal α2-helix. In addition, HJURP would prevent CENP-A-H4 association with DNA by blocking DNA wrapping near the Serine68 region. Altogether, these data suggest that HJURP disfavors CENP-A association with chromatin by preventing DNA binding and CENP-A/CENP-A interactions. Therefore, the HJURP–CENP-A–H4 trimer may be considered as an intermediate structure for CENP-A loading before its incorporation and the addition of H2A–H2B dimers. This hypothesis is in harmony with fission yeast studies showing the release of HJURP from chromatin in early mitosis when CENP-A is stably associated with centromeres (Pidoux et al. 2009; Williams et al. 2009). In addition, in vitro CENP-A nucleosome reconstitution on centromere sequences is inefficient in the absence of HJURP, suggesting the essential role of this histone chaperone in CENP-A loading (Xiao et al. 2011). It is possible that HJURP plays a comparable role to the histone chaperone NAP1 (Torigoe et al. 2011). Using purified histones and NAP1, the in vitro study of nucleosome assembly kinetics revealed a sequential mechanism. First, a non-canonical histone–DNA intermediate is formed and the histone chaperone is released. Then, the DNA is stably wrapped around the octamer in an ATP-dependent mechanism. HJURP may similarly play a dual role of loading and regulating the kinetics of CENP-A nucleosome formation.
However, the exact molecular mechanism for the targeted loading of CENP-A and conversion from a soluble complex to an octameric structure needs to be investigated further in the context of chromatin. The microenvironment at centromeres, such as H3K4me2 and centromeric factors Mis18BP1 (MIS18 binding protein 1) and CENP-C, likely contributes to HJURP recruitment in a manner not well understood, but which presumably involves the reorganization of centromeric chromatin (Barnhart et al. 2011; Bergmann et al. 2011; Moree et al. 2011; Pidoux et al. 2009; Williams et al. 2009).
Outside the centromeric box: rogue CENP-A
Despite all these complexes regulating CENP-A assembly, this histone variant can be incorporated easily outside of centromeres (Camahort et al. 2009; Van Hooser et al. 2001; Lefrancois et al. 2009; Heun et al. 2006). Low levels of endogenous or overexpressed CENP-A have been detected at euchromatin, at Ty-transposable elements, at telomeres, and at ribosomal DNA elements (Camahort et al. 2009; Van Hooser et al. 2001; Lefrancois et al. 2009; Heun et al. 2006). In fly, a genome-wide map of ectopically expressed CENP-A location revealed the preferential deposition and maintenance of this histone at euchromatin–heterochromatin boundaries (Olszak et al. 2011). An accumulation of this histone variant has also been shown at DNA double-strand breaks (Zeitlin et al. 2009).
In yeast, a specific mechanism restricts ectopic accumulation of CENP-A. The E3 ubiquitin ligase Psh1 ubiquitinates CENP-A, leading to its proteolysis (Hewawasam et al. 2010; Ranjitkar et al. 2010). Interestingly, CENP-A ubiquitination is blocked in the presence of HJURP because both HJURP and Psh1 recognize and interact with the CATD (Hewawasam et al. 2010; Foltz et al. 2009). Thus, HJURP plays a dual role by targeting CENP-A at centromeres, and by protecting the histone from ubiquitin-dependent proteolysis. Still, how HJURP specifically protects incorporated CENP-A from Psh1 activity at centromeres, but not at ectopic loci, remains mysterious.
Challenging the role of CENP-A nucleosome as the sole epigenetic mark of centromere
As we described previously, the centromere is a complex domain composed of nucleosomes and centromeric proteins whose integrity is essential for kinetochore formation. CENP-A is important for the recruitment of kinetochore proteins. Indeed, either CENP-A or HJURP is sufficient to seed centromeres when tethered to ectopic loci (Barnhart et al. 2011; Mendiburo et al. 2011). However, at native centromeres, CENP-A is not sufficient by itself for the establishment of a functional complex (Gascoigne et al. 2011; Van Hooser et al. 2001). Therefore, the importance of CENP-A as a sole epigenetic mark of centromeres has been questioned. Several laboratories have expanded their investigation to kinetochore proteins such as CENP-C, CENP-N, and the CENP-T/CENP-W complex (Hori et al. 2008). Recent studies reveal that the ectopic tethering of CENP-C and CENP-T, in the absence of CENP-A, triggers the assembly of a functional kinetochore (Gascoigne et al. 2011). In addition, CENP-C localization to the centromere is partially independent of CENP-A. CENP-C serves as an intermediary complex between inner and outer kinetochores through the direct recruitment of several kinetochore proteins in the KMN network (KNL1/Spc105, the Mis12 complex, and the Ndc80 complex; Milks et al. 2009; Przewloka et al. 2011; Tanaka and Tanaka 2009).
