SUMMARY
Binding between DIP and Dpr neuronal-recognition proteins has been proposed to regulate synaptic connections between lamina and medulla neurons in the Drosophila visual system. Each lamina neuron was previously shown to express many Dprs. Here, we demonstrate, by contrast, that their synaptic partners typically express one or two DIPs, with binding specificities matched to the lamina neuron-expressed Dprs. A deeper understanding of the molecular logic of DIP/Dpr interaction requires quantitative studies on the properties of these proteins. We thus generated a quantitative affinity-based DIP/Dpr interactome for all DIP/Dpr protein family members. This revealed a broad range of affinities and identified homophilic binding for some DIPs and some Dprs. These data, along with full-length ectodomain DIP/Dpr and DIP/DIP crystal structures, led to the identification of molecular determinants of DIP/Dpr specificity. This structural knowledge, along with a comprehensive set of quantitative binding affinities, provides new tools for functional studies in vivo.
eTOC Blurb
DIP/Dpr interactions help to pattern the Drosophila nervous system. Cosmanescu et al. quantify their interactions and map DIP expression in medulla neurons. Structural studies identify specificity determinants of DIP/Dpr interactions and reveal a conserved architecture for DIP/DIP homodimers.
INTRODUCTION
Brains from flies to humans comprise vast numbers of different types of neurons interconnected by networks of precisely patterned synaptic connections. Currently, the molecular mechanisms underlying the specification of neural circuit assembly are poorly understood. The predominant model, based on Roger Sperry’s “chemoaffinity hypothesis”, postulates that neurons make specific connections with their targets based on interactions between specific cell surface molecules (Sperry, 1963). Cell-cell recognition proteins are often members of families diversified in evolution by gene duplication to yield numerous members, each bearing a canonical binding interface characteristic of the family (Himanen and Nikolov, 2003; Patel et al., 2003; Siebold and Jones, 2013). For such protein families, binding between members is often promiscuous, and it is the distinctive strength of binding, or binding affinity, that underlies the differential biological functions of each protein (Brasch et al., 2018; Goodman et al., 2016; Harrison et al., 2012; Katsamba et al., 2009). Understanding the logic underlying the patterning of neural networks will require determination of the binding affinities of cell recognition proteins, their expression patterns, their signaling properties, and gain and loss-of-function genetic analyses.
In Drosophila, two families of cell-recognition proteins of the immunoglobulin superfamily (IgSF), the 21-member Dpr (Defective proboscis extension response) and the 11-member DIP (Dpr Interacting Proteins) families, have many of the properties expected of proteins controlling synaptic specificity. Members of each family are expressed in subsets of neurons throughout the developing nervous system (Carrillo et al., 2015; Tan et al., 2015). Within the Drosophila visual system, the five lamina monopolar neurons, L1-L5, as well as the R7 and R8 photoreceptor cells, each express unique combinations of Dpr proteins. Cognate DIPs were found to be expressed in some of their synaptic partners in the medulla, suggesting a potential role in synaptic targeting (Carrillo et al., 2015; Tan et al., 2015). It remains unclear from these earlier studies, however, whether each medulla neuron type expresses many DIPs as observed for Dpr expression in lamina neurons or only a more limited repertoire of them.
In the accompanying paper, single Dm12 neurons with DIP-α null mutations exhibit robust defects in target-layer specificity in a wild-type background and misexpression of cognate Dpr ligands dramatically re-specify these connections (Xu et al, submitted). In addition to targeting phenotypes, DIP/Dpr interactions also play a role in cell survival. Loss of DIP-γ (Carrillo et al., 2015) as well is its binding partner Dpr11 in R7 neurons (Xu et al., submitted), leads to a reduction in the number of Dm8 neurons. In both cases, cell loss results from apoptosis during development (Xu et al., submitted), consistent with the idea that DIP/Dpr interactions may influence the regulation of apoptosis. The number of DIP and Dpr paralogs, their patterns of expression within the brain and the complexity of the DIP/Dpr interactome allude to a widespread and complex role in patterning neural circuitry.
High-throughput in vitro binding experiments using an ELISA-based assay revealed a heterophilic interaction network between members of the two families, where all but two members of the DIP family were found to interact with individual or subsets of Dprs (Carrillo et al., 2015; Ozkan et al., 2013). While such assays are effective at identifying heterophilic binding, technical constraints of the method often select against the detection of homophilic interactions (Bushell et al., 2008). Furthermore, these assays utilized multimerized chimeras to increase binding affinities so as to enable robust detection; as a consequence, however, this method inherently obscures the native molecular binding affinities, yielding binary results that provide a yes/no answer as to whether an interaction takes place.
Do binding affinities of adhesion proteins significantly impact interactions between cells? Differential affinities can have clear effects on signaling between adherent cells: for example, T cells bearing receptors with different affinities for peptide-MHC complexes on antigen-presenting cells adopt different developmental fates (Stone et al., 2009). With respect to selectivity of cellular interactions, type I classical cadherin family proteins provide a typical example of the role of affinity: each type I cadherin family member binds to all other type I family members, yet the differences in affinity of each pair-wise interaction dictate their distinct adhesive and cell-patterning functions (Katsamba et al., 2009; Vendome et al., 2014). Thus, for protein families with promiscuous binding in which selectivity is dictated mainly through the differential pair-wise binding affinities of different family members, quantitative measures are required to understand their function.
In the nervous system, binding affinities of cell-cell recognition proteins have been shown to control the targeting of neurites to their appropriate partners. For example, members of the two-protein family of Ig-like sidekick (Sdk) proteins are expressed (Yamagata and Sanes, 2008, 2012; Yamagata et al., 2002) in specific layers within the inner plexiform region of the mouse retina during synapse formation. In vitro, Sdk1 and Sdk2 bind heterophilically through a canonical interface, but their homophilic affinities are stronger (Goodman et al., 2016). Despite their heterophilic binding, the higher affinity of the respective homophilic interactions appears to determine their synaptic targeting activities (Krishnaswamy et al., 2015; Yamagata and Sanes, 2008). By contrast, within this same region of the retina the type II cadherin family members cadherin-8 and cadherin-9, which show distinctive heterophilic and homophilic affinities to other type II cadherin family members (Brasch et al., 2018) appear to rely on heterophilic rather than homophilic binding for proper layer-specific targeting (Brasch et al., 2018; Duan et al., 2014) Thus, differential molecular binding affinities of both cadherins and Ig superfamily proteins contribute to synaptic patterning.
DIP-Dpr binding specificity is controlled by interactions between their immunoglobulinlike extracellular domains (Carrillo et al., 2015). The extracellular regions of Dpr family members consist of two tandem Ig-like domains, while the extracellular region of DIP family members consists of three tandem Ig-like domains (Ozkan et al., 2013). The crystal structure of a two-domain fragment of DIP-α in complex with the membrane-distal Ig1 domain of Dpr6 revealed the Ig1 -Ig1 interaction to be characterized by a buried core of hydrophobic residues and an extensive network of hydrogen bonds (Carrillo et al., 2015). The interaction topology of this complex shares a strong resemblance to other complexes of Ig-like cell adhesion molecules, including those of vertebrate nectins and C. elegans SYGs, both of which have roles in nervous system development (Carrillo et al., 2015; Harrison et al., 2012; Okabe et al., 2004; Ozkan et al., 2014; Togashi et al., 2006). Interestingly, members of the nectin and Syg-related protein families exhibit both homophilic and heterophilic binding.
Here, as a step towards understanding how DIP and Dpr protein families contribute to neural circuit assembly, we sought to extend understanding of both the binding affinities of DIPs/Dprs and the neuron-specific localization of DIPs in the Drosophila visual system. We used the multi-color flip out (MCFO) technique (Nern et al., 2015) to provide a more extensive map of DIP expression in the medulla. To assess the biophysical properties of interactions between protein family members we used surface plasmon resonance (SPR) to determine binding affinities for all DIP-Dpr interactions, identified DIPs and Dprs that form homodimers, and identified specificity determining residues in DIP-Dpr interfaces that had not previously been noticed. We used this new knowledge to design site-directed mutants with defined intermolecular binding affinities for in vivo functional experiments reported in the accompanying paper (Xu et al., submitted). Our biophysical studies raise the intriguing possibility that DIP/Dpr interactions function over a wide range of affinities to regulate neural circuit assembly throughout the Drosophila nervous system.
RESULTS
Medulla neurons express DIPs in a highly cell-type specific fashion
Using Minos-mediated integration cassette (MiMIC) insertions and derivatives of them, we demonstrated that DIPs are expressed in many medulla neuron types (Tan et al., 2015), but are largely absent in lamina neurons. Using a candidate approach, we showed that, indeed, some DIPs are expressed in synaptic partners of lamina neurons, which expressed cognate Dpr proteins. It was unclear from these studies, however, what fraction of medulla neuron types express DIPs and whether each of these medulla neuron types also expresses multiple DIPs or a more restricted set of them. Here we set out to address these issues.
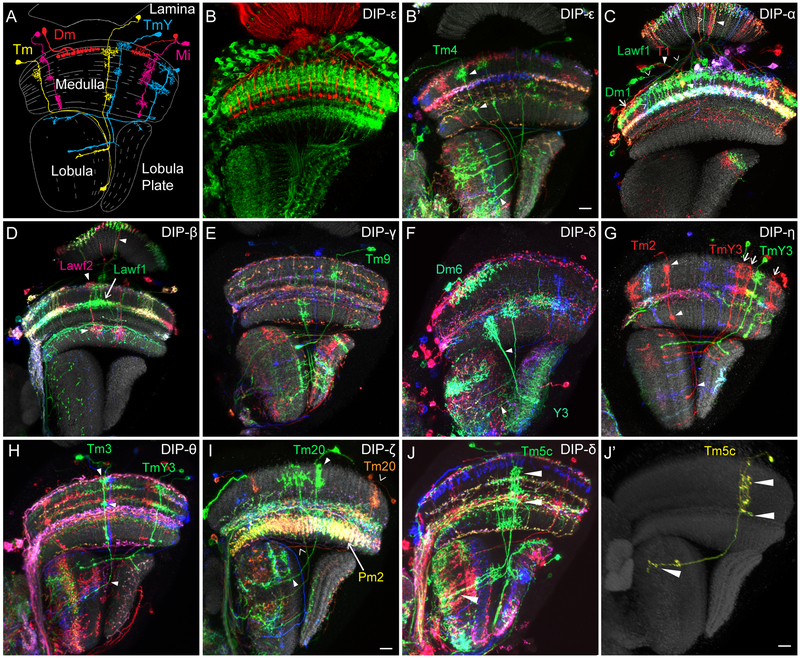
Determining the expression of Dprs using MiMIC insertions into Dpr loci was facilitated by co-staining experiments with well-characterized antibodies to nuclear proteins specific for each lamina neuron type (Tan et al., 2015). By contrast, only a few cell types in the medulla can be identified in this way, due to the paucity of appropriate antibodies. We therefore sought to correlate patterns of DIP expression with the morphologies of different medulla neurons (Figure 1A). This was done using GAL4 transcription traps inserted into different DIP loci to drive expression of a membrane-bound epitope tagged protein or a fluorescent protein that highlights the entire morphology of these neurons (Figure 1B) and a recombination-based method (i.e. MCFO) to generate sparsely labeled populations of these cells to more readily assess their morphologies (Figure 1B’-J). In some cases, the density of staining precluded a simple reconstruction of the morphology of a single neuron. In these preparations we were able to identify single neurons by comparing them to reference neurons from sparsely labeled samples (e.g. compare Figure 1J and1J’). Additional examples of cells identified in this way are shown in Figures S1-S3.
Figure 1: Identification of medulla neuron types expressing different DIPs using MCFO.
(A) Schematic of different classes of medulla neurons, Dm, Mi, Tm and TmY. (B, B’) Cell types express DIP-ε can be identified by MCFO. It is very difficult to identify cell types expressing DIPs by driving membrane-bound GFP using DIP gene traps, as shown in B. In contrast, individual cell types can be identified by morphology using MCFO to generate sparsely labeled cells, as shown in B’. White triangles, Tm4 (green). Scale bar: 10μm. (C-F) Examples of medulla neuron types identified by MCFO for the remaining seven DIPs. Colors for the lettering of cell types are the same as the single cells labeled in the images. (C) DIP-α: arrow Dm1 (green); arrowheads, Lawfl (green); triangles, T1 (red). (D) DIP-β: arrow, Lawfl (green); triangles, Lawf2 (magenta). (E) DIP-γ: Tm9 (green). (F) DIP-δ: Dm6 (Cyan); triangles, Y3 (Cyan). (G) DIP-η: arrows, TmY3 neurons (red) and another TmY3 is in green; triangles, Tm2 (red); (H) DIP-θ: triangles, Tm3 (green) and TmY3 (green) without triangles. (I) DIP-ζ: triangles, Tm20 (green); arrowheads, Tm20 (orange); Arrow, Pm2 (yellow). Pm2 neurons are always labeled in the entire layer in different colors. Scale bar: 10μm. (J, J’) An example of identifying single labeled medulla neuron in a densely labeled environment (J) by comparing its morphology to a single labeled cell in a reference image (J’). A green Tm5c is labeled in J, but it partially overlaps with another cell (described in Figure S1A, A’). By comparing patterns of arborization in specific layers in medulla and lobula between J and J’ (triangles), we can identify the cell in J. Scale bar: 10μm. See also Figures S1-S3.
