Abstract
BACKGROUND
Allogeneic hematopoietic stem-cell transplantation is the only curative treatment for patients with myelodysplastic syndrome (MDS). The molecular predictors of disease progression after transplantation are unclear.
METHODS
We sequenced bone marrow and skin samples from 90 adults with MDS who underwent allogeneic hematopoietic stem-cell transplantation after a myeloablative or reduced-intensity conditioning regimen. We detected mutations before transplantation using enhanced exome sequencing, and we evaluated mutation clearance by using error-corrected sequencing to genotype mutations in bone marrow samples obtained 30 days after transplantation. In this exploratory study, we evaluated the association of a mutation detected after transplantation with disease progression and survival.
RESULTS
Sequencing identified at least one validated somatic mutation before transplantation in 86 of 90 patients (96%); 32 of these patients (37%) had at least one mutation with a maximum variant allele frequency of at least 0.5% (equivalent to 1 heterozygous mutant cell in 100 cells) 30 days after transplantation. Patients with disease progression had mutations with a higher maximum variant allele frequency at 30 days than those who did not (median maximum variant allele frequency, 0.9% vs. 0%; P<0.001). The presence of at least one mutation with a variant allele frequency of at least 0.5% at day 30 was associated with a higher risk of progression (53.1% vs. 13.0%; conditioning regimen–adjusted hazard ratio, 3.86; 95% confidence interval [CI], 1.96 to 7.62; P<0.001) and a lower 1-year rate of progression-free survival than the absence of such a mutation (31.3% vs. 59.3%; conditioning regimen–adjusted hazard ratio for progression or death, 2.22; 95% CI, 1.32 to 3.73; P = 0.005). The rate of progression-free survival was lower among patients who had received a reduced-intensity conditioning regimen and had at least one persistent mutation with a variant allele frequency of at least 0.5% at day 30 than among patients with other combinations of conditioning regimen and mutation status (P≤0.001). Multivariate analysis confirmed that patients who had a mutation with a variant allele frequency of at least 0.5% detected at day 30 had a higher risk of progression (hazard ratio, 4.48; 95% CI, 2.21 to 9.08; P<0.001) and a lower 1-year rate of progression-free survival than those who did not (hazard ratio for progression or death, 2.39; 95% CI, 1.40 to 4.09; P = 0.002).
CONCLUSIONS
The risk of disease progression was higher among patients with MDS in whom persistent disease–associated mutations were detected in the bone marrow 30 days after transplantation than among those in whom these mutations were not detected. (Funded by the Leukemia and Lymphoma Society and others.)
PATIENTS WITH MYELODYSPLASTIC SYN-drome (MDS), the most common myeloid cancer in adults in the United States, have highly variable outcomes. Allogeneic hematopoietic stem-cell transplantation is the only curative therapy, but disease progression after transplantation remains a problem. Identification of individualized prognostic risk factors for progression of MDS after transplantation could allow for early initiation of preventive or salvage treatments to improve outcomes.
Before transplantation, prognostic risk factors that are associated with the outcomes of MDS include the patient’s age and performance status, the percentage of blast cells in bone marrow, detection of MDS cells with the use of multiparameter flow cytometry, and the presence of cytopenias, cytogenetic abnormalities, and specific gene mutations.1–9 In addition, the use of a reduced-intensity conditioning regimen has been associated with a risk of relapse of MDS after transplantation that is higher than that associated with a myeloablative regimen.10 Identification of patients who have received a reduced-intensity conditioning regimen and who are at highest risk for disease progression could help prioritize patients who are most likely to benefit from maintenance therapy after allogeneic hematopoietic stem-cell transplantation.
Studies have shown that residual disease detected after transplantation with the use of morphologic analysis, the presence of mixed chimerism, and transcripts detected by means of quantitative polymerase-chain-reaction (PCR) assay are all associated with a risk of relapse of MDS.11–14 Monitoring measurable residual disease immediately after transplantation may have a greater advantage than testing before transplantation, because tumor cells detected after treatment indicate both the cell-intrinsic biologic properties of a tumor and its response to chemotherapy.
Next-generation sequencing to monitor for measurable residual disease by detecting and quantifying mutations provides an objective, tumor-specific biomarker for tumor burden in hematologic cancers. This testing is especially valuable in MDS, in which the fraction of tumor cells present in a sample is frequently underestimated when the percentage of blast cells in the bone marrow is determined with the use of morphologic analysis.15,16 In this exploratory study, we detected residual tumor cells 30 days and 100 days after transplantation with analysis of gene mutations. Our objective was to determine whether the persistence of cells with MDS-associated mutations in the early period after allogeneic hematopoietic stem-cell transplantation was associated with outcomes.
METHODS
PATIENTS
The study was approved by the institutional review board at Washington University in St. Louis and was conducted in accordance with the provisions of the Declaration of Helsinki. A total of 94 consecutive patients with a history of MDS who had undergone allogeneic hematopoietic stem-cell transplantation at Washington University in St. Louis between 2002 and 2015 and who had sufficient DNA available for sequencing studies were included in this study. All patients had samples of bone marrow and skin obtained before transplantation and samples of bone marrow obtained 30 days after transplantation. Some patients had a second sample of bone marrow obtained before transplantation and 100 days after transplantation. Four patients were not included because they did not meet technical requirements for analysis (3 patients had mismatched skin and bone marrow samples and in 1 patient no mutations were detected by means of enhanced exome sequencing before transplantation).
RESPONSE CRITERIA AND END POINTS
Disease progression was predefined as at least 5% myeloblasts in the bone marrow, evidence of extramedullary disease, reemergence of pretrans-plantation cytogenic abnormalities, or intervention by the treating physician for loss of donor chimerism or reemergence of pretransplantation morphologic abnormalities. The reviewers were unaware of the results of the sequencing studies.
