Abstract
Epistasis (gene-gene interaction) detection in large-scale genetic association studies has recently drawn extensive research interests as many complex traits are likely caused by the joint effect of multiple genetic factors. The large number of possible interactions poses both statistical and computational challenges. A variety of approaches have been developed to address the analytical challenges in epistatic interaction detection. These methods usually output the identified genetic interactions and store them in flat file formats. It is highly desirable to develop an effective visualization tool to further investigate the detected interactions and unravel hidden interaction patterns. We have developed EINVis, a novel visualization tool that is specifically designed to analyze and explore genetic interactions. EINVis displays interactions among genetic markers as a network. It utilizes a circular layout (specially, a tree ring view) to simultaneously visualize the hierarchical interactions between single nucleotide polymorphisms (SNPs), genes, and chromosomes, and the network structure formed by these interactions. Using EINVis, the user can distinguish marginal effects from interactions, track interactions involving more than two markers, visualize interactions at different levels, and detect proxy SNPs based on linkage disequilibrium. EINVis is an effective and user-friendly free visualization tool for analyzing and exploring genetic interactions. It is publicly available with detailed documentation and online tutorial on the web at http://filer.case.edu/yxw407/einvis/.
Keywords: epistasis, networks, visualization, gene-gene interaction, software
Introduction
Epistasis (gene-gene interaction) detection is receiving an increasing amount of research attention in large-scale genetic association studies [Cordell, 2009]. Interactions between genes are fundamentally important to understand the structure and function of genetic pathways of complex genetic systems [Moore and Williams, 2009; Phillips, 2008] and have been suggested potentially to uncover the “missing heritability” in genetic association studies [Manolio et al., 2009]. Detecting epistatic interactions in the genome-wide scale is both statistically and computationally challenging. The large number of possible interactions between single nucleotide polymorphisms (SNPs) poses a serious multiple testing problem and demands intensive computational resources [Steen, 2012]. A variety of methods have been developed for epistasis detection. Please refer to Shang et al. [2011], Wang et al. [2011] for excellent review and performance study of the existing methods.
The outputs of an epistasis detection method usually include a list of SNP pairs and their test statistics or significance levels. There is a lack of effective visualization tool to further analyze and explore the discovered interactions. Moreover, most of the current study designs for searching epistatic interactions at the genome-wide level have low power because of the large number of tests. Therefore, it is interesting to systemically examine many interactions with relatively weak statistical evidence. An intuitive approach to explore the discovered genetic interactions is to use network built by linking SNPs whose pairwise interactions are stronger than a threshold [Hu et al., 2011; McKinney et al., 2009]. Representing epistatic interactions visually by networks is an effective way to demonstrate the interplay between different SNPs and genes.
Visualization tools, such as Cytoscape [Shannon et al., 2003] and VisANT 3.5 [Hu et al., 2009], are mainly designed for biomolecular network visualization. MizBee [Meyer et al., 2009] exerts a separate circular layout and other linked views to visualize syntenic data. Circos [Krzywinski et al., 2009] focuses on circular layouts and is created for comparative genomics. D3 [Bostock et al., 2011] is a general purpose visualization toolkit. Despite the success of these excellent visualization tools, none of them can be directly used for exploring the epistatic interaction network (EIN) [Doncheva et al., 2012; Pavlopoulos et al., 2008; Suderman and Hallett, 2007].
Visualizing genetic interaction network has its unique requirements and properties that can be used to enhance the visualization effectiveness. In a genetic interaction network, the relationship between SNPs, genes, and chromosomes can be represented using a hierarchical tree structure. This hierarchical structure can be utilized to enhance the effectiveness of network visualization. Circular layout is a natural choice to visualize the hierarchical structure and network structure simultaneously [Holten, 2006]. Moreover, when studying gene-gene interactions, it is important to distinguish marginal effects from interactions. For example, it is interesting to identify SNPs that have weak marginal effects but interact with many other SNPs. Tracking interacting SNPs and visualizing interactions at different levels, e.g., SNP-SNP, SNP-gene, and gene-gene interactions, are also important functions that can enhance the effectiveness of the visualization tools.
We have developed a visualization tool EINVis (Epistatic Interaction Network Visualization) for the exploration and analysis of genetic interactions. Utilizing EINVis, the user can directly see the connection pattern among SNPs, genes, and chromosomes. The interaction strength between two nodes is encoded by the edge color. The user can also provide single-locus test statistics (encoded by the node color), which help to reveal the relationship between marginal and interacting effects. EINVis allows users to observe how a set of selected nodes interact with other nodes by highlighting the connected nodes and induced edges. The user can also expand or collapse nodes, which not only help to focus on the set of nodes of interest, but also enable to observe interactions at different levels. EINVis also provides summary statistics of the visualized interaction network.
Material and Methods
EINVis Software
EINVis is created for visualization of small or moderate genetic interaction network with hundreds of edges or nodes. EINVis is open source and implemented in JavaScript language. Canvas 2D and control elements in HTML5 (the fifth revision of the HTML standard) are exerted to draw graphics and provide user-friendly interfaces. EINVis runs on all major browsers as long as they provide support for HTML5.
