Abstract
A potent and selective inhibitor of protein SUMOylation, a ubiquitin-like post-translational modification, has been developed, shedding light on the potential for developing new classes of anticancer therapeutics.
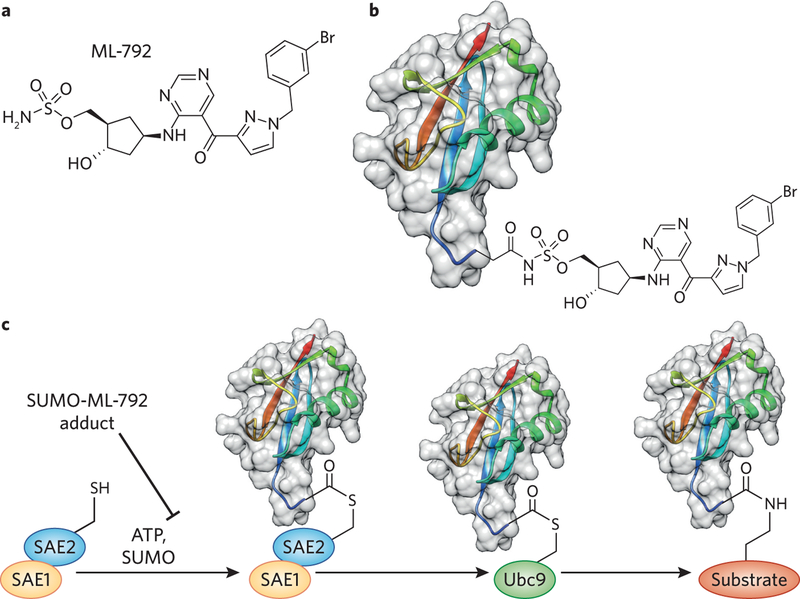
Post-translational modification is a major mechanism for regulating protein structural and functional diversity. Ubiquitin-like post-translational modifications involve the attachment of small protein tags to substrate proteins, and they are linked to major regulatory and signal transduction pathways within the cell. Small ubiquitin-like modifier (SUMO) is one example of a regulatory, protein-based post-translational modification. To date, it has proven highly challenging to identify small-molecule inhibitors of SUMO conjugation. He et al. now report the discovery of ML-792, a potent and selective inhibitor of the E1 SUMO-activating enzyme (SAE)1 (Fig. 1a).
Figure 1 |.
Inhibition of protein SUMOylation with a small molecule. (a) Chemical structure of ML-792. (b) The covalent adduct formed between the C terminus of SUMO and the sulfamate ester of ML-792. (c) Chemical inhibition of the SUMO conjugation cascade by binding of the SUMO-ML-792 adduct directly to SAE.
As with all ubiquitin-like proteins, SUMO proteins are conjugated to surface-exposed lysine side chains by an isopeptide bond. SUMOylation is regulated by an E1–E2–E3 cascade of enzymes (Fig. 1c), and SUMO tags are cleaved by SUMO-specific protease (SENP) enzymes. Although ubiquitination serves primarily as a signal for induced protein degradation, it is difficult to predict the consequences of the SUMOylation of a given substrate. However, SUMOylation events have been shown to cause changes in subcellular localization, protein–protein interactions, and enzymatic activity of substrates2,3. Recent efforts have also suggested a link between SUMOylation and cancer4,5.
ML-792 was discovered as part of a broad medicinal chemistry effort at Takeda aimed at discovering E1 inhibitors in ubiquitin-like signaling. Furthermore, it is related to a series of mechanism-based NEDD8-activating enzyme (NAE) inhibitors such as pevonedistat, which was originally reported by some of the same authors6,7. ML-792 contains a sulfamate ester that forms a covalent adduct with SUMO at its C terminus (Fig. 1b). This covalent conjugate binds tightly to, and inhibits the activity of, the catalytic subunit of SAE, known as SAE2 or UBA2 (Fig. 1c). Sulfamate esters are therefore a mechanistically intriguing and unusual class of mechanism-based small-molecule inhibitors.
Many of the reported inhibitors of SUMOylation display weak activity or polypharmacology, or have uncharacterized selectivity. In contrast, ML-792 is active in biochemical assays at single-digit nanomolar concentrations. Although mechanistically related to pevonedistat, ML-792 has a unique chemical structure that allows it to inhibit SAE without inhibiting any other ubiquitin-activating enzymes or various other ATP-using enzymes such as NAE. Furthermore, ML-792 displays potent activity in cell-based assays, wherein it dramatically decreases levels of sumoylated proteins at nanomolar concentrations.
It had been previously thought that an inhibitor of SUMOylation might be unacceptably toxic because of the essential nature of SUMO ligases (genetic knockout of the SUMO E2 enzyme Ubc9 causes embryonic lethality8). However, ML-792 is substantially more toxic to cells when Myc is overexpressed in a tetracycline-inducible system. It also is highly toxic to Myc-amplified cell lines, highlighting the potential for chemically inhibiting SUMOylation in Myc-driven cancers. These results dovetail with previous reports indicating that SAE drives synthetic lethalityin Myc-addicted tumors4. Furthermore, the compound has increased activity in clonogenicity and anchorage-independent growth assays, emphasizing the role of SUMOylation in mitotic progression and contact dependence.
Although ML-792 dramatically suppresses SUMOylation in cells, it does not have large effects on the transcriptome or on the accumulation of damaged DNA. Given that many transcription factors and DNA-damage response proteins are established SUMOylation substrates, these results came as a surprise. Thus, this work provides a stark example of the differences between using a small-molecule inhibitor and knockdown, knockout, or overexpression to study protein function. This issue had been suggested to be relevant to SUMOylation because a low percentage of a given substrate is modified at any particular time9, though applicable experiments had not been possible before the development of a potent, selective inhibitor. Though SUMO ligases are clearly essential genes, this work demonstrates that many of the effects thought to be general functions of SUMOylation may be cell-type or context dependent.
The work described here provides substantial evidence for the inhibition of SUMOylation as a potential anticancer strategy because of the role of SUMOylation in mitotic progression and SUMO dependence in Myc-amplified tumors. Furthermore, it provides a long-sought chemical tool to probe SUMO-associated signaling. ML-792 will aid greatly in efforts to unravel the complex biology linked to SUMOylation and its role in cancer biology.
Footnotes
Competing financial interests
The author declares no competing financial interests.
References
- 1.He X et al. Nat. Chem. Biol 13, 1164–1171 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR & Lima CD Nat. Rev. Mol. Cell Biol 11, 861–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R & Melchior F Nat. Rev. Mol. Cell Biol 8, 947–956 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Kessler JD et al. Science 335, 348–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B et al. Proc. Natl. Acad. Sci. USA 112, E1724–E1733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell JE et al. Mol. Cell 37, 102–111 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Soucy TA et al. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Demarque MD et al. Gastroenterology 140, 286–296 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Becker J et al. Nat. Struct. Mol. Biol 20, 525–531 (2013). [DOI] [PubMed] [Google Scholar]