We chose to assess the expression in a well-characterized population of medulla neurons in which fluorescently labeled single neurons have been analyzed in detail. These include two large sets from two separate studies (Nern et al., 2015; Takemura et al., 2013), as well as several additional cells from other analyses (Gao et al., 2008; Mauss et al., 2015; Takemura et al., 2017; Tuthill et al., 2013). Many, though not all, of these cell types were also described in the Golgi studies of Fischbach and Dittrich (1989). In total, we assessed expression of eight different DIPs in 60 cell types (see Figure 2 for a summary of expression; GAL4 insertions were not available for the remaining 3 DIPs). Of these, 26 expressed a single DIP, 12 expressed two DIPs, and one expressed four DIPs. Assuming that these 60 medulla neuron types are likely to express the three remaining DIPs for which we do not have MiMIC insertions in a similar way, we estimate that some 54 of these 60 medulla neuron types (or 90%) express one or, less frequently, two different DIPs. We present examples of each type of neuron labeled in Figures 1B’-J and Figures S1-S3.
Figure 2: Summary of DIP expression in medulla neuron types.
Expression of eight DIPs was assessed in 60 well-characterized cell types (see text). A reference for each cell type is listed in the second column. Ref 1: (Takemura et al., 2013), 2: (Tuthill et al., 2013), 3: (Nern et al., 2015), 4: (Gao et al., 2008), 5: (Takemura et al., 2017), 6: (Mauss et al., 2015). Blue, no labeled cell of the indicated type; orange, labeled cell of the indicated type. Summary of DIP-expression in each medulla neuron type is listed in the last two columns. Note that the cell types from Takemura et al. (2013) are shown in Supplementary Table 2 (see https://media.nature.com/original/nature-assets/nature/journal/v500/n7461/extref/nature12450-s1.pdf) and the Dm and Pm cells are described in Nern et al. (2015). References for a few additional cell types are as indicated.See also Figures S1-S3.
Recent studies from (Davis et al., 2018) using sequencing of nuclear RNAs (i.e. the INTACT procedure) from many different medulla neuron types come to a similar conclusion about DIP expression. There is a marked overlap in expression between these sequencing studies and our studies using the MCFO labeling method. The differences observed may reflect limitations in the DIP-GAL4 reporters or the MCFO method (e.g. different sensitivity of recombinase to heat-shock induction in different cell types, the possibility that the insertion of GAL4 within a DIP locus disrupts a subset of control elements regulating expression, or that transcripts from DIP loci are under translational control (i.e. the GAL4 mRNA is chimeric containing putative 5’ UTR translational regulatory sequences from the endogenous locus). Alternatively, differences may reflect limitations in the INTACT method (e.g. low levels of expression or contamination from other cell types through the purification of tagged nuclei). Importantly, both methods reveal limited expression of DIPs in medulla neurons, by contrast to the far broader expression of Dprs in lamina neurons. We consider the significance of these patterns in forming neural circuits in the Discussion.
In summary, the expression of many different DIPs and Dprs in processes of overlapping neurons, the requirement for some cognate pairs of these for patterning medulla circuits (see accompanying paper, Xu et al., submitted), and the potential for these proteins to mediate interactions between neurites of many different neurons in the developing medulla led us to explore in further detail the biophysical properties of the interactions between different paralogs of these two protein families.
Some DIPs and Dprs form homodimers
We used both an HEK293 and an S2 cell expression system to produce soluble whole ectodomains of 19 of the 21 Dprs and 8 of the 11 DIPs. The remaining Dprs and DIPs: Dpr9, Dpr15, DIP-δ and the two DIP-family members previously shown to have no Dpr interacting partners - CG31814 which we have named DIP-κ, and CG40378 which we have named DIP-λ - were either unstable or expressed poorly. Since structural studies show that trans-interaction specificity is contained within the Ig1 domain (Carrillo et al., 2015), for biophysical studies we produced these poorly expressed proteins as chimeras, with Ig1 of Dpr9 fused to Ig2 of Dpr8, Ig1 of Dpr15 fused to Ig2 of Dpr11, Ig1 of DIP-δ fused to Ig2-Ig3 of DIP-ε, Ig1 of DIP-κ fused to Ig2-Ig3 of DIP-α and Ig1 of DIP-λ fused to Ig2-Ig3 of DIP-θ.
We assessed the homophilic binding properties of all native and chimeric proteins with the exception of DIP-ι, using sedimentation equilibrium analytical ultracentrifugation (AUC). The results from these experiments are reported in Table 1 and Table S1, with a subset of experimental curves shown in Figure S4A. We found that at least 3 Dprs and 4 DIPs exist in a monomer-dimer equilibrium in solution. Dpr8, 12 and 21 have homophilic KD values ranging from 39.0μM – 71.3μM, while DIP-α, -η and -ζ have homophilic KD values that are similar, ranging from 22.2μM – 35.4μM. DIP-θ was found to homodimerize as well, however an accurate KD could not be determined. Further analysis of DIP-θ by multi angle light scattering (MALS) following size-exclusion chromatography (SEC) confirmed DIP-θ exists in a monomer-dimer equilibrium (Figure S4B). MALS-SEC was also used to determine that Dpr18 behaved as a monomer (Figure S4B). DIP and Dpr homodimers had not previously been reported.
Table 1:
Analytical ultracentrifugation analysis of DIP and Dpr homodimers
| Protein | Monomer MW from Mass Spectrometry (kDa) |
Apparent MW in AUC (kDa) |
Oligomeric state |
KDdimerization (μM) |
RMSDs |
|---|---|---|---|---|---|
| Dprs | |||||
| Dpr8 | 29.2 | 43.0 ± 0.05 | Dimer | 39.0 ± 0.2 | 0.00772 ± 0.00034 |
| Dpr12 | 28.9 | 35.0 ± 0.75 | Dimer | 71.3 ± 7.6 | 0.00598 ± 0.00004 |
| Dpr21 | 28.9 | 39.9 ± 0.93 | Dimer | 49.1 ± 4.0 | 0.00767 ± 0.00057 |
| DIPs | |||||
| DIP-α | 36.4 | 54.0 ± 0.19 | Dimer | 23.9 ± 0.03 | 0.00746 ± 0.00055 |
| DIP-ζ | 40.9 | 59.0 ± 0.47 | Dimer | 22.2 ± 2.1 | 0.00759 ± 0.00102 |
| DIP-η | 40.5 | 56.2 ± 0.05 | Dimer | 35.4 ± 0.4 | 0.00848 ± 0.00030 |
| DIP-θ | 44.0 | ND | Dimer* | ND | ND |
MW, Molecular Weight. ND, Not Determined. RMSDs represent the error of the global fit.
AUC data are presented as the mean of two independent measurements, ± the difference of each of these from the mean
DIP-θ was determined to be a dimer by SEC-MALS (See Figure S4B)
DIP/Dpr interactions determined by SPR identify distinct affinity binding groups
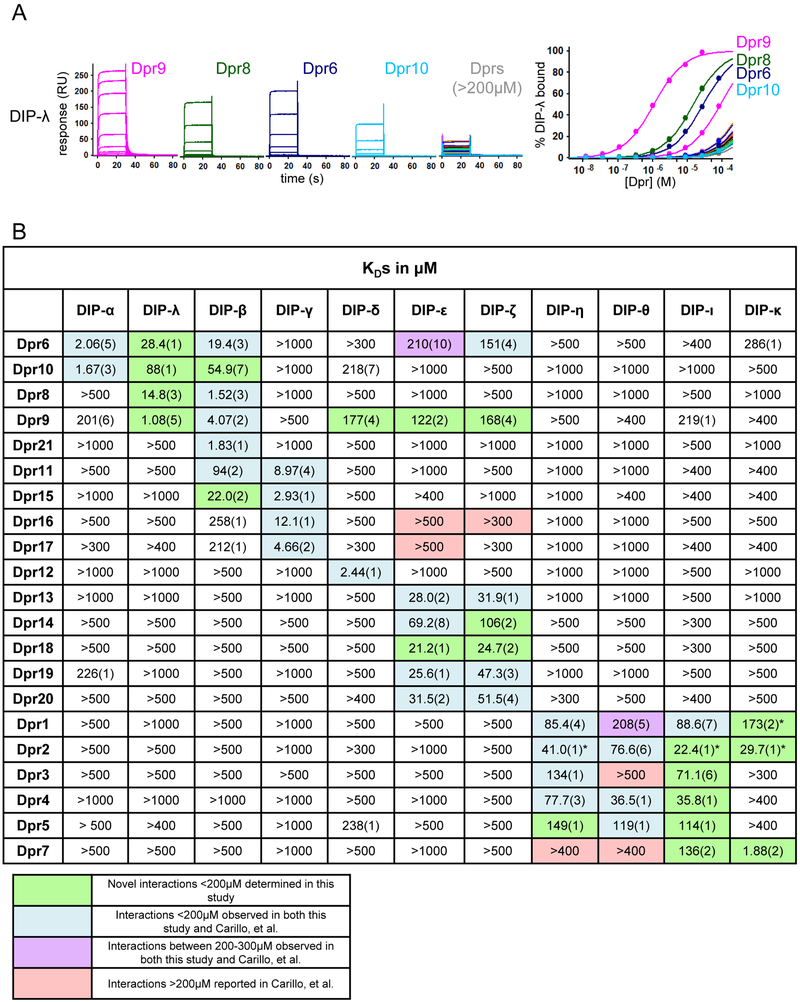
To define interactions of DIPs with Dprs and characterize the relevant binding affinities, we performed surface plasmon resonance (SPR) experiments with the purified recombinant proteins. Each DIP molecule was covalently coupled to the dextran layer of an SPR sensor chip surface using amine coupling chemistry. Twenty-one Dprs were passed over each DIP surface, and binding responses were measured and used to calculate equilibrium binding constants for interactions (Figure 3, Figures S4C and S5). These experiments revealed 21 novel interactions not previously reported, and failed to detect binding affinities stronger than 300μM for 6 interactions reported from the high-throughput studies (Carrillo et al., 2015; Ozkan et al., 2013). Figure 3B highlights the differences between the early and revised DIP/Dpr interactomes; interactions we found to be stronger than 200μM are shown in Figure 4A.
Figure 3: SPR binding analysis of DIP-Dpr interactions.
(A) An example of SPR sensorgrams of 21 Dpr analytes binding over a DIP-λ-immobilized surface and the fit of the binding data to 1:1 binding isotherms to calculate KDs. Sensorgrams of Dprs 9, 8, 6 and 10, which bind with KDs lower than 200μM are shown individually and sensorgrams for all other Dprs with KDs above 200 μM are overlaid in a single panel. The concentrations for each experiment are listed in the Star Methods section. See also Figures S4C and S5 for sensorgrams and binding isotherms for the 21 Dprs binding to the 10 other DIP-immobilized surfaces. (B) Equilibrium-binding KDs of DIP-Dpr interactions determined by SPR. The Dprs are tabulated according to their DIP binding preference. “*” indicates apparent KDs that are likely to be overestimates due to the presence of some nonspecific binding. The number in brackets represents the error of the fit.
Figure 4: DIP-Dpr quantitative interactome and phylogeny.
(A) Heterophilic and homophilic interaction network according to SPR and AUC experiments, respectively. The interactions highlighted have KDs lower than 200μM. Lines are color-coded according to the affinity of the binding pairs while dashed lines correspond to interactions between 150-200μM. Dpr2 interactions with DIP-η, -ι and -κ are represented as estimates in the 40-95 μM range and DIP- κ/Dpr1 binding is represented as an estimate in the 150-200μM range due to some non-specific binding observed in SPR sensorgrams. Color-coded self-pointing arrows highlight DIPs or Dprs that homodimerize. A black self-pointing arrow is used for DIP-θ, which homodimerizes but an accurate affinity could not be determined. (B) Phylogenetic trees of Dprs and DIPs based on Ig1 domain similarity. The scale bar denotes protein distances estimated by Jones-Taylor-Thornton model (Jones et al., 1992). Dprs are colored according to primary DIP binding preference(s). “*” indicate Dprs with binding preferences deviating from group: Dpr1 and Dpr3 do not bind to DIP-θ and Dpr21 does not bind to DIP-λ with affinities lower than 200μM (see also text and Figure 3B).