MOLECULAR ANALYSIS
Initial paired samples of bone marrow and normal skin (as a source of control DNA) obtained before allogeneic hematopoietic stem-cell transplantation were evaluated with enhanced exome sequencing to identify somatic mutations. Enhanced exome sequencing involved a combination of exome-sequencing reagents (NimbleGen, version 3) with additional probes for 285 genes that are recurrently mutated in patients with MDS and acute myeloid leukemia (AML) (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Sequencing on an Illumina HiSeq 2500 platform achieved a mean depth of 239× (1299× for the recurrently mutated genes).
To provide high sensitivity and specificity for detection of mutations after allogeneic hematopoietic stem-cell transplantation, an error-corrected sequencing approach with the use of unique molecular identifiers was combined with high-coverage depths (>30,000×). Multiple probes (mean, 4.6 per mutation) were designed to target all somatic single-nucleotide variants detected by means of enhanced exome sequencing in the pretrans-plantation samples, yielding a total of 2517 validated, trackable, single-nucleotide variants (mean, 29 per patient; range, 1 to 482). The abundance of these mutations 30 and 100 days after transplantation was determined by measuring the variant allele frequency with the use of a custom analysis pipeline with unique molecular identifier–based error correction and base-level background error-rate corrections (see the Supplementary Methods section in the Supplementary Appendix).
A total of 86 initial banked bone marrow samples, 58 pretransplantation bone marrow samples, 86 post-transplantation (day 30) bone marrow samples, 58 post-transplantation (day 100) bone marrow samples, and 86 skin samples were sequenced and analyzed. Bone marrow samples obtained 30 and 100 days after transplantation were considered to be positive if the maximum variant allele frequency was at least 0.5% (data on the threshold are provided in the Supplementary Methods section in the Supplementary Appendix).
STATISTICAL ANALYSIS
The two primary outcomes were progression-free survival (i.e., the time from transplantation to disease progression or death, whichever occurred first) and the cumulative incidence of disease progression. The association between patient and disease characteristics and an adverse outcome (disease progression or death) was assessed with the use of proportional-hazards models of time to death (overall survival) and time to progression or death (progression-free survival). To tease out effects on disease progression independent of death, association with the cumulative incidence of progression was assessed with the use of Fine–Gray subdistribution hazard models. Death without disease progression was considered as a competing risk, and data on patients who were alive and did not have disease progression at the end of the study were censored. P values in figures and hazard ratios were calculated with the use of the Cox proportional-hazards or Fine–Gray model. Survival or disease progression was measured from the time of transplantation to either death or disease progression, respectively, or to the time of censoring. Covariate associations were explored graphically and with correlation or concordance indexes.
Multivariable proportional-hazards and cumulative incidence models based on information criteria (such as the Akaike information criterion, a method to assess the quality of statistical models) were used to select covariates. All two-way interactions among the variables remaining in the model were evaluated. The fit of models (the proportional-hazards assumption and the functional form of the continuous predictor variable) was assessed with the use of martingale residuals and Kolmogorov-type supremum tests. Cumulative incidence models were evaluated graphically with the use of Schoenfeld residuals. The maximum variant allele frequency at day 30 and at day 100 was described with the use of medians and quartiles.
RESULTS
PATIENTS
We sequenced samples obtained from patients who had a history of MDS, including secondary AML or therapy-related MDS, and who had undergone allogeneic hematopoietic stem-cell transplantation at Washington University in St. Louis. All the patients provided written informed consent. We included 90 consecutive patients who had bone marrow samples that had been obtained before transplantation and 30 days after transplantation, as well as skin samples that were used as a source of normal tissue. Validated somatic mutations were detected in the bone marrow samples obtained from 86 of the 90 patients before transplantation and were evaluated further.
The clinical characteristics of the 86 patients are listed in Table 1. Allogeneic hematopoietic stem-cell transplantations from related and unrelated donors involved myeloablative and reduced-intensity conditioning regimens (Table 1, and Table S2 in the Supplementary Appendix). Of the 86 patients, 35 had disease progression after transplantation, with a median of 141 days to progression (range, 27 to 1308), and 51 did not have progression, with a median follow-up of 356 days (range, 45 to 2786).
Table 1.
Characteristics of the Patients at Initial Banking and Univariate Associations with Disease Progression.*
| Variable | All Patients (N = 86) | Disease Progression (N = 35) | No Disease Progression (N = 51) | Disease Progression or Death (N = 60) | Disease Progression | Disease Progression or Death | ||
|---|---|---|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | P Value† | Hazard Ratio (95% CI) | P Value‡ | |||||
| number of patients | ||||||||
| Age at transplantation | ||||||||
| ≤60 yr | 48 | 16 | 32 | 28 | 1.60 (0.83–3.08) | 0.16 | 1.77 (1.05–2.97) | 0.03 |
| >60 yr | 38 | 19 | 19 | 32 | ||||
| IPSS-R score | ||||||||
| Very low | 1 | 0 | 1 | 0 | 1.55 (1.04–2.30) | 0.03 | 1.20 (0.92–1.57) | 0.18 |
| Low | 14 | 4 | 10 | 7 | ||||
| Intermediate | 13 | 3 | 10 | 9 | ||||
| High | 30 | 11 | 19 | 21 | ||||
| Very high | 19 | 12 | 7 | 15 | ||||
| Missing data | 9 | 5 | 4 | 8 | ||||
| Type of MDS | ||||||||
| Primary MDS | 40 | 15 | 25 | 23 | ||||
| Therapy-related MDS | 25 | 11 | 14 | 18 | 1.19 (0.56–2.55) | 0.65 | 1.48 (0.80–2.76) | 0.22 |
| Secondary AML§ | 21 | 9 | 12 | 19 | 1.08 (0.48–2.45) | 0.85 | 1.98 (1.07–3.64) | 0.03 |
| Poor-risk gene or pathway¶ | ||||||||
| RAS pathway | 14 | 7 | 7 | 11 | 0.99 (0.51–1.92) | 0.97 | 1.03 (0.54–2.00) | 0.92 |
| TP53 | 16 | 11 | 5 | 14 | 2.77 (1.41–5.44) | 0.003 | 1.98 (1.09–3.63) | 0.03 |
| RUNX1 | 17 | 9 | 8 | 11 | 1.67 (0.79–3.56) | 0.18 | 0.98 (0.51–1.88) | 0.94 |
| ASXL1 | 19 | 8 | 11 | 13 | 1.21 (0.55–2.64) | 0.64 | 1.13 (0.61–2.10) | 0.70 |
| Conditioning regimen | ||||||||
| Myeloablative | 50 | 15 | 35 | 34 | 0.40 (0.21–0.78) | 0.007 | 0.73 (0.44–1.22) | 0.23 |
| Reduced intensity | 36 | 20 | 16 | 26 | ||||
| Day 30 variant allele frequency | ||||||||
| <0.5% | 54 | 14 | 40 | 33 | 3.65 (1.88–7.09) | <0.001 | 2.07 (1.24–3.46) | 0.005 |
| ≥0.5% | 32 | 21 | 11 | 27 | ||||
| Day 100 variant allele frequency in 58 patients | ||||||||
| <0.5% | 40 | 9 | 31 | 21 | 6.04 (2.53–14.44) | <0.001 | 2.46 (1.24–4.88) | 0.01 |
| ≥0.5% | 18 | 13 | 5 | 14 | ||||
| NA | 28 | 13 | 15 | 25 | ||||
AML denotes acute myeloid leukemia, CI confidence interval, IPSS-R Revised International Prognostic Scoring System, MDS myelodysplastic syndrome, and NA not applicable.
