SUMMARY
TGFβ signaling controls diverse normal developmental processes and pathogenesis of diseases including cancer and autoimmune and fibrotic diseases. TGFβ responses are generally mediated through transcriptional functions of Smads. A key step in TGFβ signaling is ligand-induced phosphorylation of receptor-activated Smads (R-Smads) catalyzed by the TGFβ type I receptor kinase. However, the potential of Smad dephosphorylation as a regulatory mechanism of TGFβ signaling and the identity of Smad-specific phosphatases remain elusive. Using a functional genomic approach, we have identified PPM1A/PP2Cα as a bona fide Smad phosphatase. PPM1A dephosphorylates and promotes nuclear export of TGFβ-activated Smad2/3. Ectopic expression of PPM1A abolishes TGFβ-induced antiproliferative and transcriptional responses, whereas depletion of PPM1A enhances TGFβ signaling in mammalian cells. Smad-antagonizing activity of PPM1A is also observed during Nodal-dependent early embryogenesis in zebrafish. This work demonstrates that PPM1A/PP2Cα, through dephosphorylation of Smad2/3, plays a critical role in terminating TGFβ signaling.
INTRODUCTION
Members of the TGFβ superfamily activate a broad range of cellular responses in metazoa, including cell proliferation, differentiation, extracellular matrix (ECM) remodeling, and embryonic development (Whitman and Raftery, 2005). They play key roles in the pathogenesis of cancer and fibrotic, cardiovascular, and autoimmune diseases (Akhurst, 2004; Waite and Eng, 2003). Smads are essential intracellular transducers for TGFβ signals (Feng and Derynck, 2005; ten Dijke and Hill, 2004) and also serve as a point of crosstalk with other signaling pathways (Derynck and Zhang, 2003; Moustakas and Heldin, 2005). In response to TGFβ, receptor-activated Smads (R-Smads) are phosphorylated in their C-terminal SXS motif by type I receptors. Phosphorylated R-Smads form a complex with Smad4 and are transported into the nucleus, where Smads cooperate with specific DNA binding transcription factors to regulate gene transcription in a context-dependent manner (Feng and Derynck, 2005).
Signal transduction pathways are often regulated by dynamic interplay between protein kinases and phosphatases. Ligand stimulation results in R-Smad phosphorylation, inhibition of nuclear export, and thus persistent accumulation of Smad complexes in the nucleus (Schmierer and Hill, 2005). Recent studies imply that R-Smads may require dephosphorylation to be exported from the nucleus (Inman et al., 2002b; Schmierer and Hill, 2005; Xu et al., 2002). Although this implicates the existence of a Smad-specific phosphatase (or phosphatases), the identity of the phosphatases responsible for dephosphorylating the SXS motif remains unknown.
Protein serine/threonine phosphatases (PS/TPs) control cellular functions through cleavage of phosphate from phosphorylated serine and threonine residues (pS/Ts) in proteins. PS/TPs are classified into three structurally distinct families: PPM, PPP, and FCP/SCP (Cohen, 2003; Gallego and Virshup, 2005). The PPM family, including PPM1A/PP2Cα, comprises metal-ion-dependent phosphatases that are mostly monomeric. Members of the PPP family, including PP1 and PP2A, are oligomeric holoenzymes with structurally related catalytic subunits and distinct regulatory subunits. The FCP/SCP family is represented by FCP1, a phosphatase that dephosphorylates the carboxy-terminal domain of RNA polymerase II. In addition to PS/TPs, dual-specific phosphatases (DUSPs) also catalyze dephosphorylation of pS/Ts.
To identify the long sought-after Smad phosphatase (or phosphatases), we took a functional genomic approach to search for those PS/TPs whose expression reduces SXS phosphorylation of Smad2/3. Here, we present the biochemical, genetic, and functional evidence that clearly identify PPM1A/PP2Cα as a bona fide phosphatase that dephosphorylates the SXS motif of Smads and terminates TGFβ signaling.
RESULTS
Dephosphorylation of the TGFβ Signal Transducers Smad2 and Smad3
We first studied the kinetics of Smad2/3 phosphorylation. In immortalized human keratinocyte HaCaT cells, levels of phosphorylated Smad2/3 (P-Smad2/3) peaked at 1 hr after TGFβ treatment and gradually declined in the presence of continuous TGFβ (Figure 1A). To accurately analyze the dephosphorylation of Smad2/3, HaCaT cells were treated with TGFβ (2 ng/ml, 30 min) to generate a pool of P-Smad2/3 and then with TβRI kinase inhibitor SB431542 to prevent rephosphorylation of dephosphorylated Smad2/3 (Inman et al., 2002a) (Figure 1B). The results showed that prephosphorylated Smad2/3 lasted for <2 hr in the presence of SB431542 and absence of proteasomal inhibitor MG-132 (Figures 1C and 1D). Addition of MG-132 (20 μM) slowed down the disappearance of P-Smad2/3 but did not completely restore the P-Smad2/3 levels (Figures 1C and 1D; see also Figure S1 in the Supplemental Data available with this article online), even though it effectively prevented SnoN degradation (Figure S1). This indicates that a protein phosphatase (or phosphatases), not a proteasome, is primarily responsible for P-Smad2/3 loss.
Figure 1. Dynamic Phosphorylation and Dephosphorylation of Smad2/3.
(A) Time course of Smad2/3 phosphorylation. HaCaT cells were treated with 2 ng/ml of TGFβ1 for the indicated time periods. P-Smad2/3 and total Smad2/3 were analyzed by Western blotting. Left: Line graph representing three independent experiments. Intensity of P-Smad2/3 relative to total Smad2/3 from each experiment was quantified by NIH Image software. Values are the mean of at least two independent experiments; error bars are ± standard deviation of the mean. Right: A representative Western blot.
(B) A schematic representation of treatments designed for (C)–(F).
(C) Analysis of Smad2 dephosphorylation. HaCaT cells were treated with 2 ng/ml of TGFβ1 for 30 min, followed by TGFβ washout and simultaneous addition of 5 μM SB431542 and 20 μM MG-132. Graph shows relative P-Smad2 level (over total Smad2) from three independent experiments, with values and error bars representing mean and standard deviation. The insert is one representative experiment.
(D) Experiments equivalent to (C) analyzing Smad3 dephosphorylation in HaCaT cells.
(E) Effect of okadaic acid (OA) on TGFβ-dependent phosphorylation of Smad2/3. HaCaT cells were treated with TGFβ1 and SB431542 as in (C); in a separate group of cells, 5 nM OA was added 3.5 hr before TGFβ and continued with TGFβ and SB431542 treatment (for a total of 8 hr). Graphical analysis of relative P-Smad2/3 levels is presented here (line graph) and in Figure S2A. Values are mean of three independent experiments; error bars are ± standard deviation of the mean.
(F) Effect of OA on TβRI(T204D)-induced phosphorylation of Smad2/3. HaCaT cells were infected with AdTβRI(T204D) adenoviruses at moi of 200. After 48 hr, cells were treated with 5 μM SB431542. Five nanomolar OA was added 3.5 hr before SB431542 addition and remained with SB431542. Graphical analysis of relative P-Smad2/3 levels is presented here (line graph) and in Figure S2B. Values and error bars are mean and standard deviation of three independent experiments.
We next tested whether accumulation of P-Smad2/3 is sensitive to okadaic acid (OA), an inhibitor widely used to implicate the involvement of PPP phosphatases in regulating cellular function (Cohen, 2003; Gallego and Virshup, 2005). OA treatment increased the level of P-Smad2/3, while it did not affect total levels of Smad2/3 (Figure 1E; Figure S2A), suggesting an involvement of OA-sensitive phosphatases in regulating P-Smad2/3 levels. To precisely determine whether OA-sensitive phosphatases directly dephosphorylate P-Smad2/3, we took advantage of TβRI(T204D), a gain-of-function mutant of human TGFβ type I receptor that constitutively phosphor-ylates Smad2/3 and thus avoids potential effects of dephosphorylation on the receptors. OA did not have any effects on P-Smad2/3 induced by TβRI(T204D) (Figure 1F; Figure S2B), suggesting that dephosphorylation of P-Smad2/3 is controlled by an OA-resistant phosphatase (or phosphatases) at the level upstream of Smad2/3 activation.