When analyzing Dpr binding partners and KDs, four distinct DIP groups defined by shared Dpr binding partners emerge (Figure 4A). Each Dpr, with the exceptions of Dpr6 and Dpr9, binds to one or multiple DIPs within only one group. Group I consists of DIP-α, -β, -λ and γ (DIP group I) and contains some of the strongest affinities among all DIP/Dpr interaction pairs. DIP-α bound to Dpr6 and Dpr10 with KDs ranging from 1.7μM - 2.1μM while DIP-γ bound to Dpr11, 15, 16 and 17 with KDs ranging from 2.9μM - 12.1μM. DIP-β interacts strongly with Dpr8, 9 and 21 with KDs of 1.5 - 4.1μM. In addition to these strong interactions, DIP-β interacts with Dprs that can bind DIP-α and a subset of Dprs which bind DIP-γ, although with weaker affinities: 19.4μM and 54.9μM to Dpr6 and Dpr10 respectively, and affinities of 22.0μM to Dpr15 and 94.0μM to Dpr11. While DIP-γ shows a higher sequence divergence from the other DIP members of this group, it shares two Dpr binding partners with DIP-β. Interactions of Dpr10 and Dpr15 with DIP-β were not previously observed in high-throughput experiments (Carrillo et al., 2015; Ozkan et al., 2013), nor had Dpr interactions with DIP-λ (previously CG40378) been identified. In contrast, we observed binding of DIP-λ to Dpr9 with an affinity of 1.1μM, the strongest interaction determined in this study. DIP-λ also bound Dpr6, Dpr8 and Dpr10 with affinities of 28.4μM, 14.8μM and 88μM respectively.
DIP-δ comprises its own group (DIP group II) and is the sole DIP to interact with Dpr12, binding with a strong affinity of 2.4μM. DIP group III consists of DIP-ε and DIP-ζ which each bound to the same set of Dprs, a result not seen in the previous high-throughput experiments (Carrillo et al., 2015; Ozkan et al., 2013). This group is characterized by having Dpr affinities of moderate strength when comparing affinities of all four DIP groups. Dpr13, 18, 19 and 20 bound to DIP-ε and DIP-ζ with affinities ranging from 21.2μM - 51.5μM, while Dpr14 interactions were weaker with an affinity of 69.2μM to DIP-ε and 106μM to DIP-ζ. Interactions of Dpr6 and Dpr9 with DIP-ε and DIP-ζ were also detected, ranging from 122μM −210μM. The previous high-throughput studies reported Dpr18 to be the only Dpr protein that did not interact with any DIP (Carrillo et al., 2015; Ozkan et al., 2013). However, we found that Dpr18 has the strongest binding affinity among Dprs to DIP-ε and DIP-ζ with KDs of 21.2μM and 24.7μM respectively. We also failed to detect meaningful affinities between DIP-ε and Dpr16 or Dpr17, and DIP-ζ with Dpr16, interactions that had been previously reported (Ozkan et al., 2013).
The final DIP group (DIP group IV) consists of DIP-η, -θ, -ι and DIP-κ (previously CG31814). In general, DIP-Dpr interactions in this group are significantly weaker than the interactions seen in the previously discussed groups with most measured KDs ranging from 35.8μM - 149μM. Previous studies found no interacting partners for DIP-κ, however we determined a strong binding affinity of 1.9μM to Dpr7, the strongest interaction within this subgroup. Unlike previous studies, we did not detect binding between Dpr7 and DIP-η or DIP-θ, and determined a binding affinity of 136μM with DIP-ι, revealing DIP-κ to be the primary interacting partner of Dpr7. We also observed DIP-κ binding to Dpr1 and Dpr2 with calculated affinities of 173μm and 29.7μM, respectively, however these are likely to represent overestimates since non-specific binding was observed in the SPR binding profiles. Non-specific binding was also observed in binding profiles between Dpr2 and DIP-η and DIP-ι, which have calculated affinities of 41.0μM and 22.4μM, respectively. DIP-ι was the only DIP to bind to all Dprs that interacted with this subgroup: Dpr1, 2, 3, 4, 5, and 7. This result differs significantly from previous studies, which observed DIP-ι interacting only with Dpr1 (Ozkan et al., 2013).
Inspection of our updated interactome revealed that the primary DIP binding specificities of Dprs are correlated with the grouping of Dprs in phylogenetic analysis (Figure 4B), an observation that was made previously (Ozkan et al., 2013). Similarly, we show here that the four DIP groups we characterized from our SPR experiments, which bind non-overlapping sets of Dprs, correlate with DIP phylogeny with the exception of DIP-γ, which has high sequence divergence from DIP-α, -β, and -λ of the first binding group (Figure 4B). DIP-ε and DIP-ζ are closely related and DIP-η, -θ, and -ι are all clustered together as well (Figure 4B). Taken together, these results indicate that both DIPs and Dprs have binding specificities that overlap with closely related family members.
Crystal structures of DIP-α and DIP-θ homodimers reveal a conserved homophilic interface
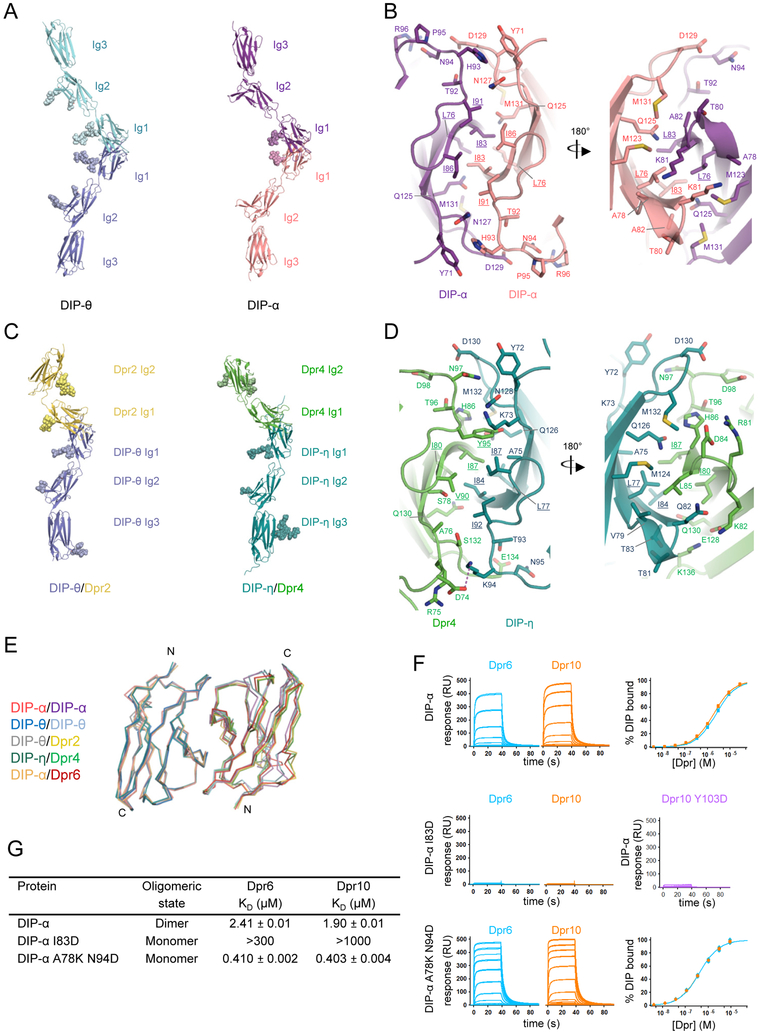
The discovery that many DIPs exist as homodimers in solution (Table 1) prompted us to investigate the structural basis of such interactions. We therefore determined the crystal structure of whole ectodomains from the homodimeric DIPs, DIP-α and DIP-Θ, to 2.9 and 3.5Å, respectively. Crystallographic statistics are summarized in Table S2. Both structures revealed highly similar homodimer interactions formed between membrane-distal Ig1 domains (Figure 5A). In each case, these interfaces are mediated by the CC’C’’FG strands of the immunoglobulinfold Ig1 domain. The orientation of the interacting Ig1 domains is highly similar to that observed for the previously published heterophilic DIP-α/Dpr6 complex (Carrillo et al., 2015). Both the DIP-α and DIP-θ homodimers bury ~1670-1750Å2 of surface area in the interface. The central core of the interface is dominated by hydrophobic interactions, where side chains of DIP-α residues Leu76, Ile83, Ile86 and Ile91 (Leu164, Ile171, Ile174, and Ile179 in DIP-θ) from apposing protomers intercalate with one other (Figure 5B; Figure S6A). Within this core, DIP-θ has an additional hydrophobic residue, Ala162 which is Gly74 in DIP-α. In addition to these hydrophobic interactions, the DIP-α homodimer has 7 unique hydrogen bond interactions while DIP-θ has 5 (Table S3). Due to the symmetrical nature of the interface, this leads to a total of 14 hydrogen bonds for DIP-α and 10 for DIP-θ. All of the hydrogen bonds are main chain to side chain, with the exception of one unique main chain/main chain hydrogen bond in the DIP-α homodimer (His93-Asn127), and one unique side chain/side chain hydrogen bond in the DIP-θ homodimer (Asn182-Asp217).
Figure 5: Structure of DIP homodimer and DIP-Dpr complexes.
(A) Ribbon representation of the full Ig ectodomain of DIP-θ and DIP-α homodimers. Individual protomers of DIP-θ are in blue and light blue, and DIP-α protomers are in pink and purple. N-linked glycans that were visible in electron density maps are represented as colored shaded spheres. (B) Structural details of DIP-α homodimer interface with side-chains contributing to interface shown as sticks. Residues comprising the hydrophobic core are underlined. See also Figure S6A (C) Ribbon representation of DIP-θ/Dpr2 and DIP-η/Dpr4 complexes rotated 30° counter clockwise in relation to structures in (A). (D) Structural details of DIP-η/Dpr4 complex interface. Dashed purple line highlights the salt bridge between Asp74 and Lys94. See also Figures S6B-C. (E) Ig1-Ig1 superposition of DIP homodimers and DIP-Dpr heterophilic complexes reported here and DIP-α/Dpr6 complex (PDB ID: 5E09). Shown as carbon-α traces and superposed on the Ig1 of the left DIP protomer. (F) SPR sensorgrams for Dpr6 and Dpr10 binding over wild-type DIP-α, I83D and A78K N94D point mutants designed to disrupt heterophilic and/or homophilic interactions. Binding of Dpr10 Y103D to wild-type DIP-α is also shown. (G) Binding KDs from SPR analysis as well as oligomeric state determined by AUC for DIP-α wild-type and mutants.
DIP/DIP and DIP/Dpr dimers are remarkably similar. Superposition of the two DIP homodimers with the DIP-α/Dpr6 complex all showed RMSD values of less than 0.8Å between 177-182 aligned Cα atoms (Figure 5E). The amino acids corresponding to intercalating residues in the central core are mostly hydrophobic across DIP and Dpr families while 8 of the hydrogen bonds are observed in both the heterophilic and homophilic DIP-α interface (Figure 6A-B; Table S3).
Figure 6: DIP-Dpr binding specificity.
(A) DIPs are grouped based on similar binding preference. Residues highlighted show variability among DIPs with different Dpr binding specificity and their positions are denoted SI1-SI4. Shaded boxes below alignment indicate interfacial residues. Yellow residues highlight residue positions of the hydrophobic core seen in crystal structures. (B) Dprs are grouped based on binding specificity with specificity residues labeled SR1-SR3. PR labels an additional residue position that is highly conserved among Dpr groups and is potentially involved in binding specificity. (C) and (D) Structural details of DIP-Dpr interaction region with specificity residues in DIP-η/Dpr4 and DIP-α/Dpr6 shown as sticks. The N102 N-glycan present in the DIP-α/Dpr6 structure is shown as grey spheres. (E) and (F) SPR sensorgrams of different Dpr4 and Dpr6 SR mutants used as analytes over DIP-η and DIP-α immobilized surfaces. Labels indicate which SR position(s) were mutated for Dpr4 and Dpr6. See also Figure S6D-E.
With the DIP-α homophilic and heterophilic interactions occurring through the same surface of the Ig1 domain, we designed mutations that could disrupt both the heterophilic and homophilic interactions together, as well as a mutant that could selectively abolish only the homophilic interaction. We set out to design these mutants to provide constructs that could define the roles of heterophilic and homophilic DIP-α interactions in an in vivo context. Genetic experiments analyzing the phenotypes of animals with these mutations are discussed in the accompanying paper (Xu et al., submitted). DIP-α I83D introduces an unpaired negative charge in the hydrophobic core shared by both the hetero and homodimer complexes. AUC experiments with this mutant showed that it behaved as a monomer, while SPR experiments showed that this mutant could not support heterophilic binding to Dpr6 or Dpr10 (Figures 5F-G). Complementary to this mutant, we designed and tested Dpr10 Y103D, which also introduces a negative charge into the hydrophobic core of the heterocomplex and abolishes binding to DIP-α (Figures 5F).