EINVis provides three different views: an EIN tree ring view, an EIN matrix view, and a linkage disequilibrium (LD) tree ring view. The EIN tree ring view is the main view in EINVis. It adopts the circular layout and looks like a tree ring. A wide range of controls and mouse interactions are included to let user interact with the visualization results. The EIN matrix view is an auxiliary view. It can help the user observe the adjacency matrix of the interaction network. The LD tree ring view is another auxiliary view. It can help the user observe the LD information among SNPs.
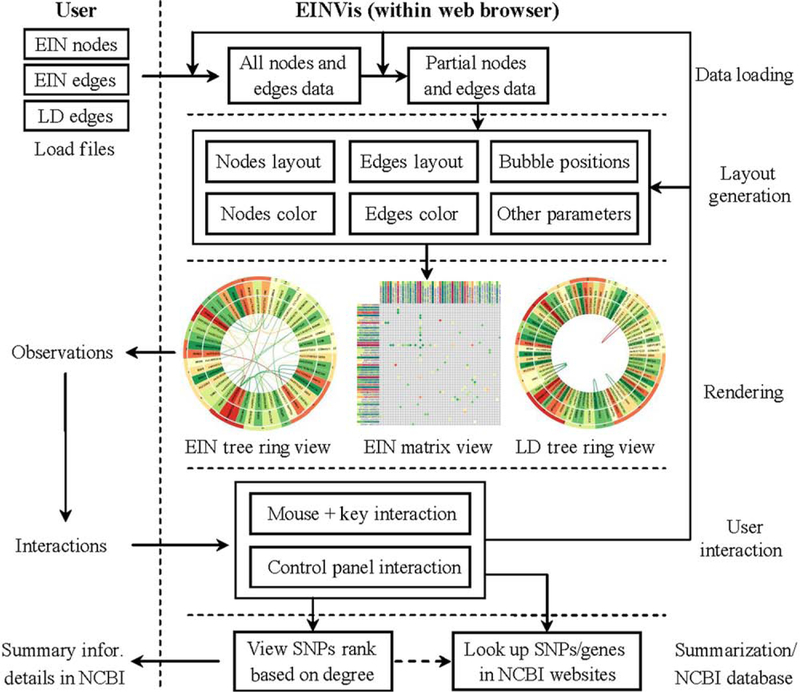
The underlying implementation architecture of EINVis is shown in Figure 1. It includes three main modules: layout generation, rendering, and user interaction modules.
Figure 1.
Software architecture of EINVis.
After the user inputs the node and edge files, EINVis will generate the layout and other rendering parameters. Then, EINVis will begin to render the tree ring views and matrix view. EINVis provides a wide range of user interactions, which can help the user to explore the visualized results. EINVis also provide SNPs rank summary information. User can link from EINVis to NCBI website and look up the interested SNPs or genes.
Layout Generation Module
The layout generation module generates the graph layout according to the user input files. All rendering parameters for the tree ring views and matrix view are generated by this module. The input files include the EIN node file (SNPs), EIN edge file (interactions between SNPs), and LD edge file (LD information between SNPs). In the EIN tree ring view, each node is rendered as a ring sector, which includes four parameters: inner radius, outer radius, begin angle, and end angle degree. The node color is mapped from the single-locus test statistic. The edge layout is generated by B-spline interpolation. There is a virtual hierarchical grid [Holten, 2006] built in the inner circle region of tree ring view. We find the shortest path between two nodes, which have an edge linking them through this virtual grid. The points on this path will be used to calculate the edge curve. The edge color is mapped from two-locus test statistic. When there is user interaction, this module will update the layouts and rendering parameters, such as node color and bubble positions. After updating, it will call the rendering module to update the visualization results.
Rendering Module
The rendering module renders graphics on the canvas elements (please refer to the official W3C specification about Canvas element). The canvas element provides scripts with a resolution-dependent bitmap canvas, which can be used for rendering graphics and other visual images on the fly. The rendering module reads the parameters generated by the layout generation module and calls the figure drawing function to render the figure on the canvas. When the user interacts with EINVis, the rendering module can also update the visualization results. For example, when the user changes the edge color, this module will update the edge color (instead of beginning rendering from scratch). The tree ring views and matrix view are rendered as bubbles [Bragdon et al., 2010; Li et al., 2011]. The extension of static multiple views to a metaphorical interface of bubbles enables a flexible layout to support analysis. The user can move the bubbles or change their layers. After this module renders the graphics, the user can observe and interact with the visualization results.
User Interaction Module
This module enables various user interactions. There are two types of interactions provided by EINVis: the mouse and keyboard interaction and the control panel interaction. The user can use mouse click and key pressing operations to expand nodes, collapse nodes, highlight nodes, and so on. When user points mouse to a SNP node or an edge between two SNPs, the corresponding single-locus test statistic, two locus test statistic, or LD information will show up. The user can also move the bubbles using mouse operations. Through the control panel, the user can change color, font size, and other rendering parameters. These interactions are essential for the user to explore the visualized results effectively. None of the existing network visualization tools provide such customized functions for exploring genetic interaction networks.