Functional Genomic Screen Identifies PPM1A as a Smad Phosphatase
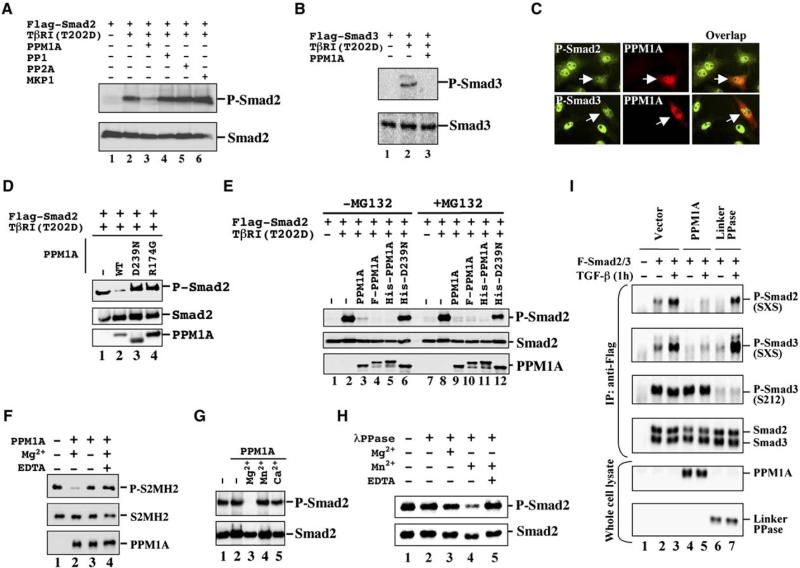
The hypothetical Smad phosphatase (or phosphatases) targeting the SXS motif should belong to either the PS/TP or the DUSP family. In a search to identify Smad phosphatase (or phosphatases), we focused on the PS/TP family. With the availability of human genome databases, we generated expression plasmids for 39 phosphatases (catalytic subunits), including 14 PPMs, 13 PPPs, 5 FCP/SCPs, 1 PH-domain phosphatase (PHLPP), 4 DUSPs, and 2 pyrophosphatases. We then examined the effects of each phosphatase on the P-Smad2/3 level induced by constitutively active rat TβRI(T202D) and found that only PPM1A/PP2Cα potently reduced the level of P-Smad2 accumulation (Table S1). These phosphatases are untagged to ensure no interference with their activities, but a separate Flag-tagged set of phosphatases were also made to examine their expression in cells (Figure S3A). PPM1A clearly reduced the level of P-Smad2/3 induced by TβRI(T202D) (Figures 2A and 2B) or TGFβ1 ligand (data not shown), whereas other phosphatases such as PPP1CA/PP1, PPP2CA/PP2A, and DUSP1/MKP1 had no effect (Figure 2A). PPM2C/PDP1 and PDP2, mammalian homologs of Drosophila PDP, which was recently identified as a Mad phosphatase (Chen et al., 2006), also exhibited no effects on Smad2 dephosphorylation (Table S1 and Figure S3B). Immunofluorescence staining also supports the notion that PPM1A promotes dephosphorylation of Smad2/3, as less P-Smad2/3 was observed in cells expressing exogenous PPM1A in HaCaT cells (Figure 2C).
Figure 2. PPM1A Dephosphorylates Smad2 and Smad3.
(A) Smad2 dephosphorylation by PPM1A. In transfected 293T cells, Flag-Smad2 was anti-Flag immunoprecipitated, and levels of P-Smad2 and total Smad2 were assessed by Western blotting.
(B) PPM1A dephosphorylates Smad3 in 293T cells.
(C) PPM1A reduces endogenous P-Smad2/3 levels. HaCaT cells were transfected with PPM1A, treated with TGFβ (2 ng/ml, 1 hr), and immunostained using anti-PPM1A (Texas red) and P-Smad2 or P-Smad3 (FITC). Arrow indicates reduced P-Smad2/3 in PPM1A-transfected cells.
(D) Smad2 dephosphorylation requires the catalytic activity of PPM1A.
(E) Smad2 dephosphorylation occurs in the presence of MG-132. Flag-PPM1A or His-PPM1A is as efficient as PPM1A.
(F) PPM1A dephosphorylates P-Smad2 MH2 domain (P-S2MH2). Dephosphorylation of P-S2MH2 required 20 mM Mg2+ (lane 2) and was abolished by 40 mM EDTA (lane 4). Equal amounts of semisynthetic recombinant P-S2MH2 (100 ng, lanes 1–4) and recombinant PPM1A (100 ng, lanes 2–4) were loaded.
(G) Smad2 dephosphorylation by PPM1A depends on 20 mM Mg2+ (lane 3), but not 2 mM Mn2+ (lane 4) or 20 mM Ca2+ (lane 5).
(H) Smad2 dephosphorylation by λ phosphatase (λPPase).
(I) PPM1A specifically dephosphorylates the SXS motif, but not pS212 of Smad3. Linker phosphatase specifically dephosphorylates pS212, but not the SXS motif.
To confirm whether the PPM1A phosphatase activity is essential for Smad2 dephosphorylation, we generated two catalytically inactive PPM1A mutants, D239N and R174G (Jackson et al., 2003). We found both D239N and R174G mutants lost the ability to dephosphorylate Smad2 (Figure 2D, lanes 3 and 4).
We next examined the effect of PPM1A on Smad2 dephosphorylation in the presence or absence of MG-132. Results showed that MG-132 treatment did not block the PPM1A-induced decrease in P-Smad2 level (Figure 2E, lanes 9–11), suggesting that PPM1A reduces the level of P-Smad2 independently of the 26S proteasome.
To rule out the possibility that PPM1A activates another phosphatase in cells that could be the direct Smad phosphatase, we carried out a cell-free dephosphorylation assay. Semisynthetic recombinant phospho-MH2 domain of Smad2 (P-S2MH2) (Wu et al., 2001) could be effectively dephosphorylated by recombinant PPM1A (Figure 2F, lane 2). PPM1A activity required Mg2+, as EDTA abolished dephosphorylation of P-S2MH2 (Figure 2F, lane 4), consistent with the property of PPM1A as a metal-ion-dependent phosphatase (Tamura et al., 2003). PPM1A failed to dephosphorylate Smad2 in the presence of only Mn2+ or Ca2+ (Figure 2G). In contrast, nonspecific bacteriophage λ phosphatase had Mn2+-dependent, but Mg2+-independent, dephosphorylating activity toward P-Smad2 (Figure 2H). These results suggest that phospho-Smad2 is a direct substrate of Mg2+-dependent PPM1A.
To further examine the specificity of PPM1A, we tested whether PPM1A dephosphorylates pS212 in the linker region of Smad3. The S212 residue is phosphorylated by Cdk4 (Matsuura et al., 2004). PPM1A failed to dephosphorylate pS212 (Figure 2I) and other pS/T residues in Smad3 (data not shown). On the other hand, linker PPase dephosphorylated pS212, but not the SXS motif. Thus, the SXS motif is a direct and specific target for PPM1A.
PPM1A Physically Interacts with Smad2
To test whether PPM1A binds to Smad2, we performed coimmunoprecipitation experiments. Results showed that Flag-PPM1A interacted with HA-Smad2 (Figure 3A, lane 3). Catalytically inactive mutant D239N also retained the ability to interact with Smad2, suggesting that the phosphatase activity is not essential for Smad binding (Figure 3A, lane 4). Similarly, Smad3 interacted with PPM1A (Figure S4). Notably, the PPM1A-Smad2 interaction also occurred under physiological conditions, as TGFβ stimulated endogenous PPM1A-Smad2 coimmunoprecipitation (Figure 3B).