The P value was calculated with the use of the Fine–Gray subdistribution hazard model and Gray’s test.
The P value was calculated with the use of the Cox proportional-hazards model and chi-square test.
Three patients received a diagnosis of therapy-related MDS but were found to have secondary AML before transplantation.
Not all patients had a mutation in one of these genes or pathways.
DETECTION OF MUTATIONS
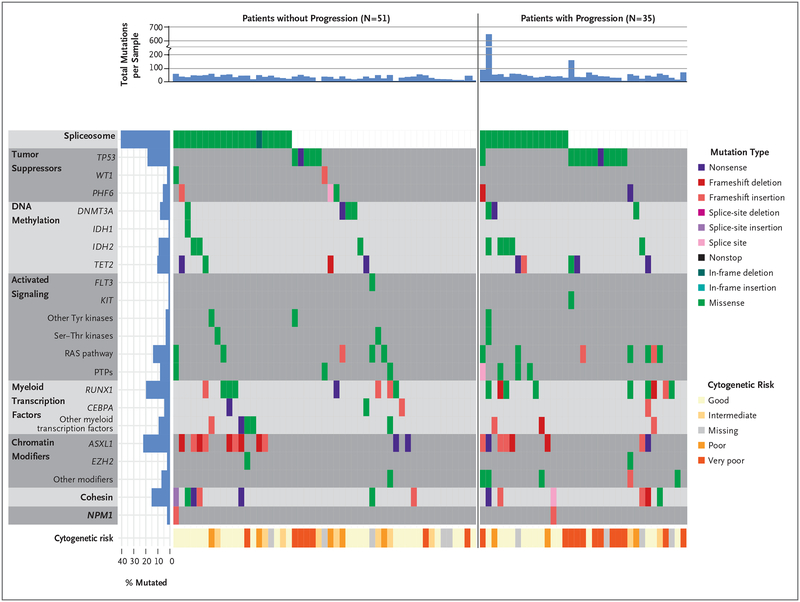
Using enhanced exome sequencing of paired samples of bone marrow and normal tissue, we identified at least one validated somatic mutation in the pretransplantation samples in 86 of the 90 patients (96%) (Fig. 1, and Table S3 in the Supplementary Appendix). We validated somatic single-nucleotide variant mutations using an error-corrected sequencing platform (Table S4 in the Supplementary Appendix). We detected the expected distribution and frequency of mutated genes, as well as known co-mutation and mutually exclusive relationships (Fig. 1). The number of validated mutations in samples at initial banking and shortly before transplantation (i.e., the pretransplantation sample) was similar in patients with disease that progressed or who died and in those who survived without disease progression at 1 year (Fig. S1A and S1B in the Supplementary Appendix). The distribution of variant allele frequencies of mutations at these time points (Fig. S1C and S1D in the Supplementary Appendix) tended to be slightly higher in patients who had disease that had progressed or who had died at 1 year than in those who had survived without disease progression (excluding Patient UPN 147457, whose bone marrow had 482 mutations).
Figure 1. Distribution of Mutations before Allogeneic Hematopoietic Stem-Cell Transplantation in Patients with and Patients without Disease Progression.
Shown is the distribution of mutations in genes and pathways of interest detected by means of enhanced exome sequencing at initial sampling and grouped according to patients with and without disease progression after transplantation. PTPs denotes protein tyro-sine phosphatases, Ser–Thr serine–threonine, and Tyr tyrosine.
The Revised International Prognostic Scoring System (IPSS-R) score (https://www.mds-foundation.org/ipss-r-calculator) is based on peripheral-blood counts, cytogenetic abnormalities, and blast-cell counts in bone marrow (Table S5 in the Supplementary Appendix). IPSS-R scores range from 0 to 10 and define five risk categories (very low, low, intermediate, high, and very high), with higher scores indicating a worse prognosis (Table S5 in the Supplementary Appendix).17 Univariate analyses of patient-related factors and gene mutations detected before transplantation indicated that variables associated with the cumulative incidence of progression were the IPSS-R score (P = 0.03), TP53 mutation status (P = 0.003), and conditioning regimen (P = 0.007). Variables associated with progression-free survival were age at transplantation (P = 0.03), type of MDS (P = 0.03), and TP53 mutation status (P = 0.03) (Table 1).