Example Hypertension Study Data Set
To illustrate the software, we applied it to the hypertension data of African American cohorts from the Candidate Gene Association Resource (CARe) study [Fox et al., 2011; Zhu et al., 2011]. We downloaded both phenotype and geno-type data from dbGaP database. We analyzed the genotype data genotyped by the 50K cardiovascular gene-centric array (ITMAT-Broad-CARe [IBC] array) [Keating et al., 2008], which consists of 47,815 gene centric SNPs after quality con-trols (QCs) in approximately 2,500 genes. Analysis was restricted to unrelated 5,493 individuals to remove the effect of population structure. The phenotype is systolic blood pressure (SBP). A total of 10 mm Hg SBP was added for individuals who were taking antihypertensive medication. SBP residual for each individual was then obtained by adjusting gender, age, age2, and body mass index. We next applied F-test to calculate the single- and two-locus test statistics. Note that this step is independent of the EINVis tool. Any existing method that is appropriate for the study under consideration can be used to generate the single- and two-locus test statistics. We select the top 200 SNP pairs with the largest two-locus test statistics and visualize their interactions. Single-locus test statistics are normalized to 0∼1 among all the SNPs, while two-locus test statistics are normalized to 0∼1 among the top 200 SNP pairs. The information on these SNP pairs is written into node and edge files, which will be used as the input files of EINVis. The LD information between the selected SNPs is generated from SNAP website [Johnson et al., 2008] with distance limit 500 kb. The LD value between two SNPs is the r2 statistic.
Results
In this section, we use the example data set on hypertension study to discuss the functions provided by EINVis and show how it can help to visualize and explore the genetic interaction network.
EIN Tree Ring View
We summary the key features of EINVis as follows.
Circular Layout
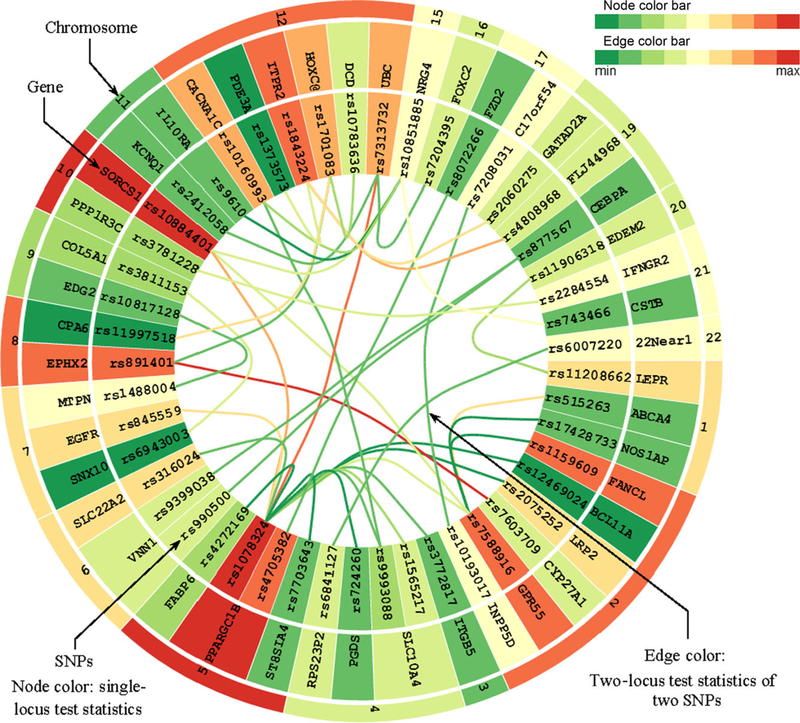
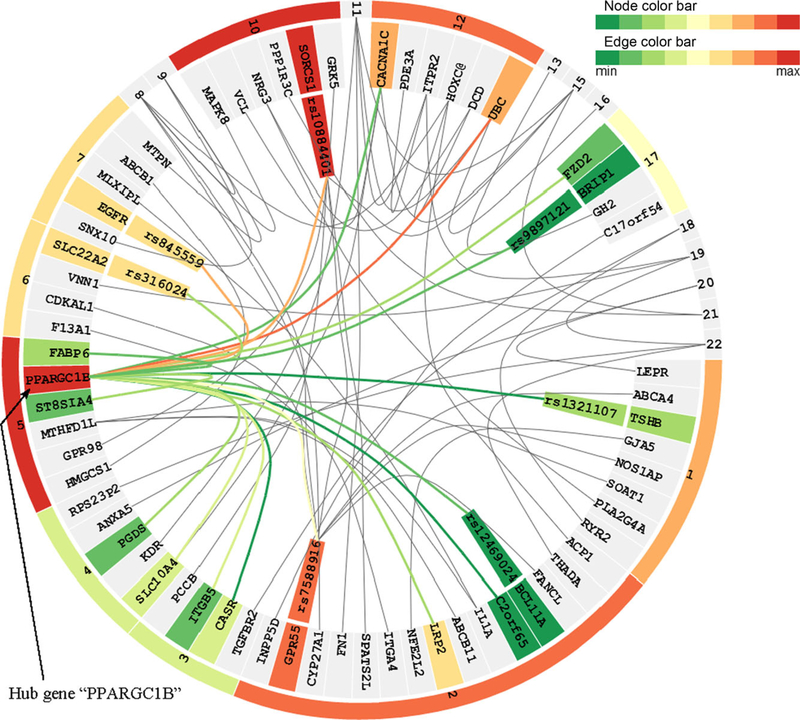
EINVis exerts a circular layout, in particular, a tree ring view [Holten, 2006], to visualize the hierarchical structure between SNPs, genes, and chromosomes, and the interactions between them. One screenshot of EINVis is shown in Figure 2. In this figure, each node is rendered as a ring sector. More specifically, the nodes in the inner ring represent the SNPs, the nodes in the middle ring represent the genes, and the nodes in the outer ring represent the chromosomes. This tree ring structure naturally represents the three-level hierarchical structure between SNPs, genes, and chromosomes. The mapping information between SNPs and genes can be found in public databases such as dbSNP
Figure 2.
