Abstract
The major capsid protein of HPV, L1, assembles into pentamers that form a T=7 icosahedral particle, but the location of the co-assembled minor capsid protein, L2, remains controversial. Several researchers have developed useful monoclonal antibodies targeting L2, but most react with linear epitopes toward the N-terminus. As a means to better define the virus capsid and better assess the localization and exposure of L2 epitopes in the context of assembled HPV, we have developed a panel of 30 monoclonal antibodies (mAbs) which target the N- terminus of L2 amino acids 11-200, previously defined as a broadly protective immunogen. Select mAbs were processed with enzymes and anti-L2 Fabs were generated. These new mAb/Fab probes will be beneficial in future studies to unravel the placement of L2 and to help better define the role of L2 in the HPV lifecycle and the nature of the broadly protective epitopes.
Keywords: HPV, antibody, mAb, L2, minor capsid protein, epitope, Fab
Introduction
Human Papillomaviruses (HPVs) are a large family of viruses with nearly 200 different strains identified and characterized. The virus is the etiological agent of all cervical cancer cases, the majority of which are the result of infection with either HPV16 or HPV18. HPV is epitheliotropic and strains are divided based upon their ability to infect mucosal or cutaneous epithelium. In addition to cervical cancer, infection with HPV results in cancer of numerous other anatomical sites. HPV is a major cause of oropharyngeal squamous cell carcinoma and the incidence of these cancers is on the rise. Unfortunately, there are no treatments available to combat active HPV infections (1).
The lifecycle of HPV is differentiation dependent and infection is initiated at the basal layer of epithelium where only early genes are expressed. As the cells differentiate within the stratified layers, the HPV lifecycle also progresses with expression of the late gene products (L1 and L2) restricted to terminally differentiated keratinocytes. By limiting expression of the structural proteins to the upper most layers of epithelium, the immune system has limited availability to these antigens as they are not present in the basal cells most accessible by the blood supply (1).
Monoclonal antibodies (mAbs) against L1 have been instrumental in the mapping and characterization of L1 epitopes as well as the overall structural characterization of the virus. The novel use of the mouse xenograft system developed by J. Kreider (2) enabled the production of authentic HPV-11 and the immunization of mice with HPV-11 for the production of mouse mAbs (3). The binding properties of antibody and their sensitivity to small changes in the AA sequence of L1 was later examined by Ludmerer et al. (4) when the group demonstrated that the dependence upon a conformational epitope is transferable. Ludmerer and colleagues were able to make an HPV11-specific mAb bind an HPV6 VLP by only changing two AAs in the HPV6 L1 protein (4). The specificity of the mAb binding to VLP was further investigated in studies which analyzed mAb reactivity against different HPV16 variants (5). Site directed mutagenesis used to change AA50 of L1 from leucine to phenylalanine enabled binding of H16.V5 to variants which H16.V5 did not previously bind. These studies stress the importance of the H16.V5 epitope in the immunogenic response and production of neutralizing HPV16 antibodies (5).
Early structural analyses utilizing imaging began with x-ray crystallography and the crystallization of small 12-pentamer L1 VLPs (6). Despite the truncation of some AAs, this crystal structure provided the first structure of L1 in a multimeric form. The crystal structure also contributed to the knowledge that L1 loops present on the capsid surface are regions of hypervariability and are sites of sequence variation among different HPV types as well as neutralizing epitopes (6). The distinct antigenic structure of HPVs was further demonstrated in a study examining the type-specificity of HPV11 neutralizing mAbs and their reactivity using HPV6 L1 VLPs (7). Despite the close phylogenic relation of HPV6 and HPV11, even these two viruses require type-specific immune responses for neutralization.
The L2 protein also elicits an immune response albeit much weaker than L1. Similar to studies with L1, the generation of anti-L2 mAbs will create useful reagents for the use in biochemical assays and assist to determine which portions of the protein are surface-exposed or buried. Importantly, unlike the type-specificity demonstrated by L1, initial studies have shown that polyclonal responses to L2 contain both type-specific and common cross-reactive epitopes that are susceptible to antibody neutralization (8-18). In a study utilizing antisera generated against a series of peptides, Heino et al. (19) concluded that HPV16 L2 AAs 32-51, 62-81, 212-231, 279-291, and 362-381 are located on the surface of HPV capsids. Numerous other studies have examined the N-terminal half of the L2 protein and have found surface-exposed sites are located between AA60-180 (8, 20-22). Cross-neutralizing epitopes have also been localized to surface-exposed AA108-120 (12, 23) as well as AA17-36 subsequent to cell surface binding and conformational changes (15). Although research has been focused at the N-terminus, neutralizing epitopes have also been mapped to the C-terminal half of CRPV L2 (24).