MUTATION CLEARANCE
To determine whether mutations detected 30 and 100 days after transplantation were associated with outcomes, we genotyped post-transplantation samples using an error-corrected unique molecular identifier–based sequencing approach for all validated single-nucleotide variants identified in a sample obtained before transplantation, as described in the Supplementary Methods section in the Supplementary Appendix.18 One to 482 somatic mutations per sample (median, 23 mutations; mean variant allele frequency, 25.3%) were sequenced in the bone marrow samples obtained 30 and 100 days after transplantation (60,930× mean total coverage [2599× unique coverage] of these variants on day 30 and 36,705× mean total coverage [3475× unique coverage] on day 100) (Fig. S2 and Table S4 in the Supplementary Appendix).
With a cutoff point of at least 0.5% for the maximum variant allele frequency of a mutation 30 days after allogeneic hematopoietic stem-cell transplantation (equivalent to 1 heterozygous mutant cell in 100 cells), 21 of 32 mutation-positive patients (66%) and 14 of 54 mutation-negative patients (26%) had disease progression after transplantation. Similarly, analysis of samples obtained 100 days after transplantation showed that 13 of 18 mutation-positive patients (72%) and 9 of 40 mutation-negative patients (22%) had disease progression. The risk of disease progression was higher among patients who had at least one mutation with a variant allele frequency of at least 0.5% at day 30 or day 100 after allogeneic hematopoietic stem-cell transplantation than among those who did not have these mutations (P<0.001) (Table 1).
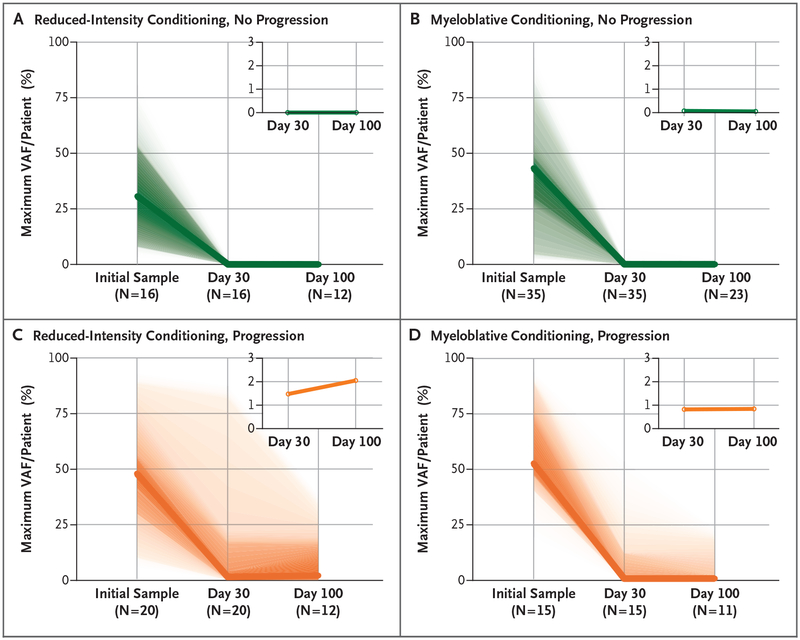
We next examined the clearance of mutations after allogeneic hematopoietic stem-cell transplantation in patients who had disease progression as compared with those who did not. When conditioning regimen was not taken into account, the maximum variant allele frequency 30 days after transplantation was higher in patients who had disease progression than among those who did not (median maximum variant allele frequency, 0.9% vs. 0%; P<0.001). In patients who did not have disease progression, similar reductions in the maximum variant allele frequencies at day 30 after transplantation were observed in those who received reduced-intensity conditioning regimens (Fig. 2A) and those who received myeloablative conditioning regimens (Fig. 2B) (median, 0% vs. 0.07%; P = 0.21). Patients who had disease progression after a reduced-intensity conditioning regimen (Fig. 2C) had a higher (but statistically insignificant) maximum variant allele frequency at day 30 after transplantation than patients treated with a myeloablative regimen (Fig. 2D) (median, 1.47% vs. 0.82%; P = 0.64).
Figure 2. Decreases in Variant Allele Frequencies after Allogeneic Hematopoietic Stem-Cell Transplantation in Patients with and Patients without Disease Progression.
Shown is the maximum variant allele frequency (VAF) detected per patient as measured by means of error-corrected sequencing at initial sampling and at day 30 and day 100 after transplantation in patients with no disease progression (green, Panels A and B) and with disease progression (orange, Panels C and D). Plots are further divided according to pretransplantation conditioning regimen (reduced-intensity conditioning in Panels A and C and myeloablative conditioning in Panels B and D). Shading represents the range of observed values (first two quartiles shown). The inset plots show the interval in the VAF from 0% to 3%; the bold line represents the median value of the maximum VAF observed in each patient. The numbers of patients with samples sequenced at each time point are indicated.