Figure 3. PPM1A Interacts with Smad2.
(A) Coimmunoprecipitation of Flag-PPM1A and HA-Smad2 in 293T cells. IP, immunoprecipitation; IB, Western blotting; WCL, whole-cell lysates.
(B) Endogenous PPM1A-Smad2 interaction is TGFβ inducible. HaCaT cells were treated with TGFβ1 (1 hr), and cell lysates were subjected to IP using anti-PPM1A (lane 5 and 6) or a control mouse antibody (lane 4).
(C) Smad2(V398R) mutant interacts with PPM1A in 293T cells.
(D) Subcellular colocalization of PPM1A and Smad2 in HaCaT cells. Smad2 and PPM1A proteins were detected by using anti-Smad2 (FITC) and anti-PPM1A (Texas red) antibodies. DNA was stained using DAPI dye.
(E) PPM1A is primarily localized in the nucleus. Left: HaCaT cells treated with TGFβ1 (2 ng/ml for 1 hr) were crosslinked with formaldehyde before fractionation and Western blotting analysis (Supplemental Data). C, cytoplasmic fraction; N, nuclear fraction. Right: NIH 3T3 cells.
(F) Direct interaction of PPM1A with Smad2/3. Purified recombinant His-PPM1A or His-D239N protein bound to recombinant GST-Smad2 (lanes 4 and 7) or Smad3 (lanes 5 and 8) but not GST alone (lanes 3 and 6) was detected by anti-PPM1A Western blotting.
(G) PPM1A exclusively binds to P-Smad2. Recombinant Smad2 MH2 (aa 241–467), phosphorylated (lane 1) or unphosphorylated (lane 2), bound to recombinant His-PPM1A protein was detected by Western blotting.
We next examined the interaction of Smad2 mutants with PPM1A. Smad2 phosphorylation-deficient mutant 2SA and phosphorylation-mimetic mutant 2SD carry Ser-to-Ala and Ser-to-Asp substitutions at the SXS motif, respectively. Smad2(V398R) has a defect in its nuclear export and thus localized in the nucleus (Xu et al., 2002). Results in Figure 3C show that whereas Smad2 wild-type and 2SA mutant interacted similarly with PPM1A, V398R mutant clearly exhibited an increased binding to PPM1A.
Endogenous PPM1A was primarily localized in the nucleus regardless of TGFβ stimulation as determined by immunofluorescence (Figure 3D). Nuclear/cytoplasmic fractionation experiments further confirmed that PPM1A was present in nuclear fraction (Figure 3E). Nuclear protein lamin A and cytoplasmic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as sample quality controls (Figure 3E). To examine whether endogenous Smad2 colocalized with PPM1A, PPM1A-stained HaCaT cells were counterstained with anti-Smad2 antibody. As expected, in the absence of TGFβ, Smad2 expression was both in the nucleus and cytoplasm, with a diffuse distribution in the cytoplasm (Figure 3D, upper left). Following TGFβ stimulation, Smad2 became localized exclusively in the nucleus (Figure 3D, lower left). Staining of PPM1A overlaps with nuclear Smad2, particularly in the presence of TGFβ.
To assess direct interaction between PPM1A and Smad2, we carried out an in vitro binding assay using purified recombinant proteins. As shown in Figure 3F, PPM1A bound to glutathione S-transferase (GST)-Smad2 fusion protein (lanes 4 and 7) but not GST protein alone (lanes 3 and 6). PPM1A also bound directly to GST-Smad3 fusion protein (lanes 5 and 8). Both wild-type and mutant PPM1A bound equally to Smad2 or Smad3. Furthermore, when assayed in parallel, phosphorylated Smad2, but not the unphosphorylated form, could bind to PPM1A (Figure 3G).
PPM1A Regulates Complex Formation and Nuclear Export of Smad2
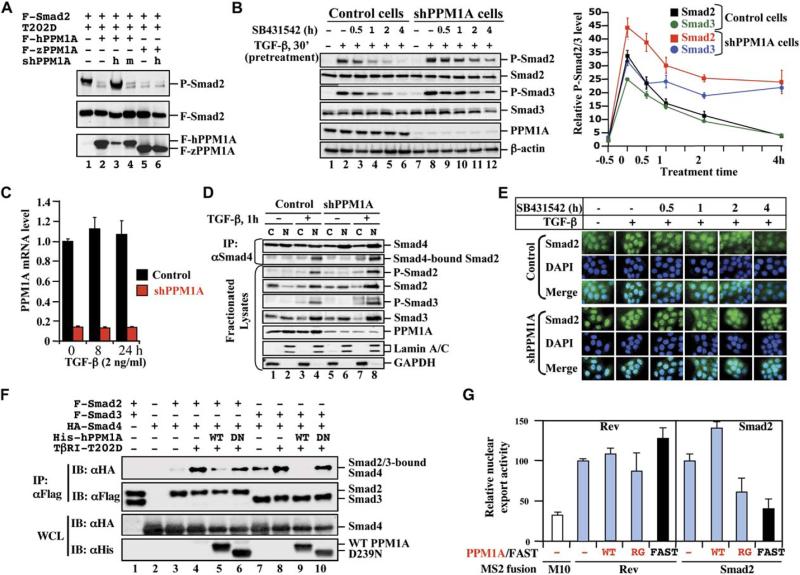
To further establish the function of PPM1A in TGFβ signaling, we investigated its loss-of-function or gain-of-functions effects on Smad phosphorylation, oligomerization, and nuclear export. We first used short hairpin RNAs (shRNAs) to knock down the expression of PPM1A (shPPM1A). Two of three shPPM1As could efficiently and specifically knock down the expression of PPM1A in transiently transfected 293T cells (Figure S5A). As shown in Figure 4A, the P-Smad2 level induced by TGFβ was diminished in the presence of human PPM1A (lane 2) but was restored by coexpression of shPPM1A494, correlating with the depleted level of PPM1A (lane 3). Similar results were observed on Smad3 dephosphorylation (Figure S5B). As a control, shmPPM1A494 targeted to the corresponding mouse PPM1A sequence (with 3 bp mismatch between the human and mouse target sequences) did not suppress the expression of human PPM1A and thus did not restore P-Smad2 accumulation (Figure 4A, lane 4). More importantly, zebrafish PPM1A (zPPM1A), which displays 75% sequence identity to human PPM1A (Figure S6), was capable of dephosphorylating P-Smad2 (Figure 4A, lane 5) and P-Smad3 (Figure S5) and retained the ability to dephosphorylate P-Smad2/3 even in the presence of shPPM1A494 (Figure 4A, lane 6; Figure S5), indicating the specificity of shPPM1A494 toward human PPM1A.
Figure 4. PPM1A Regulates Phosphorylation, Oligomerization, and Nuclear Export of Smads.
(A) Specific PPM1A knockdown in 293T cells. Flag-hPPM1A, Flag-tagged human PPM1A; Flag-zPPM1A, Flag-zebrafish PPM1A; h, shPPM1A494 against human PPM1A; m, shmPPM1A494 against mouse PPM1A. The level of P-Smad2 was inversely correlated with that of PPM1A.
(B) Reduced Smad2/3 dephosphorylation in HaCaT-shPPM1A cells. Left: Cells were treated with TGFβ and SB431542 and analyzed as described in Figure 1C. Right: Line graph showing relative levels of P-Smad2/3 over those of total Smad2/3 from three independent clones with two experiments each, with values and error bars representing the average and standard deviation.
(C) qRT-PCR analysis of PPM1A mRNA in shPPM1A cells. Values and error bars represent mean and standard deviation of two experiments (each with triplicates).
