Figure 3. ZIKV barcode viruses undergo cell-type specific reduction in barcode diversity.
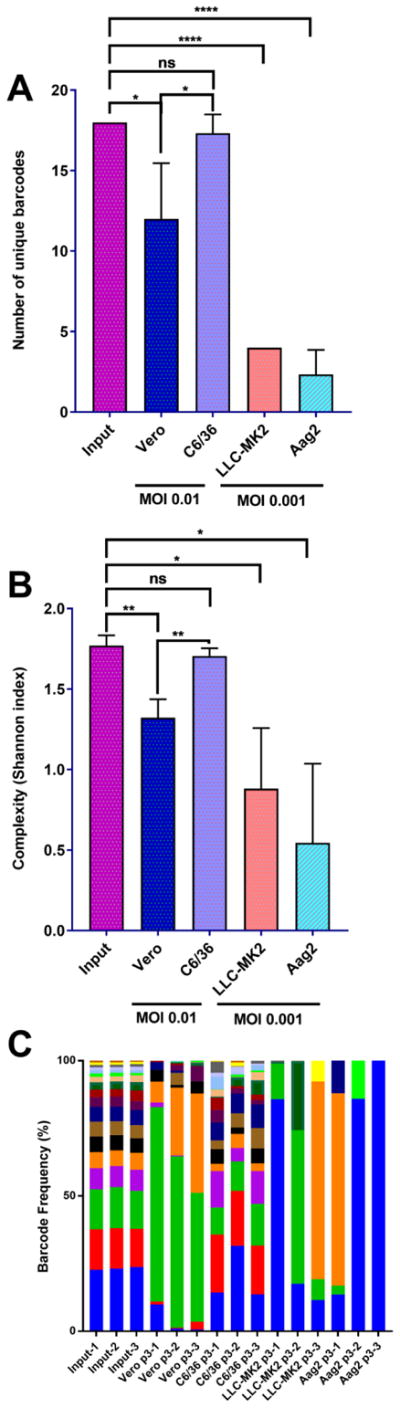
The four barcode viruses were subjected to 3 three serial passages in two mammalian (Vero and LLC-MK2) and two insect-derived (C6/36 and Aag2) cell lines. Passages were performed at MOI 0.01 for Vero and C6/36 and 0.001 for LLC-MK2 and Aag2. A) The number of unique barcodes present in each barcode virus iteration. Values were compared statistically using a one-way ANOVA with Tukey’s multiple comparisons test, * p<0.05 **** p<0.0001 ns p>0.05 B) Average genetic complexity at the barcode positions measured by Shannon’s index. Calculated as . Where ni is the frequency of each nucleotide at each barcode position and N is the total number of barcode calls at that position. Values were compared statistically using a one-way ANOVA with Tukey’s multiple comparisons test, * p<0.05 ** p<0.01 ns p>0.05 C) Frequency of individual barcodes. Each color represents a unique barcode. More bars indicate more barcode diversity. n=3 for all treatments. The number following the group label on the x-axis refers to the biological replicate.