At day 30 after transplantation, we observed differences in mutation clearance for specific genes. Mutations tended to persist at any detectable level in TP53 (15 of 20 mutations were detected; median variant allele frequency, 0.51%) and DNMT3A (8 of 9 mutations were detected; median variant allele frequency, 0.73%), whereas NRAS mutations were typically not detected at day 30 (1 of 8 mutations were detected; median variant allele frequency, 0%) (Fig. S3 and Table S4 in the Supplementary Appendix). We could not assess the prognostic significance of detecting a sole DNMT3A, TET2, or ASXL1 mutation (i.e., a DTA mutation) 30 days after allogeneic hematopoietic stem-cell transplantation because only one patient (Patient UPN 611) had a sole DTA variant allele frequency of at least 0.5% at this time point.19
PROGNOSTIC EFFECT OF MUTATION CLEARANCE
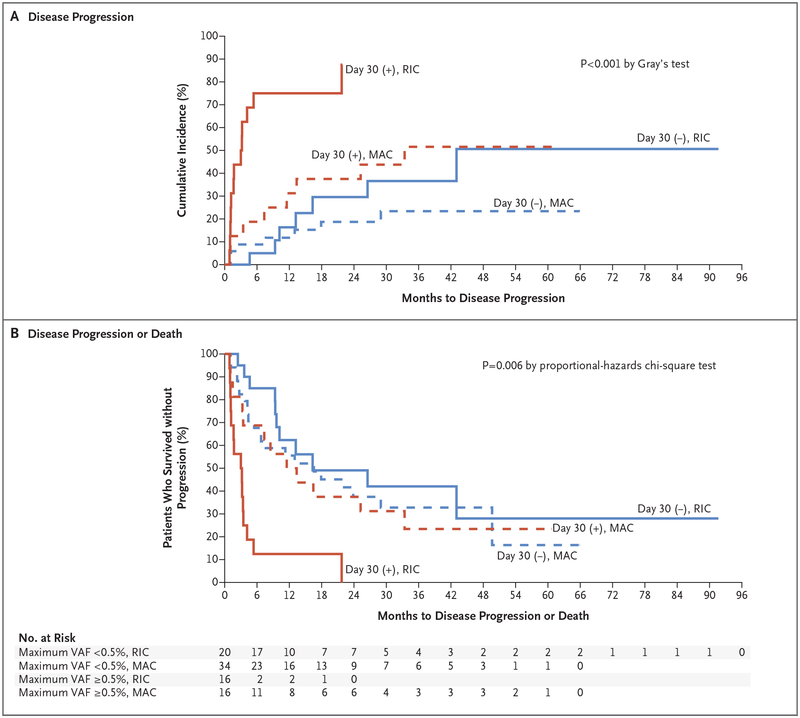
The presence of at least one mutation with a variant allele frequency of at least 0.5% 30 days after allogeneic hematopoietic stem-cell transplantation was associated with a higher risk of disease progression at 1 year than the absence of such a mutation, even after adjustment for conditioning regimen (53.1% vs. 13.0%; hazard ratio, 3.86; 95% confidence interval [CI], 1.96 to 7.62; P<0.001) (Fig. S4A in the Supplementary Appendix). Patients with at least one mutation with a variant allele frequency of at least 0.5% approximately 100 days after transplantation also had a higher risk of disease progression at 1 year than patients without such a mutation, even after adjustment for conditioning regimen (66.7% vs. 0%; hazard ratio, 6.52; 95% CI, 2.54 to 16.7; P<0.001) (Fig. S5A and S5B in the Supplementary Appendix).
The risk of progression was higher among patients who had received a reduced-intensity conditioning regimen and who had a persistent mutation with a variant allele frequency of at least 0.5% at day 30 after transplantation than among patients with other combinations of conditioning regimen and mutation status (P≤0.01 by pairwise comparisons) (Fig. 3A). The presence of at least one mutation with a variant allele frequency of at least 0.5% 30 days after transplantation was also associated with a lower 1-year rate of progression-free survival than the absence of such a mutation, even after adjustment for conditioning regimen (31.3% vs. 59.3%; hazard ratio for progression or death, 2.22; 95% CI, 1.32 to 3.73; P = 0.005) (Fig. S4B in the Supplementary Appendix). The rate of progression-free survival was lower among patients who received a reduced-intensity conditioning regimen and who had a persistent mutation with a variant allele frequency of at least 0.5% at day 30 after transplantation than among patients with other combinations of conditioning regimen and mutation status (P≤0.001 by pairwise comparisons) (Fig. 3B). Among patients who had received a reduced-intensity conditioning regimen and who had a persistent mutation with a variant allele frequency of at least 0.5% at day 30 after allogeneic hematopoietic stem-cell transplantation, the proportion of patients who were alive without disease progression at 12 months was only 12.5% (95% CI, 2.1 to 32.8), as compared with 50.0 to 62.3% in all other groups (Fig. 3B). The presence of at least one mutation 100 days after transplantation was also associated with a lower 1-year rate of progression-free survival than no mutations, even after adjustment for conditioning regimen (27.8% vs. 77.5%; hazard ratio for progression or death, 2.51; 95% CI, 1.26 to 5.01; P = 0.01) (Fig. S5C and S5D in the Supplementary Appendix). The rate of overall survival did not differ between patients with and without a detectable mutation with a variant allele frequency of at least 0.5% at 30 or 100 days after transplantation (Fig. S4C, S4D, S5E, and S5F in the Supplementary Appendix). Similar results were observed for the cumulative incidence of progression, progression-free survival, and overall survival with the use of a threshold of 0.1%, 1.0%, or 2.5% or no threshold in the maximum variant allele frequency at day 30 (Fig. S6 through S9 in the Supplementary Appendix). Although exploratory, the analyses and P values consistently indicate an association between the detection of persistent disease–associated mutations in the bone marrow after transplantation and an increased risk of disease progression.
Figure 3. Association of Mutation Clearance with Outcomes.
The VAF on day 30 after transplantation was determined with the use of error-corrected sequencing interrogating single-nucleotide variant mutations identified by enhanced exome sequencing of samples before transplantation. Patients are grouped according to the presence of positive (+) or negative (−) results for at least one mutation VAF of at least 0.5% (red lines) or all VAFs less than 0.5% (blue lines) and according to whether the patient received a reduced-intensity conditioning regimen (RIC, solid lines) or myeloablative conditioning (MAC, dashed lines). The rates of disease progression (Panel A) and disease progression or death (Panel B) are shown.
We next asked whether detection of a mutation with a maximum variant allele frequency of at least 0.5% in a bone marrow sample obtained 30 days after allogeneic hematopoietic stem-cell transplantation was predictive of progression or progression-free survival beyond known clinical and genetic risk factors, including age at transplantation, IPSS-R score, type of MDS, TP53 mutation status, and conditioning regimen. The variables that were retained in the multivariate analyses of the cumulative incidence of progression were the IPSS-R score, maximum variant allele frequency at day 30 (≥0.5% or <0.5%), TP53 mutation status, and conditioning regimen (reduced intensity or myeloablative). The presence of a mutation with a variant allele frequency of at least 0.5% at day 30 after transplantation was associated with a higher risk of progression than the absence of such a mutation (hazard ratio, 4.48; 95% CI, 2.21 to 9.08; P<0.001), even when the IPSS-R score and conditioning regimen were considered as covariates. In addition, myeloablative conditioning was associated with an approximately 73% lower hazard of progression (hazard ratio, 0.27; 95% CI, 0.12 to 0.60; P = 0.001) than a reduced-intensity conditioning regimen, and the hazard of progression increased by approximately 78% (hazard ratio, 1.78; 95% CI, 1.15 to 2.74; P = 0.01) with each 1-unit increase in the IPSS-R score.