(D) Stable PPM1A depletion increases Smad2 phosphorylation and association with Smad4. HaCaT-shPPM1A or control cells were treated with TGFβ (2 ng/ml, 1 hr). Nuclear and cytoplasmic fractions (Supplemental Data) were subjected to anti-Smad4 IP and Western blotting.
(E) PPM1A depletion increases Smad2 accumulation in the nucleus. Cells were treated with TGFβ for 1 hr and then with SB431542 for 0.5, 1, 2, or 4 hr. Smad2 was visualized using anti-Smad2 antibody (FITC). DAPI (DNA staining) and merge are indicated.
(F) Wild-type PPM1A (WT), but not D239 mutant (DN), causes Smad2/3-Smad4 dissociation.
(G) PPM1A promotes nuclear export of Smad2. An MS2-based quantitative analysis of nuclear transport assay system was used (Supplemental Data). Values and error bars represent mean and standard deviation of three experiments.
We also established HaCaT cell lines stably expressing shPPM1A. In these cells, which exhibited an at least 80% reduction in PPM1A protein level (Figure 4B, lanes 7–12) or mRNA level (Figure 4C), levels of P-Smad2/3 (relative to total levels of Smad2/3) in response to TGFβ were significantly increased (Figure 4B, lane 8). In the presence of SB431542 that blocks rephosphorylation of Smad2/3 by inhibiting TβRI, the increased P-Smad2/3 levels in shPPM1A cells remained higher than those in control cells at the same time point (Figure 4B), suggesting a loss of dephosphorylation.
Since PPM1A is a phosphatase toward P-Smad2/3 in the nucleus (Figure 3), we anticipate that depletion of PPM1A increases the levels of P-Smad2/3 and subsequently their association with Smad4 in the nucleus. Nuclear and cytoplasmic fractions were obtained from both shPPM1A and control cells with or without TGFβ treatment. In control cells, TGFβ clearly induced Smad2/3 to shift from cytoplasm to the nucleus, and P-Smad2/3 were only observed in TGFβ-treated nuclear fractions (Figure 4D, lanes 1–4). Depletion of PPM1A further increased the cytoplasmic-to-nuclear shift of Smad2/3. As a result, the level of P-Smad2/3 was higher, and consequently, a higher level of Smad2-Smad4 complex accumulated in the nucleus in shPPM1A cells (Figure 4D, lanes 5–8). Immunofluorescence also confirmed that shPPM1A cells had a higher level of Smad2 in the nucleus in the absence of TGFβ stimulation (Figure 4E). Notably, whereas SB431542 rapidly reduced nuclear accumulation of Smad2 in control cells, knockdown of PPM1A appeared to maintain a high level of nuclear Smad2 even in the presence of SB432542 (Figure 4E).
Conversely, overexpression of PPM1A abolished TGFβ-induced complex formation of Smad2-Smad4 (Figure 4F, compares lanes 4 and 5) and Smad3-Smad4 (compare lanes 8 and 9). The phosphatase-dead D239N mutant had no effects on complex formation between Smad2/3 and Smad4 (lanes 6 and 10). Considering that PPM1A dephosphorylates Smad2/3 in the nucleus, these results indicate that PPM1A causes dissociation of dephosphorylated Smad2/3 from Smad4.
We next sought to determine whether PPM1A regulates nuclear export of Smad2 since previous studies demonstrated that dephosphorylated Smad2 cycles back to the cytoplasm (Inman et al., 2002b; Xu et al., 2002). In this regard, we used a highly sensitive nuclear-export reporter assay (Coburn et al., 2001; Cullen, 2004) that was previously used to identify residues essential for Smad2 nuclear export (Xu et al., 2002) (Figure S7). As controls, HIV RNA export factor Rev, but not its mutant M10, showed clear nuclear-export activity (Coburn et al., 2001; Cullen, 2004; Figure 4G). As expected, Smad2 also exhibited nuclear-export activity, which is inhibited by FAST-1 (Xu et al., 2002; Figure 4G). We found that PPM1A further increased nuclear-export activity of Smad2, but not that of Rev, whereas the phosphatase-dead mutant R174G dominant-negatively inhibited Smad2 nuclear export. Thus, consistent with its role in dephosphorylating Smad2/3, PPM1A promotes Smad2 nuclear export.
Loss of PPM1A Expression Causes Hyperactive Antiproliferative and Transcriptional Responses to TGFβ
Having established the role of PPM1A in Smad2/3 dephosphorylation, we attempted to determine whether PPM1A-mediated Smad2/3 dephosphorylation controls the strength of TGFβ responses. We first assessed the antiproliferative effects of TGFβ by [3H]thymidine incorporation in PPM1A-depleted stable cells (shPPM1A cells) in comparison to control cells. Control cells exhibited a normal TGFβ dosage-dependent inhibitory response on DNA synthesis in the presence of 10% fetal bovine serum, as these cells remained proliferative at low doses (0.05 and 0.1 ng/ml) of TGFβ, but their proliferation was inhibited to 40% at a higher dose of TGFβ (5 ng/ml) (Figure 5A). Knockdown of PPM1A rendered cells more sensitive to TGFβ-induced growth inhibition (Figure 5A). Thus, PPM1A-mediated dephosphorylation limits the ability of TGFβ to exert its inhibitory response in DNA synthesis.
Figure 5. Loss of PPM1A Promotes TGFβ Signaling.
(A) Loss of PPM1A enhances TGFβ antiproliferative response in HaCaT cells. DNA synthesis in shPPM1A cells and control cells was determined by [3H]thymidine incorporation assay. Values and error bars represent the mean and standard deviation of two experiments.
(B) Enhanced expression of p15 in shPPM1A cells as assessed by qRT-PCR. Values and error bars represent the mean and standard deviation of two experiments (likewise for all qRT-PCR analyses below).
(C) Increased p15 promoter activity in shPPM1A cells. Values and error bars represent the mean and standard deviation of at least three experiments (likewise for all reporter analyses below).
(D) qRT-PCR analysis of p21 in HaCaT cells.
(E) Inhibitors of p38, PI3K, and GSK3 have no or minimal effect on the basal level of p21 mRNA. Cells were treated with inhibitors of p38 kinase (10 μM SB202190), PI3K (10 μM LY294002), and GSK3 (2 μM SB216763) for 3 hr before TGFβ treatment and RNA analysis.
(F) Smad2/3 mediate increased levels of both basal and inducible p21 promoter activity. pSRG-shS2/3 simultaneously express shSmad2 and shSmad3. pSRG is an empty vector.
(G) qRT-PCR analysis of c-myc in HaCaT cells.
(H) qRT-PCR analysis of PAI-1 in HaCaT cells.
(I) Increased PAI-1 promoter activity in shPPM1A cells.
(J) qRT-PCR analysis of FN in HaCaT cells.
(K) PPM1A does not influence GAPDH mRNA level.
TGFβ-induced growth arrest is partly through upregulation of the CDK inhibitors p15 (Hannon and Beach, 1994) and p21 (Datto et al., 1995) as well as downregulation of c-myc (Alexandrow et al., 1995). Smad2/3 mediate the transcriptional regulation of p15 (Feng et al., 2000), p21 (Pardali et al., 2000; Seoane et al., 2004), and c-myc (Chen et al., 2002; Frederick et al., 2004). To characterize the effect of PPM1A knockdown on TGFβ transcriptional responses, we examined the levels of p15, p21, and c-myc mRNAs in shPPM1A cells. TGFβ induced higher levels of both p15 (Figure 5B) and p21 mRNAs (Figure 5D), consistent with the increased sensitivity of shPPM1A cells to TGFβ (Figure 5A). Depletion of PPM1A also increased the basal levels of both p15 and p21 mRNAs (Figures 5B and 5D), perhaps independently of non-Smad pathways as inhibitors of p38, PI3K, and GSK3 did not reduce the high basal level of p21 mRNA (Figure 5E). Increased p15 and p21 expression correlated with increased promoter activities of these genes (Figures 5C and 5F). Simultaneous knockdown of Smad2 and Smad3 abolished TGFβ-induced p21 promoter activity (Figure 5F). With regard to c-myc expression, we found that depletion of PPM1A accelerated TGFβ-mediated repression of c-myc even though shPPM1A cells exhibited a higher basal level of c-myc mRNA. TGFβ treatment at 8 hr caused a 90% decrease in the c-myc mRNA level, in comparison to only 70% decrease in control cells (Figure 5G). Thus, sustained activation of the Smad pathway leads to enhanced p15/p21 induction and c-myc repression in response to TGFβ.