EIN tree ring view screenshot 1. EIN tree ring view with node color mapping from single-locus test statistic and edge color from two-locus test statistic.
Distinguishing Marginal Effects From Interactions
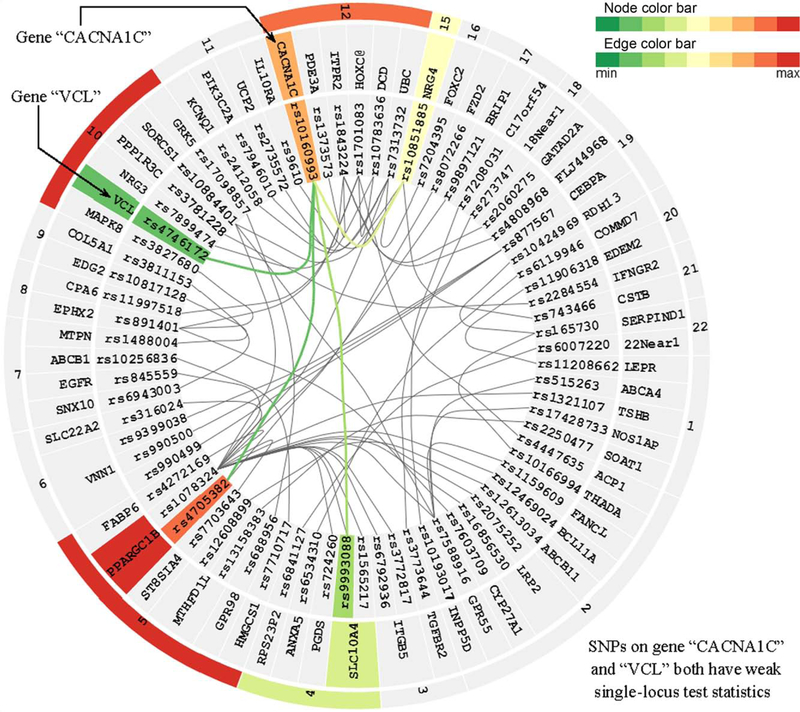
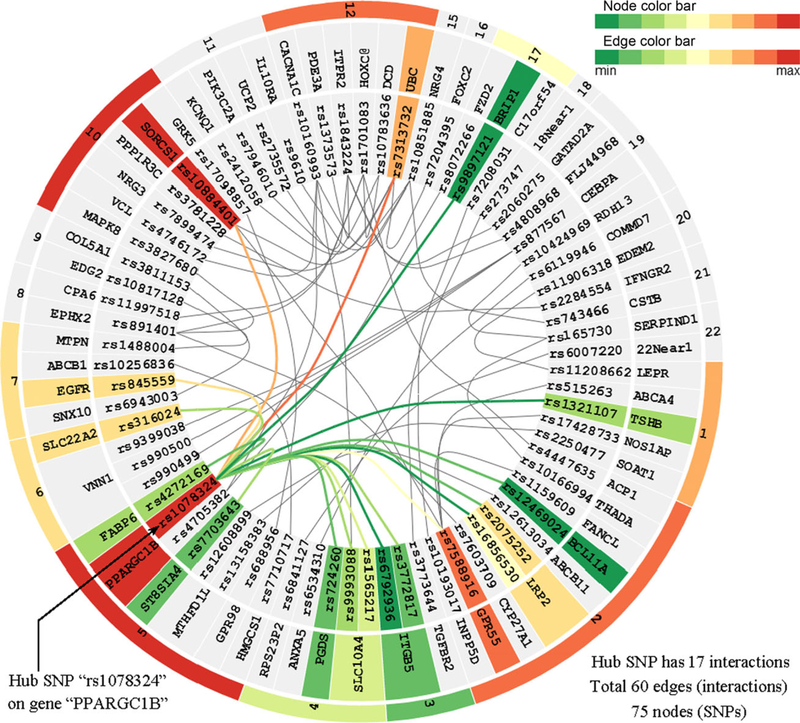
The interaction strength (represented by the two-locus test statistic) between two SNPs is encoded by the edge color. The same color mapping scheme is used to encode node color, i.e., the single-locus test. The color of a gene or chromosome node is mapped to the most significant single-locus test statistic of its child nodes. Visualizing both single- and two-locus test statistics helps to distinguish between marginal effects and interactions. For example, from Figure 3, we can see that SNP “rs10160993” on gene “CACNA1C” and SNP “rs4746172” on gene “VCL” both have weak single-locus associations, but their two-locus association is very strong. This indicates strong interaction effects. In Figure 4, we can observe that a hub SNP “rs1078324” on gene “PPARGC1B,” which interacts with many other SNPs, has strong marginal effect. Clearly, it is the marginal effect that makes this SNP a hub. EINVis allows distinguishing marginal effects from interactions. From the NCBI Genetic Association Database, we find that “CACNA1C” has been reported to have strong association with hypertension [Bremer et al., 2006; Kamide et al., 2009]. The literature shows that “CACNA1C” polymorphisms are associated with the efficacy of calcium channel blockers in the treatment of hypertension.