As for CENP-C, CENP-N depletion affects the recruitment of centromeric proteins, such as CENP-H CENP-I, CENP-K, and partially CENP-C and CENP-A, leading to kinetochore assembly defects (Carroll et al. 2009). CENP-N recognizes and interacts with CENP-A nucleosomes, but is stably associated with centromeres only in S and G2 phases, probably for re-establishing kinetochore (Hellwig et al. 2011). These data suggest that CENP-C, and CENP-N are also crucial for the establishment of a functional kinetochore (Carroll et al. 2009, 2010). Similarly, in Drosophila, the protein CAL-1, which may potentially serve as the CENP-A chaperone, interacts strongly with CENP-A and CENP-C, and they are mutually dependent on each other for their localization at centromeres (Mellone et al. 2011). In worms, the protein KNL-2 (kinetochore-null) is essential for CENP-A incorporation, and both proteins localize at centromeres in an interdependent manner and cooperate for the formation of the kinetochore and correct chromosome segregation (Maddox et al. 2007).
Another important complex is the CENP-T/ CENP-W dimer whose ectopic tethering triggers kinetochore formation in the absence of CENP-A (Gascoigne et al. 2011). Similar to CENP-A, the CENP-T/CENP-W complex binds DNA through a histone fold domain and interacts with the outer kinetochore via its N-terminal domain (Gascoigne et al. 2011). The CENP-S/CENP-X complex also contains a histone fold domain, which allows it to interact with DNA (Amano et al. 2009). Recent crystallographic studies performed by Nishino et al. (2012) reveal the existence of a novel “kineto-chore nucleosome.” (Fig. 5). CENP-T and CENP-W form a dimer via their histone fold domains, whereas the CENP-S/CENP-X complex prefers a tetrameric (CENP-S/CENP-X)2 structure. However, CENP-T and CENP-S interact together through their histone fold domains and form with CENP-W and CENP-X, a stable particle with dimensions similar to a heterotetrameric nucleosome. This ki-netochore heterotypic tetrameric nucleosome binds ~100 bp of DNA in vitro. However, unlike CENP-A, CENP-T and CENP-W are not inherited by daughter cells, suggesting that this complex assembles after replication and persists until chromosome segregation (Prendergast et al. 2011). Finally, in CENP-S or CENP-T tetramer-defective mutant cells, kinetochore formation is impaired, leading to mitotic defects and apoptosis. Thus, these complexes are essential for kinetochore formation and probably provide key structural cues for defining centromere domains.
Fig. 5.