In addition to its potent growth-inhibitory response, TGFβ strongly stimulates ECM production by inducing expression of such genes as plasminogen activator inhibitor type 1 (PAI-1) and fibronectin (FN1) through a Smad-dependent mechanism (Datta et al., 2000; Dennler et al., 1998; Itoh et al., 2003; Stroschein et al., 1999). We observed that TGFβ-induced transcription of these two genes displays distinct patterns in HaCaT cells. In control cells, PAI-1 mRNA increased by 23-fold after 4 hr of TGFβ treatment, reached 25-fold after another 4 hr, but declined to ~6-fold after 24 hr of TGFβ stimulation, indicating the termination of TGFβ signals (Figure 5H). Although it exhibited a minimal effect on PAI-1 expression at 4 and 8 hr, knockdown of PPM1A induced an even higher level of PAI-1 mRNA after prolonged TGFβ treatment, reaching a >40-fold induction after 24 hr of TGFβ stimulation (Figure 5H). The PAI-1 promoter activity was correspondingly increased in shPPM1A cells (Figure 5I). In the case of FN, we observed that TGFβ gradually induced FN mRNA, which reached a 7-fold increase after 24 hr in control cells (Figure 5J). In shPPM1A cells, the FN induction trend remained similar, but the overall TGFβ-mediated induction was more profound. At 24 hr, the FN mRNA increased by 12-fold (Figure 5J). As a control, GAPDH mRNA remained unchanged after PPM1A knockdown (Figure 5K). These data support the notion that depletion of PPM1A expression enhances global TGFβ signaling.
Inducible Expression of PPM1A Decreases TGFβ Responses
Having established that PPM1A knockdown results in hyperactive TGFβ signaling, we then tested whether overexpression of PPM1A leads to reduced TGFβ responses. To this end, we generated stable Mv1Lu cell lines that express PPM1A in a tetracycline-regulatable system (tet-off). We selected clones with increased expression of PPM1A at a level only comparable to that of endogenous PPM1A after removal of doxycycline (Figure 6A, lanes 7–12). This low-level expression of PPM1A sufficed to reduce the levels of P-Smad2/3 induced by TGFβ (Figure 6A, lane 8). On the contrary, increased expression of PPM1A (–Dox) did not exert any effects on TGFβ-induced activation of non-Smad pathways in Mv1Lu cells (Figure S8A) or NIH 3T3 cells (Figure S8B).
Figure 6. Inducible Expression of PPM1A Causes TGFβ Resistance.
(A) Reduced Smad2/3 phosphorylation in Mv1Lu cells with inducible expression of PPM1A. Mv1Lu-tet-off cells stably harboring Flag-PPM1A were grown with (+) or without (–) 10 ng/ml of doxycycline (Dox). Left: Induced expression of Flag-PPM1A (lanes 7–12). Cells were pretreated with TGFβ for 30 min (lanes 2 and 8) and then SB431542 for the indicated time periods (lanes 3–6 and 9–12). Levels of P-Smads, total Smads, and PPM1A were detected by Western blotting. Right: Line graph representing data from two stable clones with two experiments each, with values and error bars representing mean and standard deviation.
(B) PPM1A causes partial TGFβ resistance. Mv1Lu-PPM1A cells were cultured ±Dox, treated with various doses of TGFβ, and subjected to [3H]thymidine incorporation to analyze DNA synthesis.
(C) PPM1A blocks the TGFβ antiproliferative response. Mv1Lu-PPM1A cells, cultured ±Dox and treated ±TGFβ (0.2 ng/ml, 12 hr), were stained for PCNA (Zymed). PCNA-positive (black) and -negative (light gray) cells were counted and plotted.
(D) A representative field of PCNA staining as in (C).
(E) qRT-PCR analysis of PAI-1 in Mv1Lu cells. Values and error bars represent the mean and standard deviation of two experiments.
(F) PPM1A inhibits PAI-1 promoter activity. Left: Stable Mv1Lu cells were transfected with p800-luc and grown with (+) or without (–) Dox. Right: HaCaT cells were transiently transfected with p800-luc and PPM1A expression plasmids. Values and error bars represent the mean and standard deviation of at least three experiments.
The growth-inhibitory effect of TGFβ was evaluated by [3H]thymidine incorporation in the Mv1Lu tet-off cells. In the presence of Dox (+Dox), Mv1Lu cells exhibited a potent TGFβ-dependent inhibition on DNA synthesis, achieving a 90% inhibition at 0.5 ng/ml or higher doses of TGFβ (Figure 6B). Notably, expression of PPM1A (–Dox) significantly reduced TGFβ-induced inhibition on DNA synthesis, particularly at low dosages of TGFβ. At 0.05 ng/ml of TGFβ, cells expressing PPM1A were completely refractory to TGFβ, whereas control cells exhibited 30% inhibition on DNA synthesis (Figure 6B). PPM1A's effect on the TGFβ growth-inhibitory response was also demonstrated by PCNA (proliferating cell nuclear antigen) staining. Without exogenous PPM1A expression (+Dox), PCNA-positive cells decreased from 92% to 42% in response to TGFβ, whereas PPM1A expression (–Dox) blocked TGFβ-dependent inhibition on PCNA expression (Figures 6C and 6D). These results strongly suggest that inducible expression of PPM1A renders cells resistant to TGFβ antiproliferative effects.
We also determined whether increased expression of PPM1A inhibited Smad-dependent gene transcription in Mv1Lu cells. Expression of mink PAI-1 mRNA was induced by TGFβ in the presence of Dox, but this induction was markedly compromised after Dox withdrawal, suggesting that PPM1A inhibits TGFβ-induced PAI-1 transcription (Figure 6E). The PPM1A suppressive effect on endogenous PAI-1 expression correlated with decreased PAI-1 promoter activity in these cells (Figure 6F), which also coincides with loss of P-Smad2 (Figure S9). In addition, PPM1A blocked TGFβ-induced PAI-1 promoter activity in HaCaT cells (Figure 6F). Interestingly, D239N and R174G mutants appeared to enhance TGFβ-induced PAI-1 transcription (Figure 6F) as well as other gene responses (Figure S10), suggesting a dominant-negative effect of these mutants on TGFβ transcriptional responses.
PPM1A Antagonizes Dorsalizing Activity of TGFβ/Nodal Signaling In Vivo
TGFβ signaling pathways are highly conserved during evolution. We hypothesized that PPM1A-mediated dephosphorylation of Smad2/3 inhibits Nodal signaling during zebrafish early embryogenesis. Zebrafish PPM1A, which has 75% amino acid sequence identity to human PPM1A (Figure S6), displayed strong activity toward dephosphorylating P-Smad2 (Figure 4A) and P-Smad3 (Figure S5). Zebrafish PPM1A was expressed from the one-cell stage through 24 hr postfertilization (hpf) in a ubiquitous manner (Figure S11A). When injected with zebrafish PPM1A mRNA, 25.9% of embryos showed partial or complete fusion of eyes and thinner posterior notochord (Figures 7A and 7B). Since eye fusion is associated with loss of anterior prechordal plate mesoderm and is often seen in zebrafish mutant embryos defective in Nodal signaling pathway (Schier, 2003), we examined expression of the Nodal target gene goosecoid (gsc), which occurs in the embryonic shield at the onset of gastrulation and in the prechordal plate by the end of gastrulation (Schulte-Merker et al., 1994). Overexpression of PPM1A reduced gsc expression at the shield stage (Figure 7C) and further decreased or abolished gsc expression at the bud stage (Figure 7D), suggesting that PPM1A negatively regulates Nodal signaling and inhibits dorsal mesoderm development. Nodal signaling is also essential for establishment of vertebrate left-right asymmetry (Hamada et al., 2002). We noticed that lefty2 expression in the heart at 24 hpf switched to the middle or right in 27.3% of embryos injected with PPM1A mRNA (Figure 7E), further supporting a role of PPM1A in blocking Nodal-mediated regulation of left-right asymmetry.