Figure 3.
EIN tree ring view screenshot 2. SNP “rs10160993” on gene “CACNA1C” is selected. SNP “rs10160993” on gene “CACNA1C” and SNP “rs4746172” on gene “VCL” have strong interaction and low marginal effects.
Figure 4.
EIN tree ring view screenshot 3. One hub SNP “rs1078324” on gene “PPARGC1B” is selected, and related nodes and edges are highlighted with colors.
Edge Bundling and Interaction Tracking
EINVis incorporates edge bundling technique [Holten, 2006] to reduce visual clutter when there are a large number of edges. This technique models each edge as a B-spline curve and bundle adjacent edges together to form a path from one node to another. It also helps to visualize implicit connections between parent nodes resulting from the explicit connections between their child nodes. EINVis also allows the user to track the interacting nodes. When one node is selected, all its interacting nodes and corresponding edges can be highlighted. For example, Figure 4 shows that one SNP “rs1078324” on gene “PPARGC1B” is selected, and all its interacting nodes and corresponding edges are highlighted, while unrelated nodes and edges become gray.
Nodes Collapsing and Expanding
The user can collapse or expand nodes using EINVis. This allows the user not only to focus on the set of interested nodes, but also to observe interactions at different levels, i.e., SNP-SNP, SNP-gene, and gene-gene interactions. When a gene node is collapsed, all its child SNPs nodes will disappear. The edges originally connecting its child SNP nodes will be connected to the collapsed gene node. If some edges are combined into one, we map the color to the most significant two-locus test statistic among those edges before collapsing. In this way, the number of nodes and edges is reduced and the interaction pattern between nodes at different levels can also be observed. For example, Figure 5 shows that gene “PPARGC1B” has interactions with many other genes and SNPs.
Figure 5.
EIN tree ring view screenshot 4. Some nodes are collapsed. One hub gene “PPARGC1B” is selected, and related nodes and edges are highlighted with colors.
Detecting Proxy SNPs Based on LD
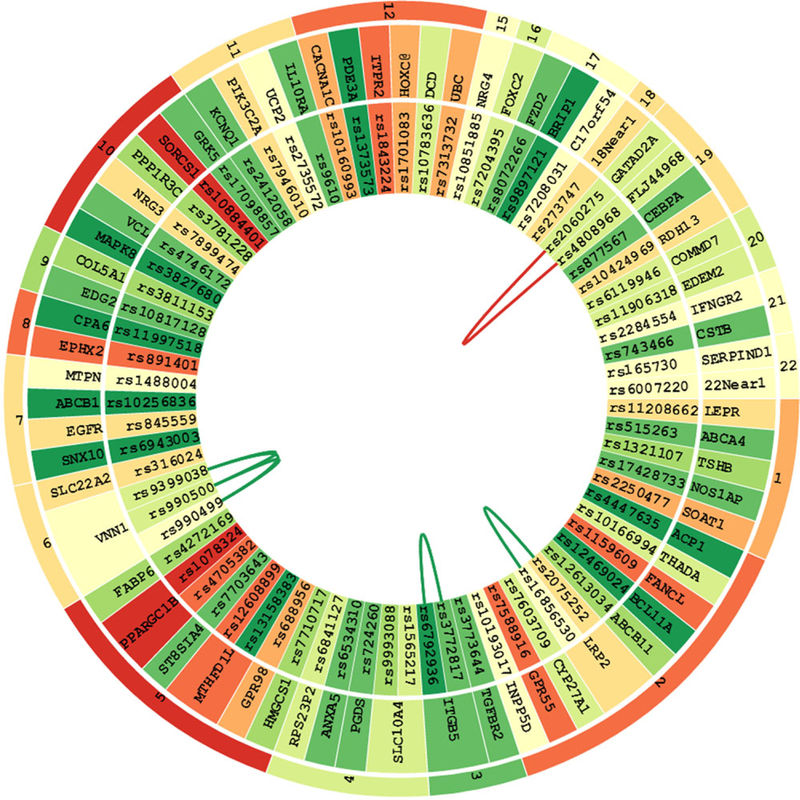
EINVis can help to detect proxy SNPs based on LD in the interaction network. LD should be eliminated when user constructs the interaction network. However, if not, we can observe some patterns in the EIN tree ring view that indicate LD between SNPs. For example, in Figure 2, we can observe that two almost parallel edges connecting SNP “rs877567” on gene “CEBPA” with two other SNPs “rs9399038” and “rs990500” on gene “VNN1.” We look up the two SNPs “rs9399038” and “rs990500” in SNAP [Johnson et al., 2008] and find that they are proxy SNPs based on LD. In Figures 3 and 4, we increase the number of edges and find that another SNP “rs990499” also has one parallel edge connecting with SNP “rs877567.” From SNAP, we find that the three SNPs “rs9399038,” “rs990500,” and “rs990499” are proxy based on LD. This LD information can also be observed in LD tree ring view as shown in Figure 7. Gene “VNN1” has been reported to be associated with hypertension [Zhu and Cooper, 2007]. Using our visualization tool EINVis, we find that this gene has strong interactions with gene “CEBPA” but itself has low marginal effect.
