Abstract
Insulin-like growth factor 2 (IGF2) is the major fetal growth hormone in mammals. We identify zinc finger protein 568 (ZFP568), a member of the rapidly evolving Kruppel-associated box-zinc finger protein (KRAB-ZFP) family linked primarily to silencing of endogenous retroelements, as a direct repressor of a placental-specific Igf2 transcript (designated Igf2-P0) in mice. Loss of Zfp568, which causes gastrulation failure, or mutation of the ZFP568-binding site at the Igf2-P0 promoter causes inappropriate Igf2-P0 activation. Deletion of Igf2 can completely rescue Zfp568 gastrulation phenotypes through late gestation. Our data highlight the exquisite selectivity with which members of the KRAB-ZFP family repress their targets and identify an additional layer of transcriptional control of a key growth factor regulating fetal and placental development.
Insulin-like growth factor 2 (IGF2) plays a key role in regulating fetoplacental development. Deletion of Igf2 or its receptor Igf1r leads to placental and fetal growth restriction in mice (1,2), and misregulation of IGF2 in humans causes the undergrowth and overgrowth conditions Russell-Silver syndrome (3) and Beckwith-Wiedemann syndrome (4), respectively. In mice, Igf2 is maternally imprinted and is differentially regulated in the placenta and fetus. It is transcribed from three promoters (designated Igf2-P1, P2, and P3) in both the fetus and placenta and additionally from a fourth placental-specific promoter designated Igf2-P0. The placental Igf2-P0 transcript is expressed in the labyrinth trophoblast and accounts for ~10% of the total placental expression of Igf2 (5). Mice lacking paternal Igf2-P0 display intrauterine growth restriction, reduced growth of the placenta (6, 7), and increased reactivity to anxietypromoting stimuli as a result of the mismatch between placental supply and fetal demand for nutrients (8). However, Igf2 is expressed at low levels in preimplantation development and in embryonic stem cells (ESCs), suggesting an important requirement for Igf2 repressors at implantation. Here we identify ZFP568, a Kruppel-associated box-zinc finger protein (KRAB-ZFP), as the direct repressor of Igf2-P0 in early development.
ZFP568 is an essential factor that regulates convergent extension during gastrulation (9–11). To determine genes that may be regulated by ZFP568, we crossed mice harboring a floxed Zfp568 allele fused to a C-terminal green fluorescent protein (GFP) tag (Zfp568-GFPFL/FL) (fig. S1A) with Rosa26-CreERT2 mice and derived ESC and trophoblast stem cell (TSC) lines from blastocysts (fig. S1B) (12). Treatment of cells with 4-hydroxytamoxifen (4-OHT), which activates the CreERT recombinase, resulted in deletion of Zfp568 and loss of ZFP568-GFP protein (fig. S1, C and D). Only two genes were significantly affected by acute Zfp568 deletion in ESCs and TSCs: Zfp568 itself and Igf2-P0, which was activated an average of eightfold (Fig. 1A; figs. S1E and S2, A and B; and table S1). Clustering analysis confirmed that differences between independently derived cell lines were greater than differences observed before and after Zfp568 deletion (fig. S2C). Furthermore, there was no misregulation of repetitive elements (fig. S2D). RNA-sequencing (RNA-seq) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) confirmed that only the placental-specific Igf2-P0 promoter and transcript were activated, whereas the fetal promoters and the Igf2 antisense (Igf2as) transcripts were not affected (fig. S2, A, B, E, and F). The increased expression of Igf2-P0 also significantly increased the IGF2 peptide levels secreted in the media in ESCs. There was no significant increase in IGF2 peptide upon Zfp568 deletion in TSCs (fig. S2G), which express high levels of Igf2-P1, P2, and P3 transcripts even in wild-type (WT) cells (fig. S2, B and F).
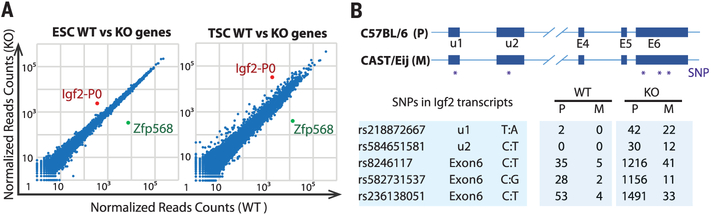
Fig. 1. Acute Zfp568 knockout leads to derepression of Igf2-P0.
(A) Scatter plots of gene expression in Zfp568 WTand KO ESCs and TSCs, as determined by RNA-seq. (B) Schematic of SNPs in CAST/Eij; C57BL/6 hybrid ESCs used to discriminate Igf2 parental alleles. SNP counts from RNA-seq in Zfp568 WT and KO CAST/Eij; C57BL/6 hybrid ESCs are indicated.