Figure 7. Function of PPM1A in Zebrafish Embryos.
(A) Lateral view of live embryos at 24 hpf (hours postfertilization). Right: Embryo injected with 300 pg of ppm1a mRNA. Eyes are indicated by arrowhead. (B) Ventral view of embryos shown in (A). Note that eyes (arrowhead) were fused in the ppm1a-injected embryo.
(C–E) Embryos injected with 400 pg of ppm1a mRNA resulted in decreased gsc expression in the shield (C), loss of gsc expression in the prechordal plate (D), and mislocalization of lefty2 expression domain (heart) in the right at 24 hpf (E). Embryos were dorsal views with the animal pole at the top (C and D) or ventral views of heads (E).
(F–M) Embryos injected with 5 ng of S2MH2 protein exhibited head enlargement (G and K) and/or tail loss (G and L) at 24 hpf. Coinjection with 400 pg of ppm1a mRNA allowed a majority of embryos to develop an abnormal tail at 24 hpf (H and M). Statistical data are shown in (I).
(N–Q) gsc expression at the shield stage. Dorsal views of embryos injected with 5 ng of S2MH2 and/or 400 pg of ppm1a mRNA are shown. Note that the gsc-inducing effect of S2MH2 was counteracted by ppm1a.
(R–U) gsc expression at the bud stage. Experiments were done similarly to in (N)–(Q).
(V and W) Statistical data for gsc expression as shown in (N)–(U). Embryos were classified, based on degree of change in gsc expression, into four categories: normal, reduction, loss (loss in the prechordal plate), and expansion.
Next we analyzed genetic interactions between Smad2/3 and PPM1A in zebrafish embryos. Embryos (93.9%) injected with recombinant MH2 domain of human Smad2 (i.e., S2MH2) were severely dorsalized, as manifested by an enlarged head and absence of the tail (Figures 7G, 7K, and 7L). When PPM1A mRNA was coinjected with the same amount of S2MH2, over 60% of embryos developed a tail, albeit with some abnormalities (Figures 7H, 7I, and 7M). We did not observe eye fusion in any coinjected embryos (Figure 7M). S2MH2-induced gsc expression was inhibited by overexpression of PPM1A (Figures 7N–7W). In addition, dorsalizing effects caused by overexpression of zebrafish Smad3b were also inhibited by PPM1A overexpression (Figures S11B–S11I). Taken together, these data suggest that PPM1A antagonizes dorsalizing activity of Smad2/3.
DISCUSSION
Stringent control of TGFβ signaling through regulation of Smads is critical for normal cellular responses. As the SXS motif's phosphorylation is essential in Smad activation, its dephosphorylation conversely serves as an important countermechanism for terminating Smad signaling. Previous studies have indicated that dephosphorylation allows disassembly of the Smad activator complex and subsequent nuclear export of R-Smads (Inman et al., 2002b; Xu et al., 2002). Here we present several lines of evidence demonstrating PPM1A-mediated dephosphorylation of Smad2/3, which provides some insights into how TGFβ signaling is terminated. First, of 39 phosphatases screened, only PPM1A dephosphorylates P-Smad2/3. Second, purified PPM1A is capable of dephosphorylating recombinant phospho-Smad2 MH2 in a cell-free assay. Third, PPM1A physically interacts with Smad2. Significantly, PPM1A exhibits a strong affinity for P-Smad2. Considering that there is no substantial pool of P-Smad2/3 in the cytoplasm (Schmierer and Hill, 2005), our data lend further support to the notion that PPM1A dephosphorylates P-Smad2/3 in the nucleus. Fourth, increased Smad dephosphorylation by PPM1A renders highly TGFβ-sensitive Mv1Lu cells refractory to TGFβ. Fifth, knockdown of endogenous PPM1A sensitizes cells to respond to TGFβ. On the contrary, PPM1A has no effect on activation of TGFβ-induced non-Smad pathways (Figure S8) and contact-inhibition-induced p27 expression (Figure S12). Lastly, ectopic expression of PPM1A in zebrafish phenocopies Nodal-deficient mutant embryos and inhibits Smad2/3-induced dorsalization, suggesting a role of PPM1A in regulating Smad2/3-mediated Nodal or Nodal-like signals during development of vertebrate embryos. Thus, we propose that PPM1A terminates TGFβ signaling through dephosphorylation of P-Smad2/3, dissociation of the Smad complex, and subsequent nuclear export of Smad2/3 (Figure S16).
An interesting question is, how selective is PPM1A toward Smad dephosphorylation? While PPM1A dephosphorylates Smad2/3 (Figure 2), PPM1A also appears to be responsible for Smad1/5/8 SXS dephosphorylation (Figure S14 and data not shown). Unlike Smad2/3, which are only targeted by PPM1A (of 39 phosphatases screened), Smad1 can be dephosphorylated by several other phosphatases besides PPM1A (data not shown), displaying certain specificity and complexity. On the same Smad2/3 molecules, PPM1A exhibits remarkable selectivity toward phosphoserine residues, as it dephosphorylates the SXS motif, but not pS/T sites in the linker region (Figure 2I and data not shown). In addition, PPM1A does not influence TβRI activation upstream of Smad2/3 (Figure S13B).
Although PPM1A was first identified nearly 20 years ago, only a limited number of substrates have been reported (Tamura et al., 2003). PPM1A specifically interacts with and dephosphorylates the phosphorylated substrates, including p38 kinase (Takekawa et al., 1998), Cdk2 (Cheng et al., 1999), phosphatidylinositol 3-kinase (PI3K) (Yoshizaki et al., 2004), and Axin (Strovel et al., 2000). Our study extends this substrate list by adding Smad2 and Smad3. Notably, p38, PI3K, and GSK3 kinases represent non-Smad targets activated by TGFβ. However, PPM1A does not influence TGFβ-mediated activation of these kinases (Figure S8), nor do these kinases mediate PPM1A-mediated inhibition of Smad signaling (Figure 5E and data not shown).
At present, it is unclear how PPM1A is regulated in cells. TGFβ seems to have no effect on the subcellular localization (Figure 3D), expression (Figure 4C), and activity of PPM1A (Figure S15). A recent study reports that G protein α2 inhibitory subunit (Gαi2) prevents Smad2 dephosphorylation in T cells; it is conceivable that PPM1A is the phosphatase inhibited by Gαi2 (Wu et al., 2005). It will be of great interest to examine how activity or subcellular localization of PPM1A is regulated to control TGFβ signaling under physiological and pathophysiological conditions.
Despite biochemical and functional evidence demonstrating specific dephosphorylating effects of PPM1A on Smad2/3, our results do not exclude involvement of additional phosphatases in Smad dephosphorylation. In our study, only 4 of 62 DUSPs have been analyzed. In addition, screening all PS/TPs through transient expression is limited by the possibility that, in the case of polymeric phosphatases, expression of the catalytic subunit may not suffice to achieve detectable phosphatase activity. A previous study reports that activity of a RARα-dependent and okadaic-acid-sensitive phosphatase limits the levels of P-Smad2/3 induced by TGFβ (Cao et al., 2003), perhaps through dephosphorylation toward molecules upstream of Smads such as TGFβ receptors (Figures 1E and 1F). Indeed, previous studies have revealed regulatory roles of the PPP-family phosphatases at the receptor level (Bennett and Alphey, 2002; Griswold-Prenner et al., 1998; Shi et al., 2004). Nonetheless, further detailed investigation is needed to determine whether additional phosphatases can directly target the SXS motif of Smad2/3 as well as other R-Smads.