Figure 7.
LD tree ring view. LD tree ring view with node color mapping from single-locus test statistic and edge color from LD information. Three SNPs “rs9399038,” “rs990500,” and “rs990499” on gene “VNN1” are in strong LD.
Control Panel
The user has great flexibility to control the visualization effects. For example, the user can select how many edges to visualize in EINVis. Dynamically changing the number of edges to visualize has the advantage to view how the network changes when adding or deleting edges. For example, when we increase the number of edges from 80 to 120, SNP “rs1078324” remains to be a hub SNP as shown in Figures 2 and 4. The user can also configure the node and edge color bar separately. There are 12 color bars taken from Color-Brewer (http://colorbrewer2.org/) website. These color bars are optimal for human to distinguish different colors. In the experiments, we find that some edges with shallow color will become invisible under the white background. Thus we add black background check box control. When the user checks this box, the tree ring view will render a black background, so that the edges in the inner circle region will be easy to observe as the contrast increases. Experiments show that the colors from ColorBrewer are much clearer than the usual continuous color bar ranging from green to red. EINVis also provides this traditional color bar for comparison purpose. The user can also control the font size of the node label text. When there are too many SNPs, user can decrease the font size to save space to show more SNPs. The user can also choose to hide the node name and focus on the node and edge color patterns. When the user finds an interesting gene or SNP, he can choose to show the node name again.
Mouse-Over Function
In the EIN tree ring view, when the user points mouse to a SNP node, the corresponding marginal statistic will be shown at the upright corner of the ring circle. When the user points mouse to an edge connecting two SNPs, the corresponding two-locus test statistic will be shown at the same place. Similarly, in the LD tree ring view, when the mouse is pointed to an edge, the corresponding LD information will be shown. This allows the user to see the exact test value in addition to the mapped color on the node and edge.
Link to NCBI Website
EINVis also allows the user to directly connect to the NCBI website, so he can get more detailed information of selected SNPs or genes. In the EIN tree ring view, the user can click on one node while pressing “L” key to link to NCBI website. In the summary statistics webpage, each SNP and gene name has a hyperlink to NCBI.
Summary Statistics
Through the “View Rank” button in the control panel on the EIN tree ring view bubble, the user can link to a new page, which shows the rank of SNPs by their degree in the interaction network. The significance of a node with certain degree is accessed as follows. Suppose that there are nv nodes and ne edges in the network. Assuming each node has the same probability to connect to an edge, the probability that a node is connected to an edge is
The number of edges connected to a node follows a binomial distribution binom(ne, Pe). If one node has a degree of d, we can test if this node has excessive number of connected edges by calculating the P-value as: P = p binom(ne, d, Pe), where “p binom” is the function to calculate the P-value using binomial distribution.
Table 1 shows the results by using the hypertension data set. From the table, we can see that SNP “rs1078324” on gene “PPARGC1B” has the largest degree of 51 with highly sig-nificant test P-value. However, its single-locus test value is relatively large. It is likely that the marginal effect makes this SNP a hub. The second SNP “rs10851885” on gene “NRG4” has a degree of 13, which is also highly significant. We notice that this SNP has a relatively low single-locus test statistic. After further examination, we found that two additional genes, “NRG1” and “NRG3,” belong to the same gene family that also appears in the top 200 SNP pairs. This suggests “NRG” gene family warrants further investigation. We did not find “NRG” gene family reported to have association with hypertension. In Table 1, we notice that gene “ADRA1A” has been reported many times to be associated with hypertension [Kitsios and Zintzaras, 2010; Zhang et al., 2009]. We notice that gene “ADRA1A” only has a 0.572 single-locus test statistic. It is possible that the genetic interactions have made this gene visible.
Table 1. Partial table of SNPs rank.