To determine whether Zfp568 deletion leads to allelic activation of Igf2, we used clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 engineering to delete Zfp568 in CAST/Eij; C57BL/6 hybrid ESCs (fig. S3, A and B), which contain single-nucleotide polymorphisms (SNPs) that allow discrimination of parental Igf2 alleles. Consistent with results from acute deletion of Zfp568, chronic deletion of Zfp568 in hybrid cells resulted in reactivation not only of Igf2-P0, but also of P1, P2, and P3 transcripts (fig. S3, C to F), suggesting that the reactivation spread to neighboring promoters over prolonged culture. Most of the Igf2-P0 transcripts were transcribed from the paternal allele (Fig. 1B and fig. S3G). We also found extensive dysregulation of additional genes in hybrid Zfp568KO/KO cells likely caused by chronic loss of Zfp568 and overexpression of IGF2, as these effects could be partially mimicked by exposing WT hybrid ESCs to IGF2 peptide (fig. S3, E and H).
To determine whether ZFP568 directly represses Igf2-P0, we performed chromatin immunoprecipitation sequencing (ChIP-seq) with antibodies against GFP on Zfp568-GFPFL/FL ESCs and TSCs. We found 137 and 86 high-confidence ZFP568-binding peaks in ESCs and TSCs, respectively (table S2), which include a peak upstream of the Igf2-P0 promoter (Fig. 2A). Motif analysis revealed a highly significant binding motif of 21 to 24 base pairs (bp) in both ESCs and TSCs, with a large fraction of the most highly enriched peaks containing the target motif, including Igf2-P0 (fig. S4A). The most-enriched 29 peaks were also shared by both ESCs and TSCs (fig. S4A). ZFP568 binding was only weakly correlated with KRAB-associated protein-1 (KAP1) and SET domain bifurcated 1 (SETDB1) binding and was not substantially marked by trimethylated histone H3 lysine 9 (H3K9me3) (fig. S4B). In contrast, the Igf2-P0 promoter contained a strong H3K9me3 signal that was completely lost upon Zfp568 deletion (Fig. 2A). Furthermore, deletion of the KRAB-ZFP corepressors Setdbl and Trim28/KAP1 resulted in derepression of Igf2-P0 and loss of H3K9me3 (fig. S4, C and D). Additionally, there was a corresponding loss of DNA methylation at an Igf2-P0 CpG island designated DMR0, but not at DMR1 or DMR2, upon loss of Zfp568 (fig. S5, A to C).
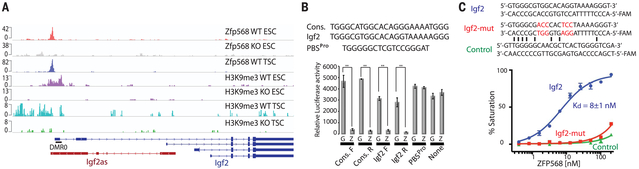
Fig. 2. ZFP568 directly targets Igf2-P0 for repression.
(A) ZFP568 and H3K9me3 ChIP-seq signals at the Igf2 locus in Zfp568 WT and KO ESCs and TSCs. DMR0 is a differentially methylated region overlapping exon 1 of the Igf2-P0 transcript. Igf2as is the Igf2 antisense transcript. (B) Relative luciferase activity in 293T cells overexpressing GFP (G) or ZFP568 (Z) and an SV40 promoter–driven luciferase plasmid containing the ZFP568 consensus (cons) or Igf2 binding site in the forward (F) or reverse (R) orientation. PBSPro is the 18-nucleotide (nt) primer binding site for proline, which is bound by ZFP809 but not ZFP568. t test: Error bars indicate standard deviation; **P < 0.01; n = 3. (C) Fluorescence-polarization binding assay of ZFP568 (ZnF1-11) protein incubated with indicated 6-carboxyfluorescein (FAM)–labeled double-stranded oligonucleotides.
To verify the binding site of ZFP568, we performed luciferase reporter assays using minimal putative ZFP568-binding sites and the full Igf2-P0 promoter (fig. S6, A and B). Expression of ZFP568 significantly repressed reporters containing the minimal Igf2-P0 binding site or with the consensus binding site (Fig. 2B). Likewise, ZFP568 repressed the full Igf2-P0 reporter (fig. S6B). Point mutation of the KRAB domain that mimics the previously described Chato mutation (11) prevented repression of the reporters (fig. S6C). Furthermore, triplet scrambling of the Igf2-P0 binding site or deletion of pairs of zinc fingers had a significant impact on transcriptional repression (fig. S6, D and E). Because the binding site contains a CpG dinucleotide, we speculated that ZFP568 binding to its target may be methylation sensitive (fig. S6B). However, bisulfite sequencing in ESCs demonstrated that these CpG sites are not methylated (fig. S6F). Furthermore, we expressed in Escherichia coli and purified the 11–zinc finger array of ZFP568 and measured its binding affinity to a 26-bp double-stranded oligonucleotide encompassing the Igf2-P0 binding site using fluorescence polarization (13). We found that the ZFP568 zinc fingers bound specifically to the Igf2-P0 binding site with a Kd of ~8 nM (Fig. 2C). Methylation of the CpG site caused a modest reduction (about threefold) in binding affinity, whereas methylation of the two CpA sites had a more significant effect, consistent with these positions being more conserved (fig. S6G).