While C-terminal SXS phosphorylation by the type I receptor is the key event in Smad activation, the linker region of Smads, which joins the MH1 and MH2 domains, has also emerged as an important regulatory platform. The S/T-rich linker of R-Smads is phosphorylated by a number of protein kinases, including MAPKs (e.g., ERK, JNK, and p38 kinases), CDKs, and other intracellular kinases (Feng and Derynck, 2005). Using a similar functional genomic method described in this study, we identified phosphatases that dephosphorylate specific sites in the linker of Smad3 (Figure 2I and unpublished data). Therefore, we anticipate that PPM1A, linker phosphatases, and phosphatases for receptors together fine tune the strength and duration of Smad signaling.
In conclusion, PPM1A is a bona fide phosphatase that directly dephosphorylates the critical SXS motif of R-Smads. PPM1A limits the duration of Smad2/3 in their activated states. Since loss of TGFβ responses through hereditary mutations, somatic mutations, and aberrant regulation of signaling components has been linked to cancers and genetic diseases (Akhurst, 2004; Roberts and Wakefield, 2003; Waite and Eng, 2003), our results could also have important clinical implication. In this regard, on the basis of its key role in antagonizing TGFβ signaling, PPM1A may represent an important target for potential therapeutic intervention in cancers and other diseases.
EXPERIMENTAL PROCEDURES
Construction of Human PS/TP cDNA Expression Library
Full-length cDNAs were synthesized from total RNAs of HaCaT cells. High-fidelity PCR was used to amplify coding regions of known or putative PS/TPs (only the catalytic subunit if polymeric) encoded by the human genome, which was then cloned into CMV-driven expression vector pRK5 (Genentech). Experimental details on PCR conditions and primer sequences of each phosphatase are in the Supplemental Data.
Expression Plasmids
Expression plasmids for epitope-tagged Smads were described previously (Feng et al., 2002). Mouse PPM1A, zebrafish PPM1A, and human PPM1A with D239N or R174G substitutions were obtained by PCR and cloned into pRK5F.
To construct shRNA plasmids, a derivative of pSUPER-retro vector called pSRG was created. We made the following shRNA constructs against PPM1A: pSRG-shPPM1A494 (target sequence GGACTTGAGACATGGTCAT), pSRG-shPPM1A881 (target sequence GCAGGGGGCTCGGTGATGA), and pSRG-shmPPM1A494 (mouse target sequence GGACTTGAATCGTGGTCAT). A new pSRG vector, modified to contain both H1 promoter- and U6 promoter-driven cassettes, was used to simultaneously express shSmad2 (target sequence GGATGAAGTATGTGTAAAC; Lin and Feng, 2005) and shSmad3 (target sequence GGATTGAGCTGCACCTGAATG; Jazag et al., 2005).
Cell Transfection, Immunoprecipitation, Western Blotting, and Immunofluorescence
Cell culture, transfection, immunoprecipitation, Western blotting, and immunofluorescence were performed essentially as described (Feng et al., 2002; Lin et al., 2003). Antibodies against Smad2 and P-Smad2 (Cell Signaling), Smad3 (Zymed), PPM1A (Abcam), and others were used per manufacturers’ instructions. Anti-P-Smad3 (SXS) and anti-pS212 sera were kind gifts from Ed Leof (Mayo Clinic) and Fang Liu (Rutgers), respectively.
Stable HaCaT cell lines with PPM1A knockdown were generated by transfection with pSRG-shPPM1A494 or pSRG-shPPM1A881, selected with puromycin (2 μg/ml), and verified by Western blotting and real-time PCR analysis.
To establish tet-off expression of PPM1A, Mv1Lu-tTA cells (Reynisdóttir et al., 1995) were stably transfected with pTRE2 vector (Clontech) carrying Flag-PPM1A and a plasmid carrying a puromycin marker. Stable clones were maintained in the presence of G418, puromycin, and Dox (10 ng/ml) and switched to Dox-free medium to achieve Flag-PPM1A expression.
In Vitro Protein Binding and Phosphatase Assays
Production of recombinant GST- and His-tagged proteins and in vitro binding assays were previously described (Feng et al., 2002; Lin et al., 2003; Supplemental Data). For comparing the binding of His-PPM1A protein with phosphorylated versus unphosphorylated Smad2 MH2 (aa 241–467), 500 ng of Smad2 MH2 polypeptide was incubated with His-PPM1A proteins immobilized on Ni-NTA agarose beads in a buffer containing 20% glycerol, 20 mM HEPES (pH 7.9), 150 mM KCl, 0.5 mM DTT, 0.2 mM EDTA, 0.2 mM EGTA, 0.2% NP-40, 60 μM imidazole, and 50 mM 1,6-hexanediol.
For phosphatase assays, recombinant PPM1A was incubated with semisynthetic recombinant phospho-Smad2 MH2 (P-S2MH2), synthesis of which was described previously (Wu et al., 2001). The phosphatase reaction was performed at 30°C for 30 min in a buffer (7 mM imidazole [pH 7.6], 20 mM magnesium acetate, 300 μg/ml bovine serum albumin, 30 μM EDTA, and 0.03% β-mercaptoethanol) and analyzed by anti-P-Smad2 Western blotting.
[3H]Thymidine Incorporation and PCNA Staining
Thymidine-incorporation-based DNA synthesis assay was performed as previously described (Feng et al., 2002; Supplemental Data). For PCNA staining, Mv1Lu-PPM1A cells (±Dox) were fixed, blocked, and immunostained using a PCNA staining kit (Zymed).
Real-Time RT-PCR (qRT-PCR)
Total RNAs were prepared using TriZol reagent (Invitrogen) from HaCaT or Mv1Lu cells treated with TGFβ (2 ng/ml) for 0, 4, 8, and 24 hr. qRT-PCR was carried out using Assay-on-Demand kits or the SYBR green method (ABI). mRNA levels of target genes were normalized against 18S RNA. Each target was measured in triplicates. Data were analyzed using Microsoft Excel.
Zebrafish Embryo Assay
Maintenance and manipulation of zebrafish embryos derived from the Tübingen strain, mRNA preparation, and in situ hybridization were carried out as previously described (Zhang et al., 2004; Supplemental Data).
Supplementary Material
ACKNOWLEDGMENTS
We thank Bryan Cullen, Kazusa DNA Research Institute, Yibing Kang, Soo-Kyung Lee, Edward Leof, Fang Liu, David Loskutoff, Joan Massagué, Kohei Miyazono, Alexandra Newton, Anita Roberts, Danny Reinberg, Zhou Songyang, Peter ten Dijke, Bert Vogelstein, Xiao-Fan Wang, Malcolm Whitman, and Lan Xu for various reagents (details are available in the Supplemental Data). This research was supported by the NIH (R01DK073932 and R21CA11293 to X.L., R01CA82171 to Y. Shi, R01GM63773 and R01CA108454 to X.-H.F.), the National Natural Science Foundation of China (Distinguished Young Scholar Fund #30428002 to X.-H.F. and #30125021 to Y.-G.C., Grant #30430360 to Y.-G.C., and Grant #90208002 to A.M.), and 973 Program (2005CB522502 to A.M. and 2004CB720002 to Y.-G.C.). K.H.W. was supported by an American Heart Association (Texas Affiliate) Postdoctoral Fellowship. C.M.D. was supported by an NIH Minority Research Supplement (via R01GM63773). X.L. is a recipient of the Baylor Breast Center SPORE Career Development Award. Y.-G.C. is a recipient of the Li Foundation Heritage Prize. X.-H.F is a Leukemia & Lymphoma Society Scholar.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, 1 table, and 23 figures and can be found with this article online at http://www.cell.com/cgi/content/full/125/5/915/DC1/.