DIP-α A78K and N94D mutations were each designed to introduce electrostatic clashes that would be present in the homodimer, but not in heterophilic complexes with Dprs. AUC experiments showed DIP-α A78K N94D to be monomeric in solution and SPR experiments showed that heterophilic interactions with Dpr6 and Dpr10 were maintained and are, surprisingly, stronger than wild-type interactions (Figures 5F-G)
Crystal structures of DIP-Dpr complexes from different DIP groups show highly conserved interaction topology
In order to characterize the molecular determinants of binding specificity, we determined crystal structures of additional DIP/Dpr heterophilic complexes: the DIP-η/Dpr4 ectodomain heterocomplex at 2.9Å and a DIP-θ/Dpr2 ectodomain heterocomplex at 3.0Å (crystallographic statistics are summarized in Table S2). These complexes are associated with a different DIP group than the previously determined DIP-α/Dpr6 structure (Carrillo et al., 2015).
Both of the new structures display the canonical Ig1-Ig1 interaction first identified in the DIP-α/Dpr6 heterodimer (Carrillo et al., 2015) showing that, as expected, DIP/Dpr interactions for DIPs from other groups form through the same Ig1 domain surface (Figures 5C-D; Figure S6B). The hydrophobic character of the residues in the core of the interface is conserved among all three DIP/Dpr complexes of known structure (Figure 5D; Figures 6A-B; Figure S6B) The DIP-η/Dpr4 complex buries a total surface area of ~1750Å2 while the DIP-θ/Dpr2 complex buries ~1830Å2. The DIP-η/Dpr4 complex has 19 hydrogen bonds while DIP-θ/Dpr2 has 20 hydrogen bonds with many of these hydrogen bonds occurring at sequence conserved positions in the DIP-α/Dpr6 complex as well as in the homodimer complexes previously discussed (Table S3). One interaction that is present in both the DIP-θ/Dpr2 and DIP-η/Dpr4 complex, but is not seen in the DIP-α/Dpr6 or either homodimer complex, is a conserved salt bridge formed between Asp135/Asp74 on the BC loop of Dpr2/Dpr4 and Lys181/Lys94 on the C’’D loop of DIP-θ/DIP-η. Instead of this electrostatic interaction, the DIP-α/Dpr6 structure has a glycan at Asn102 that contacts His93 of DIP-α (Carrillo et al., 2015).
Differing from Dpr4, the Dpr2 protomer has a significant bend at the Ig1-Ig2 interdomain region with an 81° angle between domains compared to the 142° angle between Dpr4 domains (Figure S6C). This significant difference is possible due to the 5 residue linker in Dpr2. Comparison of inter-domain linkers among all Dprs reveal only four longer than one residue: 3 of the 6 Dprs which bind members of DIP group IV (Dpr2,3,7) and an alternate isoform of Dpr10 (Dpr10A) (Figure S6C). Since the second domain is not involved in the Ig1-Ig1 interactions seen in our crystal structures, it remains unclear what role these longer linkers have in Dpr function.
An Ig1-Ig1 superposition of the DIP-θ/Dpr2 and DIP-η/Dpr4 complex, which contains DIPs and Dprs with overlapping binding partners, has an RMSD of less than 0.5Å over 176 aligned Cα atoms (Figure 5E). Superpositions of the two new complex structures with DIP-α/Dpr6, a complex from a different DIP group with no shared binding partners, reveal RMSDs between 0.6-0.7Å over 181-185 aligned Cα carbons (Figure 5E). This indicates the topology of the heterophilic interaction to be strongly conserved among DIPs and Dprs with different binding specificities.
Specificity determinants of DIP-Dpr binding interfaces
To identify specificity residues, we aligned the Ig1 domains of DIPs and Dprs, grouping the aligned sequences based on binding preferences determined by SPR, and examined sequence conservation both within and across these specificity groups (Figures 6A and6B). Potential specificity residues – interfacial residues that are highly conserved within their binding group but not conserved across groups – were identified, and are labeled in Figures 6A and 6B.
Visual inspection of these residue positions revealed a critical region at the CC’ loop of DIP Ig1, which inserts between the CC’ and FG loops of a Dpr protomer mate (Figures 6C and 6D). Three consecutive residues that begin on the DIP CC’ loop and end at the second residue of the C’ strand, labeled S11, S12, and S13 for DIP specificity residue (Figure 6A) show high sequence variability between different DIPs. Apposing SI1-3 are Dpr residues labeled SR1 and SR2, for Dpr specificity residue, which also show significant variability between specificity groups, but conserved identity within groups (Figure 6B). SR1 is a Lys or Arg in nearly all Dprs that bind DIPs-η, -θ, -ι, -ε, and -ζ, but is conserved as hydrophobic residues Leu or Met in DIP-γ-binding Dprs, and as His in Dprs that primarily bind DIP-α, -β, or -λ. Apposing SR1 is SI2, which is a conserved Lys in DIP-α, -β and -λ, and would introduce an electrostatic clash that would prevent binding with the many non-cognate Dprs that have Lys or Arg at the SR1 position.
SR2 is located in the FG loop and directly apposes SI3. SR2 is conserved as Lys in Dprs that bind DIP-η, -θ, -ι, -κ, or -γ; Val for Dprs that primarily bind DIP-α; and is predominantly Pro in Dprs that bind DIP-ε, -ζ, or -β. The significant variability of residue types between specificity groups for this set of interfacial residues indicates that this region determines DIP/Dpr interactions through either favorable van der Waals and/or electrostatic interactions, or unfavorable clashes.
In addition to this main region, SR3 located on the BC loop of Dprs, and SI4 on the DIP C’’D loop engage in a conserved salt bridge seen in both the DIP-θ/Dpr2 and DIP-η/Dpr4 complex structures and is predicted to occur in all other Dpr complexes of DIP-η, -θ or -ι with the exception of DIP-ι/Dpr7. This salt bridge is also predicted to form in complexes between DIP-ε or DIP-ζ with 3 of the 6 Dprs within their subgroup. In place of a salt bridge, Dprs that bind DIP-α, DIP-β or DIP-λ have a conserved N-glycan at the SR3 position, which contacts a His, Asn or Leu at the DIP SI4 position (Carrillo et al., 2015). Most Dprs can either form salt bridges at this position with their cognate DIP, or have a glycan, however among Dprs that bind DIP-γ, there is little conservation at the SR3 position, indicating this position may not play a significant role in determining specificity of DIP-γ interactions. We recognize an additional residue position (PR for putative Dpr specificity residue) that could play a role in binding specificity based on its strong conservation among specificity groups and its location – residing on the Dpr FG loop between the two previously discussed regions (Figures 6B-D). It is likely that residue positions, in addition to those identified here, also play a role in specificity.
Our identification of polar and charged residues as specificity determining is in contrast to the conclusions of Carrillo et al. (2015) that shape complementarity, rather than charge complementarity and polar interactions, is responsible for DIP-Dpr specificity. These researchers focused on the conserved hydrophobic core of the interface while, our analysis in this section and mutagenesis results in the next, identify and validate specificity-determining residues at the periphery of the conserved core. Carrillo et al. (2015) carried out mutagenesis experiments on the hydrophobic core and confirmed that some of these residues affected binding affinity. However they did not design mutants that switch specificity between DIP-Dpr subgroups of the type described in the next section.
Targeted mutation of Dpr specificity residues converts binding preference in SPR
We investigated whether it was possible to change the adhesive specificity of a Dpr by mutating only a few key residues implicated in DIP-binding specificity. We chose to modify Dpr4 and Dpr6 since they are members of two distinct binding groups with no shared interactions and structural data for both of their cognate-DIP complexes was available. Proteins were produced for which the residue identities of the SR1-3 positions of Dpr4 were mutated to those of Dpr6, and vice-versa. To investigate the additive effects of these mutations, and to identify the residues that needed to be mutated in order to change binding specificity, we tested three different mutants for each Dpr.
Binding of mutants was tested against wild-type proteins in SPR over DIP-α, -η and -θ surfaces (Figures 6E and 6F; Figures S6D-E). Specificity mutant Dpr4 K82H, showed a slight increase in binding response to DIP-α, however the response is so weak that the calculated KD is over 400μM. Dpr4 K82H also showed a marginal increase in binding to DIP-η and DIP-θ. Dpr4 K82H K136V, weakened binding to DIP-η and DIP-θ by at least 4-fold compared to wild-type and binds to DIP-α with a KD of 44.9μM. This remarkable result shows that we were able to swap the binding specificity of a Dpr by only mutating two specificity residues. Dpr4 D74N A76T K82H K136V, which contained an additional mutation at an interfacial residue (A76T) to introduce the N-glycosylation motif, further decreased binding to DIP-η and DIP-θ, and increased binding affinity to DIP-α with a binding KD of 16.0μM, only about 8-fold weaker than wild-type Dpr6. This ~2.5 fold increase in affinity is the result of the N-glycan and/or the A76T mutation.
A similar result was seen when measuring affinities of the Dpr6 specificity mutants. Dpr6 H110K decreased binding by 25-fold to DIP-α, but had little effect on binding to DIP-η or DIP-θ. Dpr6 H110K V164K abolished binding to DIP-α, however no significant binding was measured between this mutant to either DIP-η or DIP-θ. Dpr6 N102D H110K V164K was able to bind to wild-type DIP-η and DIP-θ with KDs of 119μM and 72.0μM respectively, both about 2-fold weaker compared to wild-type Dpr4. Taken together, our data show that SR1, SR2, and SR3 function as specificity determinants for at least two of the DIP subgroups.
DISCUSSION
Developing axons and dendrites encounter the processes of perhaps hundreds of different neuronal cell types and must select appropriate synaptic partners from a myriad of neuronal processes. RNA sequencing technologies have revealed that developing neurons express hundreds of cell surface proteins, many of which bind in vitro to proteins known to be expressed on neighboring cells (Sarin et al., 2018; Tan et al., 2015, Xu et al., submitted). Identifying which interactions are important and understanding how their expression patterns and binding interactions contribute to the specificity, complexity and function of neural circuits remains a central challenge in developmental neuroscience.
Families of cell surface proteins with related ectodomains and differences in binding specificity provide one way of generating diverse patterns of connectivity. As opposed to Dscams and Pcdhs which are expressed stochastically to provide neurons with single cell identities that form the basis of self-avoidance (Hattori et al., 2008; Thu et al., 2014), we envision that selective recognition between synaptic partners relies on deterministic mechanisms of gene regulation to ensure the appropriate cell-type specific pairing of ligands and receptors. Indeed, it is the deterministic expression of matching DIP/Dpr pairs in some synaptic partners in the visual system that led to the idea that DIP/Dpr interactions might influence synaptic specification (Carrillo et al., 2015; Tan et al., 2015). In a previous study, we demonstrated through mRNA sequencing and genetic tagging methods that Dprs were expressed in a dynamic and complex way in developing lamina neurons (Tan et al., 2015). Each lamina neuron expresses a discrete combination of numerous Dprs. We also showed that some synaptic partners of lamina neurons, specific medulla neurons, express cognate interacting DIP proteins. Here we extended these observations through a systematic analysis of eight of the 11 DIPs using the MCFO technique. We find that of the 60 neuronal cell types we analyzed, 26 expressed a single DIP, 12 expressed two DIPs, and one expressed four DIPs (i.e. 39/60 or 65% of the neurons express at least one of the eight DIPs). Assuming the remaining three DIPs, for which gene-trap GAL4s are not yet available (i.e. DIP-ι, -κ and -λ), are expressed in a similar fashion, we estimate some ~90% of the 60 different medulla neuron types considered here express one or, less frequently, two DIPs.
By comparing the synaptic connectivity maps between lamina and medulla neurons, the expression patterns of DIPs and Dprs, and the DIP/Dpr interactome, we identified many DIP/Dpr pairs expressed in synaptic partners (Figure S7) (Xu et al, submitted). We find that lamina neurons form synapses on many different medulla neuron types; for instance, lamina L3 neurons express many Dprs and form synapses with over 10 different medulla neurons, many of which express DIPs which bind to Dprs expressed in L3. It appears then that lamina neuron outputs diverge to synapse with multiple partners. By contrast, medulla neurons express a more limited set of DIPs. For instance, Dm4 neurons only express DIP-α and form synapses with on the order of 20 L3 neurons, which express, among other Dprs, Dpr6 and Dpr10, high affinity ligands for DIP-α. L3 is by far the predominant input to Dm4. The inputs into Dm4, therefore, are convergent. Indeed, information from multiple lamina neurons of the same type frequently converge onto a single DIP-expressing Dm neuron (Nern et al., 2015; Takemura et al., 2013; S. Takemura, I. Meinertzhagen, and L. Scheffer, personal communication).