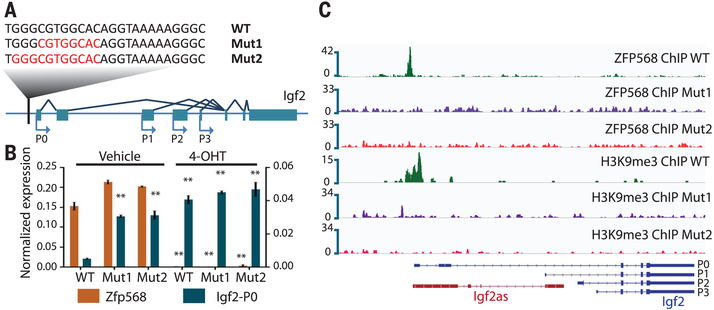
To confirm that ZFP568 was acting through its binding site within the Igf2-P0 promoter to repress Igf2-P0, we used CRISPR/Cas9 to mutate the ZFP568-binding site in Zfp568-GFPFL/FL ESCs (Fig. 3A). ESC lines harboring distinct small deletions encompassing the binding site displayed increased expression of Igf2-P0, coincident with the loss of ZFP568 binding specifically at the Igf2-P0 locus (Fig. 3, B and C, and fig. S7, A to C). Treatment of binding site-mutant ESCs with 4-OHT (to delete Zfp568) did not further increase the expression of Igf2-P0 (Fig. 3B), indicating that ZFP568 repression activity is binding site dependent. ChIP-seq and DNA methylation analysis revealed loss of H3K9me3 in ZFP568 binding site-mutant ESCs and loss of DNA methylation at DMR0 (Fig. 3C and fig. S7D). These data demonstrate that ZFP568 maintains a heterochromatin state at the Igf2-P0 promoter by direct interaction with its binding site.
Fig. 3. Mutation of the ZFP568-binding site upstream of the Igf2-P0 promoter activates Igf2-P0.
(A) Sequences of WT and two homozygous mutant ESC lines (with deleted nucleotides indicated in red) relative to the consensus binding site for ZFP568. (B) Igf2-P0 and Zfp568 levels were determined by qRT-PCR in the indicated binding site-mutant ESCs before and after Zfp568 deletion with 4-OHT. t test: Error bars indicate standard deviation; **P < 0.01; n = 3. (C) ZFP568 and H3K9me3 ChIP-seq signals at Igf2 in Zfp568 WT and binding site-mutant ESCs.
Consistent with our ESC and TSC data, we also found Igf2-P0 levels to be increased in Zfp568KO/KO embryos (Fig. 4A and fig. S8, A to D), which fail to complete gastrulation and which display phenotypes similar to the previously described Chato mutants, including convergent-extension failure and a yolk sac membrane-ruffling phenotype (that could be easily visualized and scored using autofluorescence microscopy) and arrest by embryonic day 9 (E9) (fig. S8, E and F) (9,10). Zfp568FKO/KO embryonic tissues also displayed reduced methylation specifically at Igf2 DMR0 (fig. S8, G and H). We thus reasoned that overexpression of IGF2 may contribute to the early embryonic lethality of Zfp568 mutants. To test this idea, we crossed Zfp568 knockout (KO) mice with Igf2 KO mice (14) (containing a deletion in the final exon, thus disrupting all Igf2 transcript variants) to determine if deletion of Igf2 could rescue the lethal phenotype. Loss of paternal Igf2 expression was sufficient to completely restore viability of Zfp568KO/KO embryos through mid-gestation, as Zfp568KO/KO; Igf2pat/+ embryos and fetuses were found at near Mendelian ratios and indistinguishable from Zfp568 WT littermates at E12.5 to 18.5 (Fig. 4B and fig. S9A). Despite this lack of gross morphological phenotype at late fetal stages, only two Zfp568KO/KO; Igf2KO/KO pups have been recovered after birth, both of which were found dead (fig. S9, B and C). Nonetheless, these data demonstrate a genetic interaction between Zfp568 and Igf2.