REFERENCES
- Akhurst RJ. TGFβ signaling in health and disease. Nat. Genet. 2004;36:790–792. doi: 10.1038/ng0804-790. [DOI] [PubMed] [Google Scholar]
- Alexandrow M, Kawabata M, Aakre M, Moses HL. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of TGFβ1. Proc. Natl. Acad. Sci. USA. 1995;92:3239–3243. doi: 10.1073/pnas.92.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Alphey L. PP1 binds Sara and negatively regulates Dpp signaling in Drosophila melanogaster. Nat. Genet. 2002;31:419–423. doi: 10.1038/ng938. [DOI] [PubMed] [Google Scholar]
- Cao Z, Flanders KC, Bertolette D, Lyakh LA, Wurthner JU, Parks WT, Letterio JJ, Ruscetti FW, Roberts AB. Levels of phospho-Smad2/3 are sensors of the interplay between effects of TGFβ and retinoic acid on monocytic and granulocytic differentiation of HL-60 cells. Blood. 2003;101:498–507. doi: 10.1182/blood-2002-05-1549. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen H, Shen J, Ip Y, Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Ross K, Kaldis P, Solomon M. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G, Wiegand H, Kang Y, Ho D, Georgiadis M, Cullen BR. Using viral species specificity to define a critical protein/RNA interaction surface. Genes Dev. 2001;15:1194–1205. doi: 10.1101/gad.888201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PTW. Overview of protein serine/threonine phosphatases. In: Arino J, Alexander DR, editors. Protein Phosphatases. Springer-Verlag; Heidelberg, Germany: 2003. pp. 1–20. [Google Scholar]
- Cullen BR. Assaying nuclear messenger RNA export in human cells. Methods Mol. Biol. 2004;257:85–92. doi: 10.1385/1-59259-750-5:085. [DOI] [PubMed] [Google Scholar]
- Datta P, Blake M, Moses HL. Regulation of plasminogen activator inhibitor-1 expression by TGFβ-induced physical and functional interactions between Smads and Sp1. J. Biol. Chem. 2000;275:40014–40019. doi: 10.1074/jbc.C000508200. [DOI] [PubMed] [Google Scholar]
- Datto M, Li Y, Panus J, Howe D, Xiong Y, Wang X-F. TGFβ induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y. Smad-dependent and Smad-independent pathways in TGFβ family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. Specificity and versatility in TGFβ signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGFβ. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X-H, Liang Y-Y, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGFβ-mediated induction of the CDK inhibitor p15Ink4B. Mol. Cell. 2002;9:133–143. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- Frederick J, Liberati N, Waddell D, Shi Y, Wang XF. TGFβ-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Protein serine/threonine phosphatases: life, death, and sleeping. Curr. Opin. Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Griswold-Prenner I, Kamibayashi C, Maruoka E, Mumby M, Derynck R. Physical and functional interactions between type I transforming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol. Cell. Biol. 1998;18:6595–6604. doi: 10.1128/mcb.18.11.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Hannon G, Beach D. p15Ink4B is a potential effector of TGFβ-induced cell cycle arrest. Nature. 1994;371:257–260. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Inman G, Nicolas F, Callahan J, Harling J, Gaster L, Reith A, Laping N, Hill CS. SB-431542 is a potent and specific inhibitor of TGFβ superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002a;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Inman G, Nicolas F, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGFβ receptor activity. Mol. Cell. 2002b;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Itoh S, Thorikay M, Kowanetz M, Moustakas A, Itoh F, Heldin CH, ten Dijke P. Elucidation of Smad requirement in TGFβ type I receptor-induced responses. J. Biol. Chem. 2003;278:3751–3761. doi: 10.1074/jbc.M208258200. [DOI] [PubMed] [Google Scholar]
- Jackson M, Fjeld C, Denu J. Probing the function of conserved residues in the serine/threonine phosphatase PP2Ca. Biochemistry. 2003;42:8513–8521. doi: 10.1021/bi034074+. [DOI] [PubMed] [Google Scholar]
- Jazag A, Ijichi H, Kanai F, Imamura T, Guleng B, Ohta M, Imamura J, Tanaka Y, Tateishi K, Ikenoue T, et al. Smad4 silencing in pancreatic cancer cell lines using stable RNA interference and gene expression profiles induced by TGFβ. Oncogene. 2005;24:662–671. doi: 10.1038/sj.onc.1208102. [DOI] [PubMed] [Google Scholar]
- Lin X, Feng X-H. Design and application of a versatile expression vector for RNAi in mammalian cells. J. RNAi Gene Silencing. 2005;1:38–43. [PMC free article] [PubMed] [Google Scholar]
- Lin X, Sun B, Liang M, Liang Y, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng X-H. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell. 2003;11:1389–1396. doi: 10.1016/s1097-2765(03)00175-8. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Denissova N, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Non-Smad TGFβ signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Pardali K, Kurisaki A, Moren A, ten Dijke P, Kardassis D, Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21WAF1/Cip1 regulation by TGFβ. J. Biol. Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/ Cip and Ink4 cdk inhibitors cooperate to induce cell cycle arrest in response to TGFβ. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wakefield L. The two faces of transforming growth factor β in carcinogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho K, De Robertis E, Nusslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34–PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J. Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein S, Wang W, Luo K. Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates TGFβ-induced Smad-dependent transcriptional activation. J. Biol. Chem. 1999;274:9431–9441. doi: 10.1074/jbc.274.14.9431. [DOI] [PubMed] [Google Scholar]
- Strovel E, Wu D, Sussman DJ. Protein phosphatase 2Cα dephosphorylates axin and activates LEF-1-dependent transcription. J. Biol. Chem. 2000;275:2399–2403. doi: 10.1074/jbc.275.4.2399. [DOI] [PubMed] [Google Scholar]
- Takekawa M, Maeda T, Saito H. Protein phosphatase 2Cα inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 1998;17:4744–4752. doi: 10.1093/emboj/17.16.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Li G, Komaki K, Sasaki M, Kobayashi T. Roles of mammalian protein phosphatase 2C family members in the regulation of cellular functions. In: Arino J, Alexander DR, editors. Protein Phosphatases. Springer-Verlag; Heidelberg, Germany: 2003. pp. 91–105. [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGFβ-Smad signaling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Waite K, Eng C. From developmental disorder to heritable cancer: it's all in the BMP/TGFβ family. Nat. Rev. Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- Whitman M, Raftery L. TGFβ signaling at the summit. Development. 2005;132:4205–4210. doi: 10.1242/dev.02023. [DOI] [PubMed] [Google Scholar]
- Wu J, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti D, Kyin S, Muir T, Fairman R, et al. Crystal structure of a phosphor-ylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGFβ signaling. Mol. Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- Wu J, Jin Y, Edwards R, Zhang Y, Finegold M, Wu MX. Impaired TGFβ responses in peripheral T cells of Gai2−/− mice. J. Immunol. 2005;174:6122–6128. doi: 10.4049/jimmunol.174.10.6122. [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, Col S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- Yoshizaki T, Maegawa H, Egawa K, Ugi S, Nishio Y, Imamura T, Kobayashi T, Tamura S, Olefsky JM, Kashiwagi A. Protein phosphatase-2Cα as a positive regulator of insulin sensitivity through direct activation of phosphatidylinositol 3-kinase in 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:22715–22726. doi: 10.1074/jbc.M313745200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou H, Su Y, Sun Z, Zhang H, Zhang Y, Ning Y, Chen YG, Meng A. Zebrafish Dpr2 inhibits mesoderm induction by promoting degradation of nodal receptors. Science. 2004;306:114–117. doi: 10.1126/science.1100569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.