- Number of nodes (SNPs): 218
- Number of edges (SNP-SNP interactions): 200
- Number of independent experiments: 200 (number of edges in EIN)
- Probability to yield success: 0.0092 (2/number of nodes in EIN)
| Index | Chrome ID | Gene name | SNP ID | Single-locus test statistics | Number of related edges | Binomial dist. P-value |
|---|---|---|---|---|---|---|
| 1 | 5 | PPARGC1B | rs1078324 | 0.947 | 51 | 0.0000 |
| 2 | 15 | NRG4 | rs10851885 | 0.552 | 13 | 7.4 × 10−9 |
| 3 | 2 | GPR55 | rs7588916 | 0.859 | 12 | 6.0 × 10−8 |
| 4 | 10 | SORCS1 | rs10884401 | 1.000 | 10 | 3.2 × 10−6 |
| 5 | 12 | UBC | rs7313732 | 0.725 | 9 | 2.0 × 10−5 |
| 6 | 12 | ITPR2 | rs1843224 | 0.852 | 9 | 2.0 × 10−5 |
| 7 | 12 | HOXC@ | rs1701083 | 0.772 | 9 | 2.0 × 10−5 |
| 8 | 19 | MTHFD1L | rs12608899 | 0.835 | 7 | 0.0006 |
| 9 | 5 | PARGC1B | rs4705382 | 0.809 | 7 | 0.0006 |
| 10 | 8 | EPHX2 | rs891401 | 0.819 | 6 | 0.0027 |
| 11 | 2 | FANCL | rs1159609 | 0.820 | 5 | 0.0109 |
| 12 | 12 | CACNA1C | rs10160993 | 0.669 | 5 | 0.0109 |
| 13 | 8 | ADRA1A | rs498194 | 0.572 | 4 | 0.0382 |
| 14 | 9 | VLDLR | rs2242104 | 0.769 | 4 | 0.0382 |
| 15 | 12 | DCD | rs10783636 | 0.398 | 4 | 0.0382 |
| 16 | 21 | IFNGR2 | rs2284554 | 0.488 | 4 | 0.0382 |
| 17 | 12 | HOXC@ | rs12309639 | 0.684 | 3 | 0.1135 |
EIN Matrix View
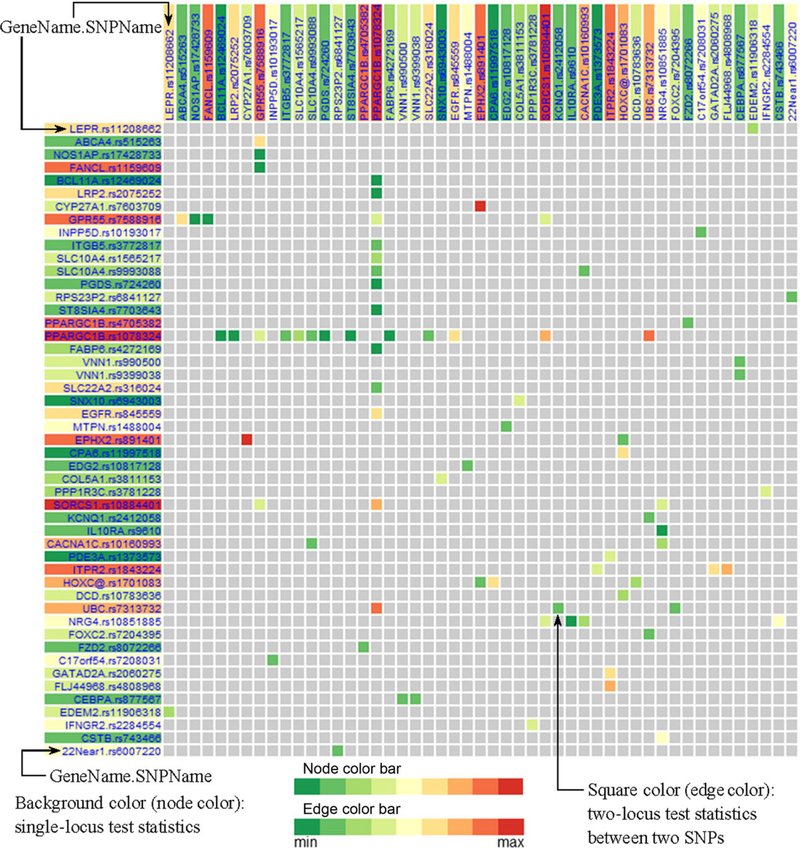
The EIN matrix view visualizes the adjacency matrix of the genetic interaction network. One screenshot is shown in Figure 6. Note that the adjacency matrix is symmetric. In the left margin, we list all the SNP names in the format of “GeneName.SNPName.” The background color of an SNP name is mapped to its single-locus test statistic. The SNP name background color bar is the same as the nodes color bar in EIN tree ring view. In the top margin, the SNPs names are shown with the same order as that in the left margin. In the matrix, the color of each entry represents the interaction strength between two SNPs on the corresponding row and column. The gray color means that there is no interaction. When the number of SNPs is too large to fit into the screen, the matrix view will visualize part of the matrix and provide a panning function to allow the user to move the window and visualize other parts of the adjacency matrix.
Figure 6.
EIN matrix view. EIN matrix view with SNP name (in left or top margin) background color mapping to single-locus test statistic, and squares (in the matrix region) color mapping to two-locus test statistic.
LD Tree Ring View
The LD tree ring view enables the visualization of the LD information between SNPs. An example is shown in Figure 7. The LD tree ring view has exactly the same layout as the EIN tree ring view. The only difference is that the edges and their weights are replaced by the LD information. The LD tree ring view is an auxiliary view and synchronizes with the EIN tree ring view. When the user interacts with the EIN tree ring view, the LD tree ring view will also have corresponding responses. When mouse is pointed to an edge between two SNPs nodes, its corresponding LD value will be shown at the upright corner of the ring circle. In the example shown in Figure 7, three SNPs “rs9399038,” “rs990500,” and “rs990499” on gene “VNN1” are in strong LD. This is consistent with the observations discussed before.