A clear pattern emerges whereby multiple Dprs on lamina neurons may promote connections to multiple targets, whereas a single DIP expressed on Dm neurons, for instance, accommodates convergence of many different neurons of the same type onto a single partner. Overall, this arrangement mirrors the interactome, where a single type of DIP tends to interact with high affinity to multiple Dprs, but in general a single Dpr exhibits high affinity binding to one type of DIP. A similar trend is seen with both Tm and TmY neurons; they typically form connections with more different types of neurons than Dms, but fewer than lamina neurons. Interestingly, about half of the Tm and TmY neurons analyzed (10/21) express more than one DIP, whereas only one of 18 Dm neurons expressed more than one DIP.
Our quantitative biophysical and structural studies enabled the identification of residues in DIPs and Dprs that control their binding specificity. Grouping DIPs and Dprs according to their cross-family binding interactions, as in Figure 6, facilitated the identification of resides at positions in the sequence that were correlated with the binding preferences of different specificity groups. Most of these specificity residues are charged or polar in contrast to the conclusion of Carrillo et al. (2015) that shape complementarity was the dominant determinant of inter-subgroup specificity. As discussed above, part of the discrepancy is due to their focus on the hydrophobic core of the interface while most distinct specificity determinants are located in the periphery.
The specificities of DIP-Dpr interactions are partially overlapping and grouped by phylogeny (Figure 4), with interaction affinities spanning approximately two orders of magnitude. Three main DIP affinity groups, and DIP-δ which forms a one-member group, emerge with cognate Dpr interactions mainly falling within a single DIP group, with sparser and weaker interactions between groups. These groupings became clear only when binding affinities were incorporated, and false positive and negative interactions removed (e.g., removal of Dpr16/Dpr17 with DIP-ε, and addition of DIP-κ and DIP-λ interactions). Quantitative binding affinities were also crucial for assigning the “primary” DIP-binding specificities [the DIP(s) with highest interaction affinity] for groups of Dprs, which we used in the identification of specificity determinants. Dprs with similar binding preferences are closely related with a few exceptions, and DIPs within each of the three main groups are also close in phylogeny (Figure 4B), with the exception of DIP-γ and DIP-κ. Indeed, single mutants in dpr6 and dpr10, which are phylogenetic nearest neighbors with similar DIP-binding profiles, show weaker phenotypes than null mutations inactivating their common binding partner, DIP-α (Xu et al., submitted).
Like other families of cell surface proteins with related ectodomains, DIPs and Dprs bind through canonical interfaces common to all family members. Since interactions between members of such diversified protein families rely on a common binding mode, many family members might be expected to bind one another, albeit with different affinities. Thus, DIP and Dpr proteins engage in promiscuous interactions, as has also been observed for other protein families implicated in targeting, e.g., type I and type II cadherins, sidekicks, nectins, syncams, and Drosophila IRM proteins (homologs of worm Syg proteins) (Bao et al., 2010; Brasch et al., 2018; Fogel et al., 2007; Goodman et al., 2016; Harrison et al., 2012; Katsamba et al., 2009). The binding properties of these protein families differ significantly from the strict homophilic recognition observed for stochastically expressed multi-domain repulsion proteins (i.e. Dscam and clustered protocadherins). These achieve recognition only when all interacting domains are matched with their cognate partners, leading to an all-or-none binding specificity (Hattori et al., 2008; Rubinstein et al., 2015). Multi-domain interfaces may be required to achieve precise fine-tuning to avoid the promiscuity that is characteristic of two-domain interfaces. In contrast, wide ranging affinities in protein families such as Dprs and DIPs may be exploited by developing neurons to sculpt neural circuitry in different ways.
Our demonstration that some DIPs and Dprs form homodimers adds another layer to the potential regulatory complexity of interactions between these proteins. DIP homodimerization affinities are in the range of 22-35μM, with Dpr homodimerization affinities ranging from 39-71μM (Table 1). The homodimerization affinity of a DIP can be significantly weaker than with its heterophilic binding to Dpr partners (DIP-α), equivalent to the strongest heterophilic interactions of its group (DIP-ζ), or stronger than its heterophilic interactions (DIP-η). For Dprs, in each case the homodimer affinities we determined were substantially weaker than their heterophilic DIP interactions. Crystal structure and mutational analyses reveal that DIP/DIP and DIP/Dpr interfaces are largely overlapping. The Dpr/Dpr dimer structure has not yet been determined. While we used AUC to identify homodimers, in principle heterophilic DIP-DIP and Dpr-Dpr interactions could also form, though we have not sought to identify such potential interactions in the current study. Indeed, Ozkan et al. (2013) detected Dpr3-Dpr7 and Dpr5-Dpr6 heterophilic interactions in their high-throughput interaction study.
In principle, some DIPs and Dprs could function in cell-cell recognition driven by homophilic rather than heterophilic interactions. In support of this possiblity, genetic rescue studies indicate that, in some contexts, homophilic interactions can substitute for heterophilic binding. For example, DIP-α overexpression in DIP-α-interacting neurons reduces Dm4 cell loss by apoptosis in Dpr6/10-null mutants (Xu et al, submitted). In some contexts, competition between homophilic and heterophilic binding partners could play a regulatory role in controlling interactions between neurons, as has been suggested for Sdks and nectins (Goodman et al., 2016; Harrison et al., 2012). Interestingly, germline knock-in mutants of a homophilic binding-deficient form of DIP-α designed in this study led to a 50% increase in synapse number for Dm4 neurons (Xu et al., submitted). These findings are consistent with the notion that complex regulatory roles may modulate DIP/Dpr interactions during circuit assembly and these, in turn, may regulate cell number and neuronal morphogenesis, as well as the distribution, number and specificity of synaptic connections (see Xu et al., submitted).
Altogether, these findings provide a firm biophysical basis for the exploration, through genetic analysis, of the role of DIP/Dpr interactions in neural circuit assembly. Moving forward we are now in a position to design DIP and Dpr mutants that abrogate, increase, or decrease homophilic and heterophilic interactions so as to allow a detailed exploration of the role of binding affinities in neural circuit assembly.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lawrence Shapiro (lss8@columbia.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster rearing and strains
Flies were reared at 25°C on standard medium. We used female flies for all MCFO studies. The following stocks were used: pBPhsFlp2::PEST;; HA_V5_FLAG (MCFO-1) (BDSC 64085), R57C10-Gal4 (BDSC 39171), and hs-Flp:PEST (BDSC 77141).
Cell lines
The FreeStyle™ 293F cell line was obtained from Thermo Fisher. Human Embryonic Kidney (HEK) 293 cell line, of which the sex is female, is the parental cell for Freestyle™ 293F. FreeStyle™ 293F cells were cultured in suspension in Freestyle™ 293 Expression medium at 37°C and 10% CO2.
Schneider 2 Cells (S2) were obtained from Expres2ion biotechnologies and derived from male late stage Drosophila melanogaster embryos. S2 cells were cultured in EX-CELL® 420 Serum-Free medium (Sigma-Aldrich) and 10% heat-inactivated fetal bovine serum (Thermo-Fisher) at 25°C.
METHOD DETAILS
Construction of transgenes and transgenic flies
DIP-GAL4 were generated from DIP-MiMIC using ΦC31 recombinase mediated cassette exchange in Hugo Bellen’s lab at Baylor College of Medicine: MI02031 (DIP-α, CG32791), MI01971 (DIP-β, CG42343), MI08287 (DIP-δ, CG34391), MI07948 (DIP-η, CG14010), MI03191 (DIP-θ, CG31646), MI03222 (DIP-γ, CG14521), MI11827 (DIP-ε, CG42368), MI03838 (DIP-ζ, CG31708).
The reference neuron images were generated using VT048653-GAL4 (TmY15) or a panneuronal driver (R57C10-Gal4). Images show resampled views (generated using Vaa3D (Peng et al., 2010) of segmented single cells together with a reference pattern (anti-Brp).
The full genotype for the 57C10 MCFO is: OL-KD (29C07-KDGeneswitch-4) in attP40; R57C10-GAL4 in attP2, tubP-KDRT>GAL80-6-KDRT> in VK00027 crossed to MCFO-1 (the genotype and the components are all described in Nern et al. (2015).
MCFO Immunohistochemistry
We crossed the MCFO-1 line with each DIP-Gal4 line. Flies with DIP-Gal4 and MCFO transgenes were raised at 25°C and receive heat-shock at 37°C for 10-20 min at mid-pupal stage, then they were dissected within two days after eclosion and the brains were stained following the MCFO immunohistochemistry protocol as described previously (Nern et al., 2015).
Colocalization Immunohistochemistry
The method is the same as in (Tan et al., 2015), lines for cell type-specific labeling are: Dm12 (R47G08-GAL4, UAS-CD8-RFP), Dm14 (R47E05-GAL4, UAS-CD8-RFP), Dm15 (R18G08-GAL4, UAS-CD8-RFP).
Microscopy and Image Analysis
Confocal images were acquired on a Zeiss LSM780 confocal microscope. The staining patterns were reproducible between samples. However, some variation on the overall fluorescence signal and noise levels existed between sections and samples. Thus, proper adjustments of laser power, detector gain, and black level settings were made to obtain similar overall fluorescence signals. Single plane or maximum intensity projection confocal images were exported into TIFF files using ImageJ software (Schindelin et al., 2012). For identification of DIP-expressing medulla neuron types, we made a pool of 60 medulla neuron types including ones that are well characterized in two large published studies (Nern et al., 2015; Takemura et al., 2013) and a few additions as indicated in Table 1. DIP-expressing cell types were identified by comparing the layer specificity and patterns of arborization of single labeled cells to the above mentioned references based on immunofluorescence staining of isolated well characterized cells, as well as Golgi staining in Fischbach and Dittrich (1989); Nern et al. (2015).
Plasmid construction and protein expression
For protein produced in human embryonic kidney cells (HEK293F), complementary DNA sequences encoding the extracellular regions listed were amplified and inserted into the mammalian expression vector VRC-8400 (Barouch et al., 2005) between the NotI and BamHI sites: Dpr1A (Tyr30-Glu261), Dpr4 (Glu32-Glu245), Dpr5A (Gln60-Glu290), Dpr6C (Trp70- Glu275), Dpr7F (Thr37-Glu269), Dpr8A (Thr39-Glu244), Dpr10D (Trp50-Glu255), Dpr11B (Leu114-Glu324), Dpr12C (Ser72-Asp285), Dpr13B (Phe171-Asp375), Dpr19A (Asp23-Glu305), Dpr20 (Arg262-Glu486), Dpr21 (Asp48-Asp253), DIP-α isoform A (Phe40-Pro341), DIP-β isoform C (Ile82-Glu408), DIP-γ (Gly22-Lys358), DIP-ε (Glu50-Ser351), DIP-ζ isoform A (Glu111-Tyr412), DIP-η isoform B (Gln31-Pro338), DIP-θ (Asp128-Pro423). For chimeras, sequences consisting of Dpr9A (Arg252-Glu362) followed by Dpr8A (Pro145-Asp244), Dpr15A (Lys188-Val290) followed by Dpr11B (Val228-Glu324), DIP-δ isoform D (Asp42-Val143) followed by DIP-ε (Pro156-Ser351), DIP-κ [CG31814] isoform A (Asp72-Val174) followed by DIP-α isoform A (Val143-Pro341) and DIP-λ [CG40378] (Ile48-Val150) followed by DIP-θ (Pro231-Pro423) were constructed and inserted into VRC-8400. All sequences were preceded by the signal sequence of human binding immunoglobulin protein BiP (MKLSLVAAMLLLLSAARA), and the kozak sequence (GCCACC). Constructs were followed by a C-terminal hexa-histidine tag. Point mutations were introduced using the QuickChange method (Agilent).
For proteins produced in S2 cells, complementary DNA sequences encoding the extracellular regions listed were amplified and inserted into a modified Expres2 vector (Expres2ion biotechnologies) between EcoRI and NotI sites: Dpr2F (Tyr103-Glu323), Dpr3B (Gln233-Glu481), Dpr14A (Thr26-Glu283), Dpr16C (Leu195-Glu451), Dpr17A (Ala403-Glu616), Dpr18 (His214-Glu478), and DIP-ι (Phe23-Ala325). EcoRI site and sequences were preceded by the signal sequence of human Binding immunoglobulin protein BiP, a 2A skip peptide(GGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAG GAGAACCCTGGACCT), EGFP and a kozak sequence (GCCGCCACC). Sequences were followed by an octa-histidine tag.