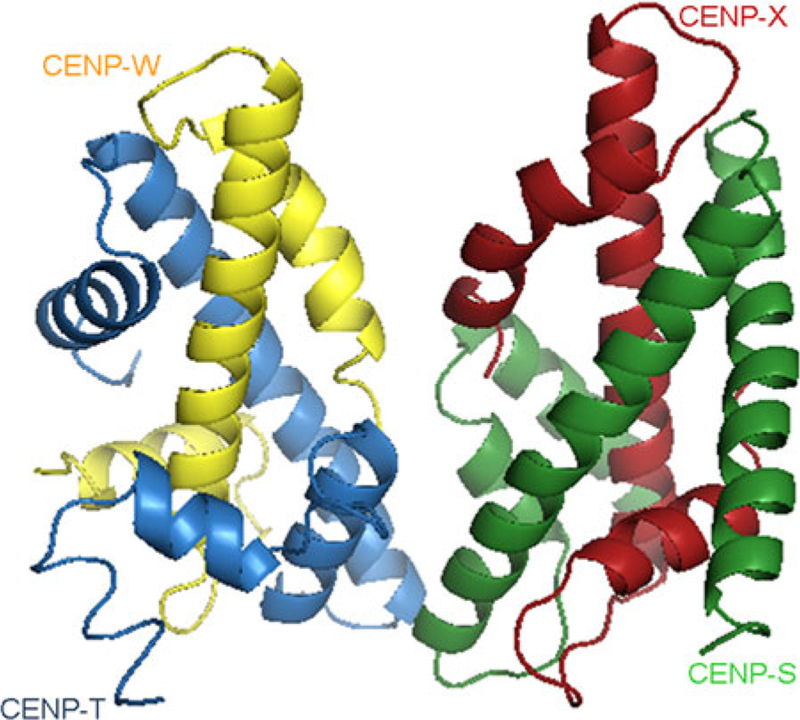
Frontal view of the crystal structure of the CENP-T-W-S-X heterotetramer (PDB 3VH6; Nishino et al. 2012). Ribbon diagram was generated in Pymol
Conclusion and future perspectives
Cell integrity depends in part on the cell’s ability to maintain faithful chromosome segregation during mitosis. This process is dependent on the centromere, wherein the histone variant CENP-A was long assumed to be the sole epigenetic mark. However, studies have revealed a more elaborate organization with alternating domains of H3- and CENP-A-nucleosomes, both essential for centromere function. In addition to the potential for an oscillating structure for the CENP-A nucleosome itself, novel kinetochore complexes may be essential for maintaining centromere identity. Why so many different soluble and chromatin-associated complexes and regulatory pathways are necessary for the correct function of the centromere remains mysterious. Expanding studies into organisms with smaller centromere complexes, and devising strategies for intact centromere purification, will allow for the establishment of a minimal spatial and temporal map of the centromere (Akiyoshi et al. 2010).
Another unexplored avenue is the role of ectopically localized CENP-A. Even though several mechanisms police CENP-A targeting to the centromere, ectopic CENP-A localization has been observed. Since two types of CENP-A particles might conceivably exist at centromeres, a logical question that has not yet been investigated is the structure of CENP-A nucleosomes at non-centromeric sites. Can CENP-A form hybrids with other histone variants that might alter the properties of chromatin in ectopic regions? Do special characteristics exist at ectopic locations that prevent such CENP-A from seeding centromeres? Such studies will become important in determining the potential role of CENP-A in cancer cells, where it is has long been noted that CENP-A is overexpressed (Tomonaga et al. 2003). Despite the difficulty in specifically isolating non-centromeric subpopulations of CENP-A nucleosomes, such structural studies will be crucial in determining the potential contribution of CENP-A to chromosome fragility. Indeed, comparing the differences between CENP-A at centromeres and at ectopic sites may reveal not just distinctive centromere-specific interactions and particular functions of this histone variant but also how neocentromeres are established de novo.
Acknowledgments
We thank Minh Bui and Marcin Walkiewicz for discussion and critical reading of the manuscript and Emmanuel Capitaine for help in the figure design. We apologize to colleagues whose work could not be cited due to space constraints.