Discussion
EINVis provides a visualization framework for exploring genetic interactions. It utilizes the tree ring view to simultaneously visualize the hierarchical SNP-gene-chromosome structure and the interacting pattern between nodes at different levels. EINVis provides a variety of functions for the user to conveniently visualize and explore the genetic interaction network. It can significantly facilitate the identification of candidate genes and novel interaction patterns embedded in the large amount of possible interactions.
Acknowledgments
We thank Marziehossadat Ayati for valuable suggestions. This work was partially supported by the US National Science Foundation grants IIS-1162374, IIS-1218036, DBI-1062057, IIS-1302755, EPS-0903234, DUE-0817106, and a Public Health Service grant (HG003054) from the US Na-tional Human Genome Research Institute.
Footnotes
The authors have no conflict of interest.
References
- Bostock M, Ogievetsky V, Heer J. 2011. D3: data-driven documents. IEEE Trans Vis Comput Graph 17(12):2301–2309. [DOI] [PubMed] [Google Scholar]
- Bragdon A, Reiss SP, Zeleznik R, Karumuri S, Cheung W, Kaplan J, Coleman C, Adeputra F, LaViola JJ Jr. 2010. Code bubbles: rethinking the user interface paradigm of integrated development environments. In Proceedings of the 32nd ACM/IEEE International Conference on Software Engineering, Vol. 1 Cape Town, South Africa: ACM; p 455–464. [Google Scholar]
- Bremer T, Man A, Kask K, Diamond C. 2006. CACNA1C polymorphisms are associated with the efficacy of calcium channel blockers in the treatment of hypertension. Pharmacogenomics 7(3):271–279. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. 2009. Detecting gene–gene interactions that underlie human diseases. Nat Rev Genet 10(6):392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva NT, Assenov Y, Domingues FS, Albrecht M. 2012. Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc 7(4):670–685. [DOI] [PubMed] [Google Scholar]
- Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Pihur V. 2011. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet 20(11):2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holten D 2006. Hierarchical edge bundles: visualization of adjacency relations in hierarchical data. IEEE Trans Vis Comput Graph 12(5):741–748. [DOI] [PubMed] [Google Scholar]
- Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C. 2009. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res 37(Suppl 2):W115–W121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Sinnott-Armstrong NA, Kiralis JW, Andrew AS, Karagas MR, Moore JH. 2011. Characterizing genetic interactions in human disease association studies using statistical epistasis networks. BMC Bioinformatics 12(1):364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. 2008. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24(24):2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamide K, Yang J, Matayoshi T, Takiuchi S, Horio T, Yoshii M, Kawano Y. 2009. Genetic polymorphisms of L-type calcium channel alpha1C and alpha1D subunit genes are associated with sensitivity to the antihypertensive effects of L-type dihydropyridine calcium-channel blockers. Circ J 73(4):732. [DOI] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, McCarthy MI. 2008. Concept, design and implementation of a cardiovascular genecentric 50 k SNP array for large-scale genomic association studies. PLoS One 3(10): e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsios GD, Zintzaras E. 2010. Synopsis and data synthesis of genetic association studies in hypertension for the adrenergic receptor family genes: the CUMAGAS-HYPERT database. Am J Hypertens 23(3):305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Bragdon AC, Pan Z, Zhang M, Swartz SM, Laidlaw DH, Zhang C, Liu H, Chen J. 2011. VisBubbles: a workflow-driven framework for scientific data analysis of time-varying biological datasets (SAP-0281). In SIGGRAPH Asia Posters Hong Kong, China: ACM. [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, Visscher PM. 2009. Finding the missing heritability of complex diseases. Nature 461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BA, Crowe JE, Guo J, Tian D. 2009. Capturing the spectrum of interaction effects in genetic association studies by simulated evaporative cooling network analysis. PLoS Genet 5(3):e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Munzner T, Pfister H. 2009. MizBee: a multiscale synteny browser. IEEE Trans Vis Comput Graph 15(6):897–904. [DOI] [PubMed] [Google Scholar]
- Moore JH, Williams SM. 2009. Epistasis and its implications for personal genetics. Am J Hum Genet 85(3):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos GA, Wegener AL, Schneider R. 2008. A survey of visualization tools for biological network analysis. Biodata Min 1(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9(11):855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Zhang J, Sun Y, Liu D, Ye D, Yin Y. 2011. Performance analysis of novel methods for detecting epistasis. BMC Bioinformatics 12(1):475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M, Hallett M. 2007. Tools for visually exploring biological networks. Bioinformatics 23(20):2651–2659. [DOI] [PubMed] [Google Scholar]
- Steen VK. 2012. Travelling the world of gene–gene interactions. Briefings Bioinform 13(1):1–19. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu G, Feng M, Wong L. 2011. An empirical comparison of several recent epistatic interaction detection methods. Bioinformatics 27(21):2936–2943. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hong X, Liu H, Huo Y, Xu X. 2009. Arg347Cys polymorphism of α1Aadrenoceptor gene is associated with blood pressure response to nifedipine GITS in Chinese hypertensive patients. J Hum Genet 54(6):360–364. [DOI] [PubMed] [Google Scholar]
- Zhu X, Cooper RS. 2007. Admixture mapping provides evidence of association of the VNN1 gene with hypertension. PLoS One 2(11):e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, Levy D. 2011. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet 20(11):2285–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]