HEK293F cells were transiently transfected with expression constructs using the Polyethylenimine method (Baldi et al., 2012). For proteins produced in S2 cells, 3mL cultures of S2 cells at 2million/mL were transfected in EX-CELL 420 Serum-Free Medium (Sigma-Aldrich) in non-shaking 6 well plates. 30μL of ExpreS2 5×TR (Expres2ion Biotechnologies) was mixed with 7.5μg of transfection-grade plasmid DNA and added to the cultures. The transfected cells were supplemented with 600μL of heat inactivated fetal bovine serum (FBS) (Life technologies) 2 hours post transfection. Zeocin (Invitrogen) was added at a concentration of 2 mg/mL 24 to 48hr post transfection to begin stable line selection. Two to three days after transfection, the supernatant was changed and replaced with media containing 10% FBS and zeocin. For the following two weeks, media was either replaced or cell cultures were split in half by dilution everyday depending on cell density. GFP expression was used to monitor and select successful transfectants.
After the 2 week selection, cells were expanded to a T25 culture flask, and after reaching confluency, were further expanded to a T75 culture flask. Once confluency was reached in a T75, antibiotic selection was complete and cells were centrifuged at 300g and resuspended in EX-CELL media with 10% FBS at a cell density of 8 million/mL into a 125ml shake flask. Cultures were continually expanded to higher volumes while maintaining a cell density of 8-40mil/ml. Before final expansion to a 1L culture at 8mil/ml, cells were centrifuged at 300g and resuspended in serum-free Excell420 media. 1L cultures were grown for 5-6 days before harvesting media.
Protein purification
Conditioned media was equilibrated to 10mM Tris-HCl pH 8.0, 500mM NaCl, 3mM CaCl2 and 5mM Imidazole pH 8.0 (or 20mM Imidazole pH 8.0 for S2 conditioned media) and incubated with Ni2+ charged IMAC Sepharose 6 Fast Flow resin (GE Healthcare) for 1 hour at 25°C. Resin was washed with at least 20 column volumes of buffer containing 10mM Imidazole pH 8.0 for HEK293 produced proteins or 20-50mM Imidazole pH 8.0 for S2 produced proteins before proteins were eluted with buffer containing 90mM Imidazole pH 8.0. Gel electrophoresis with NuPage 4-12% Bis-TRIS gels (Life Technologies) was used to detect which elutions contained desired protein.
Proteins were further purified by size-exclusion chromatography (Superdex 200 HiLoad 26/60 or Superdex S200 Increase 10/300 GL; GE Healthcare) on an AKTA pure fast protein liquid chromatography system (GE Healthcare). Most proteins were stored in a buffer of 10mM Bis-Tris pH 6.6 and 150mM NaCl. The following proteins were stored in modified buffers due to stability issues: Dpr4, Dpr8, Dpr17, DIP-α, DIP-ι, DIP-κ were stored at 10mm Bis-Tris pH 6.0, 150mM NaCl; Dpr12 was at 10mM Bis-Tris pH 6.0, 500mM NaCl; Dpr15 was at 10mM Bis- Tris pH 6.0, 300mM NaCl; and DIP-η was purified at 10mM Tris-HCl pH 8.0, 150mM NaCl. UV absorbance at 280nm was used to determine protein concentration and verification of purity was determined by gel electrophoresis. Accurate molecular weights were determined through MALDI-TOF mass spectrometry at the Proteomics Shared Resource facility at Columbia University.
Sedimentation equilibrium by analytical ultracentrifugation
Experiments were performed in a Beckman XL-A/I analytical ultracentrifuge (Beckman- Coulter, Palo Alto CA, USA), utilizing six-cell centerpieces with straight walls, 12 mm path length and sapphire windows. Protein samples were dialyzed to 10mM Bis-Tris pH 6.6, 150mM NaCl with the exceptions of: Dpr15 dialyzed to 10mM Bis-Tris pH 6.0, 300mM NaCl; Dpr17 and DIP-κ were dialyzed to 10mM pH Bis-Tris 6.0, 150mM NaCl. The samples were diluted to an absorbance at 10 mm and 280 nm of 0.65, 0.43 and 0.23 in channels A, B and C, respectively. Dilution buffer were used as blank. The samples were run at four speeds. Most proteins were run at 15000, 19000, 23000, and 27000 RPM. Dpr19, DIP-γ and DIP-ε were run at 15000, 18000, 21000, and 24000 RPM; Dpr18 and DIP-β were run at 11000, 14000, 17000 and 20000 RPM. For all runs the lowest speed was held for 20hr and then four scans were taken with a 1hr interval, the second lowest held for 10hr then four scans with a 1hr interval, and the third lowest and highest speed measured as the second lowest speed. Measurements were done at 25°C, and detection was by UV at 280 nm. Solvent density and protein v-bar were determined using the program SednTerp. (Alliance Protein Laboratories) To calculate the KD and apparent molecular weight, data was fit to a global fit model, using HeteroAnalysis software package, obtained from University of Connecticut (Cole et al., 2008) (www.biotech.uconn.edu/auf).
Size exclusion chromatography with multi-angle static light scattering
Size exclusion chromatography with multi-angle static light scattering (SEC-MALS) experiments was performed using an AKTA FPLC system with a Superdex 200 Increase 10/300 GL column (GE Healthcare). Proteins were flowed in a buffer of 10mM Bis-Tris pH6.6, 150mM NaCl and at a flow rate of 0.5 mL/min. Injection volume were 100μL, sample concentration of DIP-θ was 150μM and the concentration of Dpr18 was 32μM. UV data at 280 nm was collected using the AKTA UV detector, differential refractive index with a Wyatt Optilab TRex detector and scattering data with a Wyatt DAWN Heleos-II detector (Wyatt Technology). Molecular weights were calculated using the software Astra 6.1 (Wyatt Technologies), and calculation was done using a regular Zimm-plot.
Surface Plasmon Resonance (SPR) binding experiments
SPR binding assays were performed using a Biacore T100 biosensor equipped with a Series S CM4 sensor chip (GE Healthcare). DIPs were immobilized over independent flow cells using amine-coupling chemistry in HBS-P pH 7.4 (10mM HEPES, 150mM NaCl, 0.005% (v/v) tween-20) buffer at 25°C using a flow rate of 20 μL/min. DIP-η, which was produced in a TRIS pH 8.0 buffer, was desalted into a buffer of 10mM Bis-Tris, pH 6.6, 150mM NaCl using Zeba spin desalting columns (Thermo Scientific) prior to immobilization. Dextran surfaces were activated for 7 minutes using equal volumes of 0.1M NHS (N-Hydroxysuccinimide) and 0.4M EDC (1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide). Each DIP was immobilized at ~30μg/mL in 10 mM sodium acetate, pH 5.5 until the desired immobilization level was achieved. The immobilized surface was blocked using a 4-minute injection of 1.0 M ethanolamine, pH 8.5. Typical immobilization levels ranged between 700-900 RU. In each experiment, a BSA-immobilized surface was used as a reference control to remove bulk refractive index shifts. BSA was immobilized using a similar amine-coupling protocol with the exception of diluting it into 10mM sodium acetate, pH 4.25 and injecting it over the activated surface for 3 minutes.
All binding experiments were performed at 25°C in a running buffer of 10mM Tris-HCl, pH 7.2, 150mM NaCl, 1mM EDTA, 1 mg/mL BSA and 0.01% (v/v) Tween-20. For the DIP-Dpr interactome experiments, each of the 21 Dprs was prepared and tested in running buffer using a three-fold dilution series at nine concentrations ranging from 81 to 0.012 μM, with some exceptions: Dprs 8, 9 and 21 binding over DIP-β and Dpr12 binding over DIP-δ were tested at seven concentrations ranging from 9 to 0.012 μM while Dprs 6 and 10 binding over DIP-α, Dprs 15 and 17 binding over DIP-γ, Dprs 15 binding over DIP-ε, Dpr 17 binding over DIP-η, DIP-θ and DIP-ι, Dpr 7 binding over DIP-κ, and Dprs 8 and 9 binding over DIP-λ, were tested at eight concentrations ranging from 27-0.012 μM. During a binding cycle, the association phase was monitored for 30 seconds followed by 120-second dissociation phase, each at 50μL/min. The last step was a buffer wash injection at 100μL/min for 60 seconds. Running buffer “blanks” were used instead of a Dpr analyte sample every 3 binding cycles to double-reference the binding signals by removing systematic noise and instrument drift. Each Dpr analyte series was tested in order of increasing concentration separated by two buffer analyte binding cycles and a repeat of the same Dpr analyte series in order of increasing concentration to determine the reproducibility of the experiment. The responses between 25 and 29 seconds were plotted against the Dpr concentration and fit to an 1:1 interaction model to calculate the KD (Cooper, 2009). The binding reaction for each Dpr/DIP interaction was fitted to an independent Rmax. The data was processed and analyzed using Scrubber 2.0 (BioLogic Software).
For Figures 5F-G, DIP-α and its mutants I83D and A78K N94D were immobilized at 900-930 RU on independent flow cells. Dpr6 was tested at 8 concentrations ranging between 9-0.004 μM and Dpr10 was tested at 9 concentrations ranging between 27-0.004 μM respectively over all three surfaces. Dpr10 Y103D was also tested at the same concentration range as wild-type Dpr10 over the wild-type DIP-α surface only. The binding analysis was performed under similar conditions as described for the interactome except a longer association phase of 40s was used. In these experiments, the responses between 35 and 39 seconds were plotted against the Dpr concentration and fit to an 1:1 interaction model to calculate the KD.
For Figures 6E-F and S6D-E, DIP-η, DIP-α and DIP-θ were immobilized over independent flow cells at 770-900 RU. All Dpr4 and Dpr6 analytes used in this experiment were tested at 8 concentrations ranging from 81 to 0.037 μM using the same experimental conditions as previously described for the DIP-α binding assay shown in Figures 5F-G.
Crystallization and structure determination
Sparse matrix screening was performed in sitting drop assays at 22°C. For crystallization of complexes, a 1:1 volume ratio of Dpr and DIP purified protein samples was mixed and incubated on ice for at least 30 minutes before added to crystallization conditions. For DIP-η/Dpr4 crystals, the 1:1 volume ratio was of 12mg/mL (332μM) DIP-η with 11mg/ml (449μM) of Dpr4 and for DIP-θ/ Dpr2 crystals, 12.8mg/mL (377μM) of DIP-θ with 9.5mg/ml (371μM) of Dpr2. DIP-θ was crystallized at 12.8mg/mL concentration and DIP-α at an 8.8mg/mL concentration. 96 well sitting drop assays were set up using a Mosquito robotic crystallization system (TTP lab tech) with 200nl drop volumes consisting of 100nl of protein mix and 100nl of screening condition. Crystallization drops were incubated and imaged using an automated Rock Imager (Formulatrix).
Successful hits were further optimized in 24 well plates using a vapor diffusion method with 1-2μl hanging drops at 22°C. Protein mix to mother liquor ratios of 1:1, 2:1, and 1:2 ratios mix was tested during optimization with a 2:1 ratio usually producing better crystals. DIP-η/ Dpr4 crystallized in conditions from the Morpheus screen (Molecular Dimensions) and the condition that produced diffracting crystals was: 28% ethylene glycol-PEG8000 (Molecular Dimensions), 0.1M Morpheus Buffer 2 pH 7.5 (Molecular Dimensions), and 10% Morpheus Carboxylic Acid mix (Molecular Dimensions). DIP-θ/Dpr2 crystals were grown in 18% PEG 3350, 0.2M TriNH4 Citrate pH 6.5. DIP-α crystals were grown in 2% PEG3350, 17% Tacsimate pH 7.0 (Hampton Research), 0.1M Hepes pH 7.0. DIP-θ crystals were grown in 12% PEG4000, 0.3M AmSO4, 0.08M Sodium Acetate pH 4.6.
Crystals were harvested using nylon loops of various sizes mounted to metal bases (Hampton Research) and were transferred and immersed in a cryoprotectant before being flash frozen in liquid nitrogen for long term storage and data collection. Cryoprotectants consisted of the crystallization condition with an additional 15% (2R,3R)-(−)-2,3-Butanediol (Sigma-Aldrich) for DIP-θ/Dpr2 or an additional 30% glycerol for DIP-α and DIP-θ crystals. DIP-η/Dpr4 crystals were flash frozen in crystal mother liquor.
X-ray diffraction data was collected from single crystals at 100K at Northeastern Collaborative Access Team (NE-CAT) beamlines 24ID-C and 24ID-E at the Advanced Photon Source, Argonne National Laboratory. All datasets were processed using XDS (Kabsch, 2010) and AIMLESS (Evans and Murshudov, 2013) as part of the CCP4 suite (Winn et al., 2011).