The variables that were retained in the multivariable analysis of progression-free survival were the maximum variant allele frequency at day 30 (≥0.5% or <0.5%), conditioning regimen (reduced intensity or myeloablative), age at transplantation, and type of MDS. A maximum variant allele frequency of at least 0.5% at day 30 after allogeneic hematopoietic stem-cell transplantation was associated with a lower rate of progression-free survival than a maximum variant allele frequency of less than 0.5% at day 30 (hazard ratio for progression or death, 2.39; 95% CI, 1.40 to 4.09; P = 0.002). Although there was a strong interaction between a maximum variant allele frequency of at least 0.5% at day 30 and the conditioning regimen, the maximum variant allele frequency of at least 0.5% approximately doubled the risk of progression or death, even after the effects of other known risk factors were taken into account. In this analysis, a diagnosis of secondary AML was also associated with a lower rate of progression-free survival than primary MDS (hazard ratio for progression or death, 2.24; 95% CI, 1.21 to 4.15; P = 0.01).
GENE-PANEL SEQUENCING TO MONITOR MUTATION CLEARANCE
Since exome sequencing of paired samples of tumor and normal skin followed by the design of custom-targeted probes is an impractical clinical assay at this time, we evaluated whether tracking mutations in a limited set of recurrently mutated genes (i.e., a “clinical gene panel”) would also have prognostic significance. To address this possibility, we downsampled the exome data and analyzed a subset of 40 genes (Table S6 in the Supplementary Appendix) that are recurrently mutated in MDS and AML.20–23 With the use of this approach, 68 of 86 patients (79%) were found to have one to six trackable mutations in samples collected before transplantation, with an average of two mutations (excluding Patient UPN 147457). With the use of a cutoff of at least 0.5% for the variant allele frequency at day 30 after transplantation, 18 of 26 mutation-positive patients (69%) and 14 of 42 mutation-negative patients (33%) were found to have disease progression.
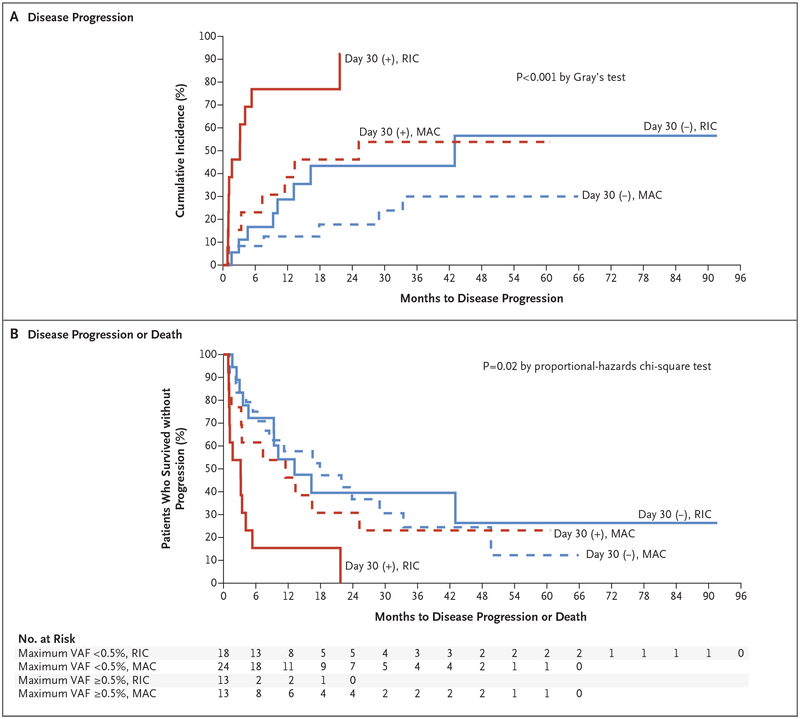
With the use of the 40-gene panel test only, the presence of at least one mutation with a variant allele frequency of at least 0.5% 30 days after allogeneic hematopoietic stem-cell transplantation was associated with a higher risk of disease progression at 1 year (57.7% vs. 19.1%; hazard ratio, 3.39; 95% CI, 1.68 to 6.83; P = 0.001) (Fig. S10A in the Supplementary Appendix) and a lower 1-year rate of progression-free survival than the absence of such a mutation, even after adjustment for conditioning regimen (30.8% vs. 57.1%; hazard ratio for progression or death, 2.09; 95% CI, 1.18 to 3.70; P = 0.02) (Fig. S10B in the Supplementary Appendix). The risk of disease progression was higher (Fig. 4A) and the rate of progression-free survival was lower (Fig. 4B) among patients who received a reduced-intensity conditioning regimen and who had a persistent mutation with a variant allele frequency of at least 0.5% at day 30 after allogeneic hematopoietic stem-cell transplantation than among patients with other combinations of conditioning regimen and mutation status. The rate of overall survival did not differ between patients with at least one mutation with a variant allele frequency of at least 0.5% 30 days after allogeneic hematopoietic stem-cell transplantation and patients without such a mutation (Fig. S10C and S10D in the Supplementary Appendix).
Figure 4. Association of Outcomes with Mutation Clearance Determined with the Use of a 40-Gene Panel.
Only the VAFs of 40 genes recurrently mutated in myeloid cancers were used to assess mutation clearance at day 30 after transplantation. VAFs were determined with the use of error-corrected sequencing interrogating single-nucleotide variants identified by enhanced exome sequences of samples before transplantation. Patients are grouped according to the presence of at least one VAF of at least 0.5% (red lines) or all VAFs less than 0.5% (blue lines) and according to whether they received a reduced-intensity conditioning regimen (RIC, solid lines) or myeloablative conditioning (MAC, dashed line). The rates of disease progression (Panel A) and disease progression or death (Panel B) are shown.