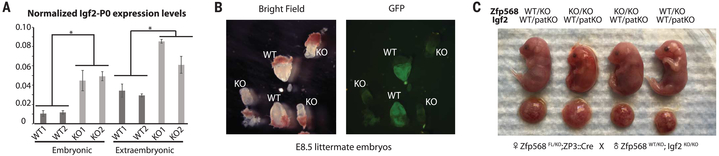
Fig. 4. Igf2 KO rescues Zfp568 KO–induced lethality.
(A) Relative Igf2-P0 levels in embryonic and extraembryonic tissues from E8.5 Zfp568-GFPFL/FL and Zfp568KO/KO littermate embryos. t test: Error bars indicate standard deviation; *P < 0.05; n = 3. (B) Bright-field and GFP-fluorescence images of Zfp568-GFPFL/FL and Zfp568KO/KO littermate embryos at E8.5. (C) Images of pups of indicated genotypes from Zfp568 KO and paternal (pat) Igf2 KO crosses at E18.5.
Our results provide biochemical and genetic evidence that the KRAB-zinc finger protein ZFP568 is a specific and direct repressor of Igf2-P0 in mice. Recent studies have revealed that the majority of KRAB-ZFPs interact with and likely restrict the expression of specific retrotransposon families, potentially facilitating the domestication of retrotransposons and the evolution of gene regulatory networks (15–19). Whether the binding site for ZFP568 at Igf2-P0 was derived from an ancient and since decayed retrotransposon or was generated by genetic drift is unknown, but our findings nonetheless demonstrate that KRAB-ZFPs can evolve essential roles in precision developmental gene silencing. Notably, ZFP568 orthologs have been detected in all eutherian mammals examined and contain a conserved zinc “fingerprint” (16), suggesting that their DNA-binding specificity is conserved. Intriguingly, Zfp57, another KRAB-ZFP that emerged in mammals, is a critical factor that maintains genomic imprints at Igf2-P0 (20,21), a process linked to viviparity (22).
Furthermore, the human Zfp568 ortholog, Znf568, is among the most-rapidly evolving human genes, with three common allele variants found within human populations that have been linked to relative head size at birth (12). Thus, KRAB-ZFP/KAP1-dependent regulation of Igf2-P0 has shaped, and likely continues to shape, the evolution of mammals.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Pfeifer and C. Gebert for lgf2 KO mice. We thank S. Coon, J. Iben, and T. Li for Next Generation Sequencing (NGS) support. This work was supported by NIH grants 1ZIAHD008933 (T.S.M.) and GM049245-23 (A.P. and X.C.), Ministry of Science and Technology (MOST) Frontier of Science Award, Academia Sinica Senior Investigator Award (C.-K.J.S.), National Natural Science Foundation of China (NSFC) 31471392, and Future Scientists Exchange Program of the China Scholarship Council (CSC) (Y.W.). NGS data have been deposited in the Gene Expression Omnibus (GEO) database (GSE84832). Zfp568-GFPFL/FL mice are available from C.-K.J.S. under a material transfer agreement with the Academia Sinica, Taipei, Taiwan, Republic of China.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Baker J, Liu JP, Robertson EJ, Efstratiadis A, Cell 75, 73–82 (1993). [PubMed] [Google Scholar]
- 2.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A, Cell 75, 59–72 (1993). [PubMed] [Google Scholar]
- 3.Gicquel C et al. , Nat. Genet 37, 1003–1007 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Weksberg R, Shuman C, Smith AC, Am. J. Med. Genet 137C, 12–23 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Moore T et al. , Proc. Natl. Acad. Sci. U.S.A 94, 12509–12514 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constância M et al. , Nat. Genet 26, 203–206 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Constância M et al. , Nature 417, 945–948 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Mikaelsson MA, Constância M, Dent CL, Wilkinson LS, Humby T, Nat. Commun 4, 2311 (2013). [DOI] [PubMed] [Google Scholar]
- 9.García-García MJ, Shibata M, Anderson KV, Development 135, 3053–3062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata M, García-García MJ, Dev. Biol 349, 331–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata M, Blauvelt KE, Liem KF Jr., García-García MJ, Development 138, 5333–5343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien HC et al. , PLOS ONE 7, e47481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel A, Horton JR, Wilson GG, Zhang X, Cheng X, Genes Dev. 30, 257–265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeChiara TM, Efstratiadis A, Robertson EJ, Nature 345, 78–80 (1990). [DOI] [PubMed] [Google Scholar]
- 15.Schmitges FW et al. , Genome Res. 26, 1742–1752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imbeault M, Helleboid P, Trono D, Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Wolf G et al. , Genes Dev. 29, 538–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecco G et al. , Dev. Cell 36, 611–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs FM et al. , Nature 516, 242–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X et al. , Dev. Cell 15, 547–557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quenneville S et al. , Mol. Cell 44, 361–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renfree MB, Suzuki S, Kaneko-Ishino T, Philos. Trans. R. Soc. London B Biol. Sci 368, 20120151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.