Abbreviations
- AFM
Atomic force microscopy
- CATD
CENP-A-targeting domain
- CBF1/3
Centromere binding factor 1/3
- CENH3
Centromeric histone H3 variant
- CENP-
Centromeric protein
- EM
Electron microscopy
- H3K4me2
Dimethylation on lysine 4 of histone H3
- HJURP
Holliday junction recognition protein
- Mis18BP1
MIS18 binding protein 1
- MNase
Micrococcal nuclease
- NAP1
Nucleosome assembly protein 1
References
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S (2010) Tension directly stabilizes reconstituted kinetochore–microtubule attachments. Nature 468 (7323):576–579. doi: 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T (2009) The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 186(2):173–182. doi: 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194(2):229–243. doi: 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30(2):328–340. doi: 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW (2004) Structural determinants for generating centromeric chromatin. Nature 430(6999):578–582. doi: 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Bloom KS, Carbon J (1982) Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29(2):305–317 [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2(3):319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. (2012) A map of nucleosome positions in yeast at base-pair resolution. Na-ture 486(7404):496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL (2009) Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell 35(6):794–805. doi: 10.1016/j.molcel.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 11(7):896–902. doi: 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189(7):1143–1155. doi: 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Harrison SC (2011) Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A 108(23):9367–9371. doi: 10.1073/pnas.1106389108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Harrison SC (2012) Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat Struct Mol Biol 19(1):48–55. doi: 10.1038/nsmb.2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ (2011) The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci U S A 108 (31):12687–12692. doi: 10.1073/pnas.1104978108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A (2007) CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol 370(3):555–573. doi: 10.1016/j.jmb.2007.04.064 [DOI] [PubMed] [Google Scholar]
- Dalal Y, Bui M (2010) Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol 22(3):392–402. doi: 10.1016/j.ceb.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S (2007) Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol 5(8):e218. doi: 10.1371/journal.pbio.0050218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K (2011) Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2:313. doi: 10.1038/ncomms1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y (2010) Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A 107(47):20317–20322. doi: 10.1073/pnas.1009563107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6(2):e1000835. doi: 10.1371/journal.pgen.1000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137 (3):485–497. doi: 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH (2011) H3.3 is depos-ited at centromeres in S phase as a placeholder for newly assembled CENP-A in G phase. Nucleus 2(2):146–157. doi: 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF (2008) Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183(5):805–818. doi: 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137(3):472–484. doi: 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S (2009) Centromeric nucleosomes induce positive DNA supercoils. Cell 138(1):104–113. doi: 10.1016/j.cell.2009.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145(3):410–422. doi: 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Carot V, Alilat M, De Lucia F, O’Donohue MF, Revet B, Prunell A (1996) Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci U S A 93(15):7588–7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, Meraldi P, Diekmann S (2011) Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci 124(Pt 22):3871–3883. doi: 10.1242/jcs.088625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiede-berg L, Erliandri I, Diekmann S (2008) Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 180(6):1101–1114. doi: 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kin etochores. Dev Cell 10 (3 ):303–315. doi: 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL (2010) Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40(3):444–454. doi: 10.1016/j.molcel.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CD, Wu Y, Yan H, Jiang J (2009) Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol 26 (12):2877–2885. doi: 10.1093/molbev/msp208 [DOI] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135(6):1039–1052. doi: 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM (2011) Structure of a CENP-A–histone H4 heterodimer in complex with chaperone HJURP. Genes Dev 25(9):901–906. doi: 10.1101/gad.2045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Chang KM, Cui H, Jayaram M (2011) Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc Natl Acad Sci U S A 108 (33):13671–13676. doi: 10.1073/pnas.1101944108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176(6):795–805. doi: 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessee B, Gargiulo G, Razvi F, Worcel A (1982) Analogous cleavage of DNA by micrococcal nuclease and a 1–10-phenanthroline-cuprous complex. Nucleic Acids Res 10 (19):5823–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR (2011) Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem 286(5):4021–4026. doi: 10.1074/jbc.M110.189340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle C, Prunell A (2007) Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle 6 (17):2113–2119 [DOI] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M (2009) Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10:37. doi: 10.1186/1471-2164-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D (2011) Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21(2):175–186. doi: 10.1016/j.gde.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389(6648):251–260. doi: 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176(6):757–763. doi: 10.1083/jcb.200701065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10(11):882–891. doi: 10.1038/nsb996 [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Marshall AT, Choo KH (2008) Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol 183 (7):1193–1202. doi: 10.1083/jcb.200804078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH (2011) Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7(5): e1002068. doi: 10.1371/journal.pgen.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P (2011) Drosophila CENH3 is sufficient for centromere formation. Science 334(6056):686–690. doi: 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF (2009) Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell 20(19):4246–4255. doi: 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C (2007) Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129 (6):1153–1164. doi: 10.1016/j.cell.2007.04.026 [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF (2011) CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194(6):855–871. doi: 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148(3):487–501. doi: 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P (2011) Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol 13(7):799–808. doi: 10.1038/ncb2272 [DOI] [PubMed] [Google Scholar]