DISCUSSION
In this exploratory study involving patients with MDS, the risk of disease progression was higher and the rate of progression-free survival was lower among patients in whom persistent mutations were detected in bone marrow samples obtained 30 days after allogeneic hematopoietic stem-cell transplantation than among those in whom these mutations were not detected. The risk of progression was higher among patients with detectable mutations who had received a reduced-intensity conditioning regimen than among those who had received a myeloablative regimen. These findings suggest that sequencing bone marrow samples at early clinical time points after transplantation could be used as an individualized risk-assessment biomarker for disease progression in patients with MDS.
The prognostic usefulness of monitoring measurable residual disease after treatment for MDS and AML has been studied with the use of a variety of approaches, including morphologic analysis, assessment of chimerism, quantitative PCR, and multiparameter flow cytometry.13,14,24–26 However, these approaches can be limited by sensitivity (e.g., in morphologic analysis and assessment of chimerism) or applicability to only a subgroup of patients (e.g., in quantitative PCR). They may also be subject to differences in interpretation among observers (e.g., in multiparameter flow cytometry). Sequencing-based tests of measurable residual disease have the potential to overcome many of these limitations. Several independent studies have shown that the presence of persistent mutations in patients with AML who have a complete morphologic remission after induction therapy has been associated with inferior outcomes.19,27,28 Separate studies have evaluated whether outcomes are affected by the early initiation of salvage therapy, including withdrawal of immunosuppression, infusion of donor lymphocytes, or the use of azacitidine after detection of measurable residual disease or early relapse after allogeneic hematopoietic stem-cell transplantation.12,29–31 Although these studies suggest that early detection and preemptive treatment of disease progression may have benefit, prospective clinical trials to establish an effect on outcomes are lacking. In our study, we observed a median of 67 days between detection of a mutation at day 30 after allogeneic hematopoietic stem-cell transplantation and disease progression, a period that may allow for the initiation of salvage therapy or planning for a second transplantation in some patients.
We observed that patients with measurable residual disease at day 30 who had been treated with a reduced-intensity conditioning regimen had the highest risk of progression. This finding may provide insight into results from a previous randomized study involving patients with MDS or AML in which outcomes were compared according to the use of reduced-intensity or fully myeloablative regimens. In that study, patients with MDS who received reduced-intensity conditioning regimens had a higher rate of relapse than patients who received myeloablative regimens, but the rate of overall survival did not differ according to conditioning regimen. (In contrast, patients with AML who received reduced-intensity conditioning regimens had a higher rate of relapse and lower rate of overall survival than patients who received myeloablative regimens.)10 It is not yet clear why we observe an effect on progression-free survival — but not overall survival — among patients with MDS: this effect may be due to biologic characteristics of the disease or to the nature of response to salvage therapy in patients with MDS. Although a larger study with longer follow-up is lacking, our results suggest that the negative effect of a reduced-intensity conditioning regimen on progression-free survival among patients with MDS may at least partially be due to persistent disease on day 30 after transplantation.
Important variables to consider for monitoring of measurable residual disease in patients with MDS and AML include choosing the best platform on the basis of sensitivity and accessibility (e.g., sequencing vs. multiparameter flow cytometry), the best time points for acquisition of samples, the use of blood or bone marrow samples, and a detection threshold that is clinically meaningful. Data are lacking regarding whether incorporation of insertion and deletion mutations, copy-number alterations, or both improves the prognostic usefulness of point mutations alone. Since high-coverage exome sequencing is not routinely available in the clinical setting, we also analyzed our data using a subset of genes that are commonly included in gene-panel sequencing assays for MDS and AML. Although we identified fewer patients with mutations with the use of this approach than with enhanced exome sequencing, the prognostic value of detection of measurable residual disease was still highly clinically significant. Although this exploratory study has limitations, our results suggest that sequencing-based detection of tumor cells and measurable residual disease after allogeneic hematopoietic stem-cell transplantation has prognostic significance for patients with MDS.
Supplementary Material
Acknowledgments
Supported by a Leukemia and Lymphoma Society Quest for Cures and Scholar Award (to Dr. Walter); grants from the Edward P. Evans Foundation (to Drs. Walter and Duncavage), the National Cancer Institute (NCI) (R33CA217700, to Drs. Walter and Duncavage), the Specialized Program of Research Excellence in AML of the NCI (P50CA171963 Career Enhancement Program, to Dr. Duncavage), and the Washington University Institute of Clinical and Translational Sciences (UL1TR002345) from the National Center for Advancing Translational Sciences of the National Institutes of Health (TL1TR002344, to Dr. Duncavage); an American Society of Hematology Scholar Award (to Dr. Jacoby); and grants from Gabrielle’s Angel Foundation (to Dr. Walter) and the Lottie Caroline Hardy Trust (to Dr. Walter). Support for procurement of human samples was provided by the Genomics of AML Program Project of the NCI (P01 CA101937, to Dr. Ley) and the Specialized Program of Research Excellence in AML (P50CA171963, to Dr. Link). Core services were provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core and Biostatistics Shared Resource Core supported by an NCI Cancer Center grant (P30CA091842).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
Eric J. Duncavage, Department of Pathology and Immunology
Meagan A. Jacoby, Department of Medicine, Division of Oncology
Gue Su Chang, McDonnell Genome Institute
Christopher A. Miller, Department of Medicine, Division of Oncology McDonnell Genome Institute.