All structures were solved by molecular replacement using PHASER (McCoy et al., 2007) in the Phenix suite (Adams et al., 2010). For the DIP-η/Dpr4 complex structure, Dpr6 and DIP-α domains from 5EO9 was used as a model for Dpr4 Ig1 and DIP-η Ig1-Ig2. An ensemble of models including SIRP alpha (4CMM) and Sdk1EC4 (5K6U) was used to model DIP-η Ig3 and Dpr4 Ig2. The DIP-α structure was solved using domains from (5EO9) and its Ig3 domain was modelled with DIP-η Ig3. The DIP-θ/Dpr2 complex was solved using models derived from the DIP-η/Dpr4 structure and the DIP-θ structure was solved using DIP-θ Ig1-3 from the DIP-θ/Dpr2 complex. Structures were refined by iterative rounds of model building in Coot (Emsley et al., 2010), automated refinement using phenix.refine (Afonine et al., 2012) and additional refinements using PDB-REDO (Joosten et al., 2014). Ig1 complex super position RMSDs were calculated using Pymol (Schrödinger). Chimera (Pettersen et al., 2004) was used to calculate angles between Dpr domains. PISA (Krissinel and Henrick, 2007) was used to determine hydrogen bonding pairs and buried surface area in complex interfaces.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics of x-ray diffraction datasets and crystal structures are reported in Table S2 and were determined using AIMLESS (Evans and Murshudov, 2013) and Phenix (Afonine et al., 2012). Phylogenetic trees of Dprs and DIPs were based on Ig1 domain similarity and generated using PHYLIP (http://evolution.genetics.washington.edu/phylip.html).
DATA AND SOFTWARE AVAILABILITY
Coordinates and structural factors for DIP-α Ig1-3, DIP-θ Ig1-3, Dpr4 Ig1-2/DIP-η Ig1-3, and Dpr2 Ig1-2/DIP-θ Ig1-3 are available from the Protein Data Bank under accession codes PDB: 6EFY, PDB: 6EFZ, PDB: 6EG0 and PDB: 6EG1 respectively.
Supplementary Material
Figure S1: Additional examples of identifying single labeled medulla neurons in densely labeled samples, Related to Figures 1,2. MCFO images (A-G) were compared to labeled cells in reference images (A’-G’). The morphologies of some of these cell types were not shown in the Golgi stains in Fischbach and Dittrich (1989). (A, A’) A green TmY15 was identified in DIP-δ MCFO by comparing A with A’. Note that this cell overlaps with Tm5c, as shown in Figure 1J. Also note that the width of TmY15 arborization in lobula and lobula plate in A is not as wide as in A’. We did not take the full stack containing this cell, due to processes of other neurons which overlap with TmY15 in the maximum intensity projection. (B, B’) A red Lpi34 was identified in a DIP-δ MCFO image by comparing B with B’. Note that another red cell has a red axon projecting into the lobula in close proximity to Lpi34 in this preparation. (C, C’) A green TmY4 is identified in DIP-γ MCFO in C by comparing it to C’. Note that there are multiple green labeled cells in close proximity to this TmY4. Nevertheless, TmY4 can be identified on the basis of its arbor pattern in the medulla, lobula and lobula plate. (D, D’) A green TmY9 is identified in DIP-γ in the MCFO images by comparing D with D’. In D, triangles in the medulla and lobula plate point to branches in each neuropil; triangle in lobula points to its axon terminal. (E, E’) A green TmY14 is identified in DIP-γ MCFO images (E) by comparing it to E’. The five triangles in each of the two panels point to the same parts of the neurons in each image. (F, F’) A red TmY13 was identified in DIP-η MCFO images by comparing F with F’. The four triangles in each of the two panels point to the same parts of the neurons in each image. (G, G’) A red Mi10 was identified in DIP-η MCFO images by comparing G with G’. The two triangles in each of the two panels point to the same parts of the neurons in each image. Scale bar: 10μm.
Figure S2: MCFO images of medulla neuron types expressing DIP-α, -β, -γ, -δ, -η and -θ, Related to Figures 1,2. The neuropil is labeled by nc82, an antibody which recognizes a synaptic protein. The names of the cell types are in the same color as the single cells labeled in the images, indicated with arrows and triangles. (A) DIP-α MCFO labels Dm4 (green; arrows). (B) DIP-β MCFO labels C2 neurons (green; triangles and arrow (right)). (C) DIP-β MCFO also labels Lawf1 (green) and Lawf2 (green; arrows) and TmY14 (green; triangles). (D) DIP-γ MCFO labels Tm5a (green; triangles). (E) DIP-γ MCFO labels three Dm8 neurons (green). (F) DIP-γ MCFO labels Tm20 (green; middle column in the medulla and lobula). C3 (red; triangles) is also labeled. (G) DIP-γ MCFO labels Mi4 (green). (H) DIP-δ MCFO labels one green and one red Pm4 neuron. (I) DIP-δ MCFO labels Dm17. (J) DIP-η MCFO labels Tm3 (red; triangles) and Tm2 (green). (K) DIP-η MCFO labels Tm5c (red; triangles). (L) DIP-η MCFO labels Mi9 (red; triangles). (M) DIP-θ MCFO labels Tm5b (red; triangles). (N) DIP-θ MCFO labels Dm15 (magenta) and TmY3 (green). (O) DIP-θ MCFO labels Dm15 (green) and Tm5a (green; triangles). (P) DIP-θ MCFO labels Tm20 (red; triangles). Scale bar: 10μm.
Figure S3: MCFO images of medulla neuron types expressing DIP-θ, -ζ, -ε, Related to Figures 1,2 (A-K), and co-localization images for DIP-α and DIP-δ (L-N). (A-K) The neuropil structure is labeled by nc82, an antibody to a synaptic protein. Cell type names are the same color as the staining highlighting cell morphology. (A) DIP-θ MCFO labels TmY3 (green) and Y3 (red; triangles). (B) DIP-ζ MCFO labels TmY10 (red) and Pm2 (magenta; arrow) which overlaps with the TmY10 in the proximal medulla. (C) DIP-ε MCFO labels Dm18 (green; arrow) and Dm13 (pink; arrow). (D) DIP-ε MCFO labels Dm13 (green). (E) DIP-ε MCFO labels Tm9 (green; triangles). (F) DIP-ε MCFO labels Tm16 (green; triangles). Note that the soma is not shown. (G) DIP-ε MCFO labels Tm20 (green; triangles). (H) DIP-ε MCFO labels Tm5Y (green; triangles). Note that the soma is not shown. (I) DIP-ε MCFO labels TmY9 (green; triangles). (J) DIP-ε MCFO labels Dm13 (blue) and TmY10 (champagne; triangles). (K) DIP-ε MCFO labels TmY13 (green; triangles). Scale bar: 10μm. (L-N) Co-localization images for DIP-α and DIP-δ protein traps and Dm-type specific markers. These images are used for Dm12, Dm14 and Dm15 here because of the difficulty in isolating single labeled neurons using MCFO. (L) DIP-α is expressed in Dm12 (R47G08-GAL4, UAS-CD8-RFP), as indicated by the colocalization in the soma. (M) DIP-δ is expressed in Dm14 (R47E05-GAL4, UAS-CD8-RFP). (N) DIP-δ is expressed in Dm15 (R18G08-GAL4, UAS-CD8-RFP). Scale bar: 10μm.
Figure S4: Biophysical characterizaion of DIP homophilic and DIP/Dpr heterophilic interactions, Related to Table 1 and Figure 3.
(A) AUC sedimentation equilibrium analysis of DIP-α and Dpr21. The blue line in the upper panels represents the global fit to the data. Residual fitting errors are indicated in the lower panel.
(B) SEC-MALS experiments for DIP-θ and Dpr18. Plots show proteins peak as refractive index changes (RI, relative scale) against elution time (min). Grey dotted line shows monomer molecular weight determined by MALDI and apparent molecular weight determined from light scattering analysis is plotted in blue at peak position. (C) Each row shows SPR sensorgrams of 21 Dpr analytes binding over an individual DIP-immobilized surface. Binding signals representing the seven strongest Dpr binders are shown in individual panels and the remaining 14 Dprs are shown as an overlay with each color representing a single Dpr. Sensorgrams of interactions with KDs lower than 200μM are enclosed within the red box.
Figure S5: Equilibrium-binding analysis of DIP-Dpr interactions using SPR, Related to Figure 3.
Binding isotherms for Dpr analytes binding to individual DIP-immobilized surfaces. Each panel shows the fit of the binding data to an 1:1 binding model to calculate the equilibrium binding KDs for 21 Dprs binding to the DIP noted in the top left. Dprs with KDs lower than 200μM are labeled. The KDs are tabulated in Figure 3B.
Figure S6: Crystal structures of DIP-θ homodimer, DIP-θ/Dpr2, and DIP-θ SPR analysis of Dpr4/Dpr6 specificity mutants, Related to Figures 5, 6.
(A) & (B) Structural details of DIP-homodimer (blue and light blue protomers) and DIP-θ/Dpr2 complex (DIP-θ blue and Dpr2 in yellow). Side-chains contributing to the interface are shown as sticks with residues contributing to hydrophobic core underlined. (C) Ig1 superposition of Dpr2 (yellow) and Dpr4 (green) highlights the difference in interdomain angles made possible by the longer interdomain linker of Dpr2. Alignments of Dprs reveal only four Dprs have interdomain linkers longer than 1 amino acid residue. (D) and (E) SPR sensorgrams of different Dpr4 and Dpr6 SR mutants used as analytes over a DIP-θ immobilized surfaces. Labels indicate which SR position(s) were mutated for Dpr4 and Dpr6. The number in brackets represents the error of the fit.
Figure S7: Convergence and divergence of L3 and its synaptic partners, Related to Figure 2. (A) Multiple L3 neurons are presynaptic to one Dm4. L3 expresses Dpr6 and Dpr10 that can interact with DIP-α expressed in Dm4. (B) A single L3 is presynaptic to 10 different medulla neuron types in each column. Seven of the ten medulla neuron types express one, two or four DIPs that interact with Dprs expressed in L3 (shown along the arrow). DIP expressing cell types are identified in MCFO experiments, Dpr expression in L3 are from RNA seq data at 40 hrs APF (Tan et al., 2015). Reads per Kilobase Million (RPKM) ranges of Dprs expressed in L3 that interacts with the DIPs expressed in synaptic partners are also shown on the side of L3 dendrites.
Table S1: Additional analytical ultracentrifugation analysis of DIPs and Dprs, Related to Table 1
Table S2: Crystallographic statistics, Related to Figure 5
Table S3: Hydrogen bonds in DIP homodimers and DIP-Dpr complexes, Related to Figure 5
Highlights.