- Orr B, Sunkel CE (2011) Drosophila CENP-C is essential for centromere identity. Chromosoma 120(1):83–96. doi: 10.1007/s00412-010-0293-6 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC (2009) Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33(3):299–311. doi: 10.1016/j.molcel.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF (2011) Premitotic assembly of human CENPs -T and - W switches centromeric chromatin to a mitotic state. PLoS Biol 9(6):e1001082. doi: 10.1371/journal.pbio.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10 (3):192–206. doi: 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM (2011) CENP-C is a structural platform for kinetochore assembly. Curr Biol 21(5):399–405. doi: 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40(3):455–464. doi: 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC (2010) A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A 107 (23):10484–10489. doi: 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D (2006) Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 16(3):336–343. doi: 10.1016/j.sbi.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH (2000) Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore- associated proteins. Hum Mol Genet 9(2):175–185 [DOI] [PubMed] [Google Scholar]
- Schroeder-Reiter E, Sanei M, Houben A, Wanner G (2012) Current SEM techniques for de- and re-construction of centromeres to determine 3D CENH3 distribution in barley mitotic chromosomes. J Microsc 246(1):96–106. doi: 10.1111/j.1365-2818.2011.03592.x [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED (2010) Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol 27(7):1585–1597. doi: 10.1093/molbev/msq043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE (2010) The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 467(7313):347–351. doi: 10.1038/nature09323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF (1997) Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol 136(3):501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol 151(5):1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11 (11):1076–1083. doi: 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T (2011) Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol 193(1):125–140. doi: 10.1083/jcb.201012050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476 (7359):232–235. doi: 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Ara-bidopsis histone H3 variant. Plant Cell 14(5):1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3 (4):18. doi: 10.1186/jbiol11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Tanaka TU (2009) Live cell imaging of kinetochore capture by microtubules in budding yeast. Methods Mol Biol 545:233–242. doi: 10.1007/978-1-60327-993-2_14 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, Yokoyama S (2005) Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. J Biol Chem 280(50):41609–41618. doi: 10.1074/jbc.M509666200 [DOI] [PubMed] [Google Scholar]
- Tatchell K, Van Holde KE (1979) Nucleosome reconstitution: effect of DNA length on nuclesome structure. Biochemistry 18(13):2871–2880 [DOI] [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD (1975a) An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A 72(7):2626–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD (1975b) Cleavable cross-links in the analysis of histone–histone associations. FEBS Lett 58 (1):353–358 [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F (2003) Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res 63(13):3511–3516 [PubMed] [Google Scholar]
- Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA 101(45):15986–15991. doi: 10.1073/pnas.0407154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT (2011) Identification of a rapidly formed nonnucleosomal histone–DNA intermediate that is converted into chromatin by ACF. Mol Cell 43(4):638–648. doi: 10.1016/j.molcel.2011.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114(Pt 19):3529–3542 [DOI] [PubMed] [Google Scholar]
- van Steensel B (2011) Chromatin: constructing the big picture. EMBO J 30(10):1885–1895. doi: 10.1038/emboj.2011.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S (2002) Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol 22(21):7553–7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dalal Y, Henikoff S, Lindsay S (2008) Single-epitope recognition imaging of native chromatin. Epigenetics Chromatin 1(1):10. doi: 10.1186/1756-8935-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33(3):287–298. doi: 10.1016/j.molcel.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C (2011) Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell 43(3):369–380. doi: 10.1016/j.molcel.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T (2000) Human centromere protein A (CENP-A) can replace histone H3 in nucleosome recon-stitution in vitro. Proc Natl Acad Sci U S A 97(13):7266–7271. doi: 10.1073/pnas.130189697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Zhu B (2012) Histone variants and epigenetic inheritance. Biochim Biophys Acta 1819(3–4):222–229. doi: 10.1016/j.bbagrm.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW (2009) Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci USA 106(37):15762–15767. doi: 10.1073/pnas.0908233106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH (2012) Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell 45(2):263–269. doi: 10.1016/j.molcel.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y (2011) Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472 (7342):234–237. doi: 10.1038/nature09854 [DOI] [PMC free article] [PubMed] [Google Scholar]