Natasha Edwin, Department of Medicine, Division of Oncology
Jin Shao, Department of Medicine, Division of Oncology
Kevin Elliott, Department of Medicine, Division of Oncology
Joshua Robinson, Department of Medicine, Division of Oncology
Haley Abel, McDonnell Genome Institute
Robert S. Fulton, McDonnell Genome Institute
Catrina C. Fronick, McDonnell Genome Institute
Michelle O’Laughlin, McDonnell Genome Institute
Sharon E. Heath, Department of Medicine, Division of Oncology
Kimberly Brendel, Department of Medicine, Division of Oncology
Raya Saba, Department of Medicine, Division of Hospital Medicine
Lukas D. Wartman, Department of Medicine, Division of Oncology
Matthew J. Christopher, Department of Medicine, Division of Oncology
Iskra Pusic, Department of Medicine, Division of Oncology
John S. Welch, Department of Medicine, Division of Oncology
Geoffrey L. Uy, Department of Medicine, Division of Oncology
Daniel C. Link, Department of Medicine, Division of Oncology
John F. DiPersio, Department of Medicine, Division of Oncology
Peter Westervelt, Department of Medicine, Division of Oncology
Timothy J. Ley, Department of Medicine, Division of Oncology
Kathryn Trinkaus, Siteman Biostatistics Shared Resource, Siteman Cancer Center
Timothy A. Graubert, Washington University School of Medicine in St. Louis, St. Louis; and Massachusetts General Hospital Cancer Center, Boston
Matthew J. Walter, Department of Medicine, Division of Oncology
References
- 1.de Witte T, Bowen D, Robin M, et al. Allogeneic hematopoietic stem cell transplantation for MDS and CMML: recommendations from an international expert panel. Blood 2017; 129: 1753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Della Porta MG, Gallì A, Bacigalupo A, et al. Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol 2016; 34: 3627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med 2017; 376: 536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshizato T, Nannya Y, Atsuta Y, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood 2017; 129: 2347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenecke C, Göhring G, de Wreede LC, et al. Impact of the revised International Prognostic Scoring System, cytogenetics and monosomal karyotype on outcome after allogeneic stem cell transplantation for myelodysplastic syndromes and secondary acute myeloid leukemia evolving from myelodysplastic syndromes: a retrospective multicenter study of the European Society of Blood and Marrow Transplantation. Haematologica 2015; 100: 400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol 2014; 32: 2691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuser M, Gabdoulline R, Löffeld P, et al. Individual outcome prediction for myelodysplastic syndrome (MDS) and secondary acute myeloid leukemia from MDS after allogeneic hematopoietic cell transplantation. Ann Hematol 2017; 96: 1361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Festuccia M, Deeg HJ, Gooley TA, et al. Minimal identifiable disease and the role of conditioning intensity in hematopoietic cell transplantation for myelodys-plastic syndrome and acute myelogenous leukemia evolving from myelodysplastic syndrome. Biol Blood Marrow Transplant 2016; 22: 1227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mo XD, Qin YZ, Zhang XH, et al. Minimal residual disease monitoring and preemptive immunotherapy in myelodysplastic syndrome after allogeneic hematopoietic stem cell transplantation. Ann Hematol 2016; 95: 1233–40. [DOI] [PubMed] [Google Scholar]
- 10.Scott BL, Pasquini MC, Logan BR, et al. Myeloablative versus reduced-intensity hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol 2017; 35: 1154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Israyelyan A, Goldstein L, Tsai W, et al. Real-time assessment of relapse risk based on the WT1 marker in acute leukemia and myelodysplastic syndrome patients after hematopoietic cell transplantation. Bone Marrow Transplant 2015; 50: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenow F, Berkemeier A, Krug U, et al. CD34(+) lineage specific donor cell chimerism for the diagnosis and treatment of impending relapse of AML or myelodysplastic syndrome after allo-SCT. Bone Marrow Transplant 2013; 48: 1070–6. [DOI] [PubMed] [Google Scholar]
- 13.Tobiasson M, Olsson R, Hellström-Lindberg E, Mattsson J. Early detection of relapse in patients with myelodysplastic syndrome after allo-SCT. Bone Marrow Transplant 2011; 46: 719–26. [DOI] [PubMed] [Google Scholar]
- 14.Christopeit M, Ocheni S, Haferlach T, et al. Evaluation of BM cytomorphology after allo-SCT in patients with MDS. Bone Marrow Transplant 2013; 48: 465–6. [DOI] [PubMed] [Google Scholar]
- 15.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012; 366: 1090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby MA, Duncavage EJ, Chang GS, et al. Subclones dominate at MDS progression following allogeneic hematopoietic cell transplant. JCI Insight 2018. March 8 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012; 120: 2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uy GL, Duncavage EJ, Chang GS, et al. Dynamic changes in the clonal structure of MDS and AML in response to epigenetic therapy. Leukemia 2017; 31: 872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med 2018; 378: 1189–99. [DOI] [PubMed] [Google Scholar]
- 20.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014; 28: 241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122: 3616–3627, 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter MJ, Shen D, Shao J, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 2013; 27: 1275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christopeit M, Miersch K, Klyuchnikov E, et al. Evaluation of BM cytomorphology after allo-SCT in patients with AML. Bone Marrow Transplant 2012; 47: 1538–44. [DOI] [PubMed] [Google Scholar]
- 25.Bornhäuser M, Oelschlaegel U, Platz-becker U, et al. Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica 2009; 94: 1613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díez-Campelo M, Pérez-Simón JA, Pérez J, et al. Minimal residual disease monitoring after allogeneic transplantation may help to individualize post-transplant therapeutic strategies in acute myeloid malignancies. Am J Hematol 2009; 84: 149–52. [DOI] [PubMed] [Google Scholar]
- 27.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 2015; 314: 811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med 2016; 374: 422–33. [DOI] [PubMed] [Google Scholar]
- 29.Platzbecker U, Wermke M, Radke J, et al. Azacitidine for treatment of imminent relapse in MDS or AML patients after allogeneic HSCT: results of the RELAZA trial. Leukemia 2012; 26: 381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroeder T, Czibere A, Platzbecker U, et al. Azacitidine and donor lympho cyte infusions as first salvage therapy for relapse of AML or MDS after allogeneic stem cell transplantation. Leukemia 2013; 27: 1229–35. [DOI] [PubMed] [Google Scholar]
- 31.Woo J, Howard NP, Storer BE, et al. Mutational analysis in serial marrow samples during azacitidine treatment in patients with post-transplant relapse of acute myeloid leukemia or myelodysplastic syndromes. Haematologica 2017; 102(6): e216–e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.