Most neurons in the Drosophila medulla express a single DIP
Some DIPs and some Dprs form homodimers
Quantified binding affinities for all DIP/Dpr interactions
Full-ectodomain crystal structures of DIP/Dpr and DIP/DIP complexes
ACKNOWLEDGEMENTS
We thank Surajit Banerjee and Igor Kourinov for help with synchrotron data collection at the APS NE-CAT 24-ID-C/E beamlines, supported by NIH P41GM103403. This work was supported by the Training Program in Neural Microcircuits from National Institute of Neurological Disease and Stroke (T32NS058280) (S.X.), NIH grant R01GM067858 (H.B.), the Harold and Leila Y. Mathers Foundation (S.L.Z.), and US National Science Foundation grant MCB-1412472 (B.H.). The Microscopy Core at Baylor College of Medicine is supported by IDDRC (U54 HD083092) from National Institute of Child Health and Human Development. H.B., B.H. and S.L.Z. are Investigators of the Howard Hughes Medical Institute
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi L, Hacker DL, Meerschman C, and Wurm FM (2012). Large-scale transfection of mammalian cells. Methods Mol Biol 801, 13–26. [DOI] [PubMed] [Google Scholar]
- Bao S, Fischbach KF, Corbin V, and Cagan RL (2010). Preferential adhesion maintains separation of ommatidia in the Drosophila eye. Dev Biol 344, 948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Yang ZY, Kong WP, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, et al. (2005). A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J Virol 79, 8828–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J, Katsamba PS, Harrison OJ, Ahlsen G, Troyanovsky RB, Indra I, Kaczynska A, Kaeser B, Troyanovsky S, Honig B, et al. (2018). Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior. Cell Rep 23, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell KM, Sollner C, Schuster-Boeckler B, Bateman A, and Wright GJ (2008). Large- scale screening for novel low-affinity extracellular protein interactions. Genome Res 18, 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo RA, Ozkan E, Menon KP, Nagarkar-Jaiswal S, Lee PT, Jeon M, Birnbaum ME, Bellen HJ, Garcia KC, and Zinn K (2015). Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell 163, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Lary JW,T, P.M., and Laue TM (2008). Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol 84, 143–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA (2009). Label-free biosensors : techniques and applications (Cambridge ; New York: Cambridge University Press; ). [Google Scholar]
- Davis FP, Nern A, Picard S, Reiser MB, Rubin GM, Eddy SR, and Henry GL (2018). A genetic, genomic, and computational resource for exploring neural circuit function. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, and Sanes JR (2014). Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF, and Dittrich APM (1989). The Optic Lobe of Drosophila-Melanogaster .1. A Golgi Analysis of Wild-Type Structure. Cell Tissue Res 258, 441–475. [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, and Biederer T (2007). SynCAMs organize synapses through heterophilic adhesion. J Neurosci 27, 12516–12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. (2008). The neural substrate of spectral preference in Drosophila. Neuron 60, 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KM, Yamagata M, Jin X, Mannepalli S, Katsamba PS, Ahlsen G, Sergeeva AP, Honig B, Sanes JR, and Shapiro L (2016). Molecular basis of sidekick-mediated cell-cell adhesion and specificity. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Vendome J, Brasch J, Jin X, Hong S, Katsamba PS, Ahlsen G, Troyanovsky RB, Troyanovsky SM, Honig B, et al. (2012). Nectin ectodomain structures reveal a canonical adhesive interface. Nat Struct Mol Biol 19, 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, and Zipursky SL (2008). Dscam-mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol 24, 597–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, and Nikolov DB (2003). Eph receptors and ephrins. Int J Biochem Cell Biol 35, 130–134. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, and Thornton JM (1992). The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Joosten RP, Long F, Murshudov GN, and Perrakis A (2014). The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. (2009). Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci U S A 106, 11594–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, and Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Mauss AS, Pankova K, Arenz A, Nern A, Rubin GM, and Borst A (2015). Neural Circuit to Integrate Opposing Motions in the Visual Field. Cell 162, 351–362. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A 112, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe N, Shimizu K, Ozaki-Kuroda K, Nakanishi H, Morimoto K, Takeuchi M, Katsumaru H, Murakami F, and Takai Y (2004). Contacts between the commissural axons and the floor plate cells are mediated by nectins. Dev Biol 273, 244–256. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, and Garcia KC (2013). An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan E, Chia PH, Wang RR, Goriatcheva N, Borek D, Otwinowski Z, Walz T, Shen K, and Garcia KC (2014). Extracellular architecture of the SYG-1/SYG-2 adhesion complex instructs synaptogenesis. Cell 156, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Chen CP, Bahna F, Honig B, and Shapiro L (2003). Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol 13, 690–698. [DOI] [PubMed] [Google Scholar]
- Peng H, Ruan Z, Long F, Simpson JH, and Myers EW (2010). V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol 28, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, Ahlsen G, Chevee M, Halim A, Clausen H, et al. (2015). Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell 163, 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Zuniga-Sanchez E, Kurmangaliyev YZ, Cousins H, Patel M, Hernandez J, Zhang KX, Samuel MA, Morey M, Sanes JR, et al. (2018). Role for Wnt Signaling in Retinal Neuropil Development: Analysis via RNA-Seq and In Vivo Somatic CRISPR Mutagenesis. Neuron 98, 109–126 e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold C, and Jones EY (2013). Structural insights into semaphorins and their receptors. Semin Cell Dev Biol 24, 139–145. [DOI] [PubMed] [Google Scholar]
- Sperry RW (1963). Chemoaffinity in the Orderly Growth of Nerve Fiber Patterns and Connections. Proc Natl Acad Sci U S A 50, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JD, Chervin AS, and Kranz DM (2009). T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology 126, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. (2013). A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Nern A, Chklovskii DB, Scheffer LK, Rubin GM, and Meinertzhagen A (2017). The comprehensive connectome of a neural substrate for ‘ON’ motion detection in Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zhang KX, Pecot MY, Nagarkar-Jaiswal S, Lee PT, Takemura SY, McEwen M, Nern A, Xu S, Tadros W, et al. (2015). Ig Superfamily Ligand and Receptor Pairs Expressed in Synaptic Partners in Drosophila. Cell 163, 1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, and Maniatis T (2014). Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell 158, 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Miyoshi J, Honda T, Sakisaka T, Takai Y, and Takeichi M (2006). Interneurite affinity is regulated by heterophilic nectin interactions in concert with the cadherin machinery. J Cell Biol 174, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, and Reiser MB (2013). Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, Kaczynska A, et al. (2014). Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proc Natl Acad Sci U S A 111, E4175–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2008). Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451, 465–469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2012). Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci 32, 14402–14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, and Sanes JR (2002). Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110, 649–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Additional examples of identifying single labeled medulla neurons in densely labeled samples, Related to Figures 1,2. MCFO images (A-G) were compared to labeled cells in reference images (A’-G’). The morphologies of some of these cell types were not shown in the Golgi stains in Fischbach and Dittrich (1989). (A, A’) A green TmY15 was identified in DIP-δ MCFO by comparing A with A’. Note that this cell overlaps with Tm5c, as shown in Figure 1J. Also note that the width of TmY15 arborization in lobula and lobula plate in A is not as wide as in A’. We did not take the full stack containing this cell, due to processes of other neurons which overlap with TmY15 in the maximum intensity projection. (B, B’) A red Lpi34 was identified in a DIP-δ MCFO image by comparing B with B’. Note that another red cell has a red axon projecting into the lobula in close proximity to Lpi34 in this preparation. (C, C’) A green TmY4 is identified in DIP-γ MCFO in C by comparing it to C’. Note that there are multiple green labeled cells in close proximity to this TmY4. Nevertheless, TmY4 can be identified on the basis of its arbor pattern in the medulla, lobula and lobula plate. (D, D’) A green TmY9 is identified in DIP-γ in the MCFO images by comparing D with D’. In D, triangles in the medulla and lobula plate point to branches in each neuropil; triangle in lobula points to its axon terminal. (E, E’) A green TmY14 is identified in DIP-γ MCFO images (E) by comparing it to E’. The five triangles in each of the two panels point to the same parts of the neurons in each image. (F, F’) A red TmY13 was identified in DIP-η MCFO images by comparing F with F’. The four triangles in each of the two panels point to the same parts of the neurons in each image. (G, G’) A red Mi10 was identified in DIP-η MCFO images by comparing G with G’. The two triangles in each of the two panels point to the same parts of the neurons in each image. Scale bar: 10μm.
Figure S2: MCFO images of medulla neuron types expressing DIP-α, -β, -γ, -δ, -η and -θ, Related to Figures 1,2. The neuropil is labeled by nc82, an antibody which recognizes a synaptic protein. The names of the cell types are in the same color as the single cells labeled in the images, indicated with arrows and triangles. (A) DIP-α MCFO labels Dm4 (green; arrows). (B) DIP-β MCFO labels C2 neurons (green; triangles and arrow (right)). (C) DIP-β MCFO also labels Lawf1 (green) and Lawf2 (green; arrows) and TmY14 (green; triangles). (D) DIP-γ MCFO labels Tm5a (green; triangles). (E) DIP-γ MCFO labels three Dm8 neurons (green). (F) DIP-γ MCFO labels Tm20 (green; middle column in the medulla and lobula). C3 (red; triangles) is also labeled. (G) DIP-γ MCFO labels Mi4 (green). (H) DIP-δ MCFO labels one green and one red Pm4 neuron. (I) DIP-δ MCFO labels Dm17. (J) DIP-η MCFO labels Tm3 (red; triangles) and Tm2 (green). (K) DIP-η MCFO labels Tm5c (red; triangles). (L) DIP-η MCFO labels Mi9 (red; triangles). (M) DIP-θ MCFO labels Tm5b (red; triangles). (N) DIP-θ MCFO labels Dm15 (magenta) and TmY3 (green). (O) DIP-θ MCFO labels Dm15 (green) and Tm5a (green; triangles). (P) DIP-θ MCFO labels Tm20 (red; triangles). Scale bar: 10μm.
Figure S3: MCFO images of medulla neuron types expressing DIP-θ, -ζ, -ε, Related to Figures 1,2 (A-K), and co-localization images for DIP-α and DIP-δ (L-N). (A-K) The neuropil structure is labeled by nc82, an antibody to a synaptic protein. Cell type names are the same color as the staining highlighting cell morphology. (A) DIP-θ MCFO labels TmY3 (green) and Y3 (red; triangles). (B) DIP-ζ MCFO labels TmY10 (red) and Pm2 (magenta; arrow) which overlaps with the TmY10 in the proximal medulla. (C) DIP-ε MCFO labels Dm18 (green; arrow) and Dm13 (pink; arrow). (D) DIP-ε MCFO labels Dm13 (green). (E) DIP-ε MCFO labels Tm9 (green; triangles). (F) DIP-ε MCFO labels Tm16 (green; triangles). Note that the soma is not shown. (G) DIP-ε MCFO labels Tm20 (green; triangles). (H) DIP-ε MCFO labels Tm5Y (green; triangles). Note that the soma is not shown. (I) DIP-ε MCFO labels TmY9 (green; triangles). (J) DIP-ε MCFO labels Dm13 (blue) and TmY10 (champagne; triangles). (K) DIP-ε MCFO labels TmY13 (green; triangles). Scale bar: 10μm. (L-N) Co-localization images for DIP-α and DIP-δ protein traps and Dm-type specific markers. These images are used for Dm12, Dm14 and Dm15 here because of the difficulty in isolating single labeled neurons using MCFO. (L) DIP-α is expressed in Dm12 (R47G08-GAL4, UAS-CD8-RFP), as indicated by the colocalization in the soma. (M) DIP-δ is expressed in Dm14 (R47E05-GAL4, UAS-CD8-RFP). (N) DIP-δ is expressed in Dm15 (R18G08-GAL4, UAS-CD8-RFP). Scale bar: 10μm.
Figure S4: Biophysical characterizaion of DIP homophilic and DIP/Dpr heterophilic interactions, Related to Table 1 and Figure 3.
(A) AUC sedimentation equilibrium analysis of DIP-α and Dpr21. The blue line in the upper panels represents the global fit to the data. Residual fitting errors are indicated in the lower panel.
(B) SEC-MALS experiments for DIP-θ and Dpr18. Plots show proteins peak as refractive index changes (RI, relative scale) against elution time (min). Grey dotted line shows monomer molecular weight determined by MALDI and apparent molecular weight determined from light scattering analysis is plotted in blue at peak position. (C) Each row shows SPR sensorgrams of 21 Dpr analytes binding over an individual DIP-immobilized surface. Binding signals representing the seven strongest Dpr binders are shown in individual panels and the remaining 14 Dprs are shown as an overlay with each color representing a single Dpr. Sensorgrams of interactions with KDs lower than 200μM are enclosed within the red box.
Figure S5: Equilibrium-binding analysis of DIP-Dpr interactions using SPR, Related to Figure 3.
Binding isotherms for Dpr analytes binding to individual DIP-immobilized surfaces. Each panel shows the fit of the binding data to an 1:1 binding model to calculate the equilibrium binding KDs for 21 Dprs binding to the DIP noted in the top left. Dprs with KDs lower than 200μM are labeled. The KDs are tabulated in Figure 3B.
Figure S6: Crystal structures of DIP-θ homodimer, DIP-θ/Dpr2, and DIP-θ SPR analysis of Dpr4/Dpr6 specificity mutants, Related to Figures 5, 6.
(A) & (B) Structural details of DIP-homodimer (blue and light blue protomers) and DIP-θ/Dpr2 complex (DIP-θ blue and Dpr2 in yellow). Side-chains contributing to the interface are shown as sticks with residues contributing to hydrophobic core underlined. (C) Ig1 superposition of Dpr2 (yellow) and Dpr4 (green) highlights the difference in interdomain angles made possible by the longer interdomain linker of Dpr2. Alignments of Dprs reveal only four Dprs have interdomain linkers longer than 1 amino acid residue. (D) and (E) SPR sensorgrams of different Dpr4 and Dpr6 SR mutants used as analytes over a DIP-θ immobilized surfaces. Labels indicate which SR position(s) were mutated for Dpr4 and Dpr6. The number in brackets represents the error of the fit.
Figure S7: Convergence and divergence of L3 and its synaptic partners, Related to Figure 2. (A) Multiple L3 neurons are presynaptic to one Dm4. L3 expresses Dpr6 and Dpr10 that can interact with DIP-α expressed in Dm4. (B) A single L3 is presynaptic to 10 different medulla neuron types in each column. Seven of the ten medulla neuron types express one, two or four DIPs that interact with Dprs expressed in L3 (shown along the arrow). DIP expressing cell types are identified in MCFO experiments, Dpr expression in L3 are from RNA seq data at 40 hrs APF (Tan et al., 2015). Reads per Kilobase Million (RPKM) ranges of Dprs expressed in L3 that interacts with the DIPs expressed in synaptic partners are also shown on the side of L3 dendrites.
Table S1: Additional analytical ultracentrifugation analysis of DIPs and Dprs, Related to Table 1
Table S2: Crystallographic statistics, Related to Figure 5
Table S3: Hydrogen bonds in DIP homodimers and DIP-Dpr complexes, Related to Figure 5