The production of anti-L2 mAbs has been more challenging but has been achieved with BPV, HPV1, HPV33, and HPV16 (15, 20, 21, 23, 25-33). Kawana et al. (21) achieved the production of HPV16 L2 mAbs by immunizing mice with HPV16 VLPs produced in insect cells. The authors characterized eighteen mAbs and notably determined that their mAbs discriminate between VLPs containing L2 and L1 VLPS. However, the binding demonstrated against VLPs by ELISA was weak, lacked positive controls for comparison, and binding was not improved upon testing reactivity with purified L2 protein. Another larger mAb panel was produced using bacterial thioredoxin as a scaffold for putative neutralizing epitopes AA20-38, 28-42, 56-75, 64-81, 96-115, and 108-120 (34). Out of forty-six mAbs examined, only four were found to have neutralizing activity and by the very design of the experiment, mAbs recognizing AAs beyond 120 were not possible (28).
Due to the limited cross protection offered by the L1 vaccine, a favorable vaccine candidate would drive an antibody response against the highly conserved minor capsid protein, L2. Several L2 vaccines are currently in production (35, 36), and a better understanding of the antigenic sites, surface exposed AAs and neutralizing L2 epitopes will only help to drive the vaccine effort. Additionally, this knowledge could help unravel the nuances of the capsid structure as well as the finer mechanics of viral entry and the infection pathway. It is known that the T=7 icosahedral capsid of HPV is composed of 360 L1 monomers assembled as 72 pentamers, but the general structure of L2, the copy number, and the relative location of L2 remain controversial. Adding to the complexity of the virus structure, previous work has demonstrated that the viral capsid is not a static structure. Conformational changes are believed to occur upon interactions with the cell surface and cellular attachment factors such as HSPGs (37-41) as well as within the endosome upon acidification (42). Characterization of these changes and the exposure of the L2 protein has been difficult due to the limited number and variety of precise probes available to assess L2.
Therefore, here we have developed a unique panel of monoclonal antibodies against HPV16 L2 AA11-200 to function as unbiased probes in the assessment of antigenic sites, surface exposed regions, neutralizing epitopes and overall capsid structure. Through the characterization of this mAb panel, we have gained valuable information regarding conformational/linear epitopes of previously untargeted portions of L2.
Materials and Methods
Recombinant Particle Production
HPV Pseudovirus (PsV) particles were produced and purified as previously described (43). 293TT cells were transfected with Lipofectamine 2000 (LifeTechnologies) and plasmids encoding the HPV capsid proteins (a kind gift from the Schiller lab and Addgene.org) and a pseudo-genome encoding secreted alkaline phosphatase (pSEAP). Following a 48-hour incubation, cells were harvested and lysed with detergent. Particle maturation was achieved by incubating cell lysates for 24 hours at 37°C. Particles were purified from cell lysates by Optiprep ultracentrifugation.
Monoclonal Antibody Production
Mouse mAbs were produced as previously described (24, 44). Balb/c mice were immunized subcutaneously with 50μl HPV16 L2 peptide AA11-200 (1mg/mL) (10) in Sigma adjuvant. Mice were immunized twice, one month apart and boosted without adjuvant one month after the second immunization. Spleen cells were harvested for hybridoma production 3 days post-booster. The resultant hybridoma supernatants were screened for reactivity against the L2 protein. Hybridomas were cloned by limiting dilution.
Monoclonal Antibody Isotyping and Purification
The cloned antibody supernatant was assessed by the Rapid ELISA Mouse mAb Isotyping Kit (Thermo Fisher Scientific) to determine the mAb isotype. Purification of hybridoma supernatants was achieved by applying supernatants to either Protein A or Protein G purification columns (Pierce).
Epitope Mapping
mAb epitopes were mapped by examining by ELISA the reactivity of hybridoma supernatants against overlapping HPV L2 peptides purchased from China Peptides. Peptides were added to the wells of a 96-well microtiter plate at 1μg/well diluted in 50mM Na2CO3 pH9.6 protein binding buffer. Hybridoma supernatants were added to the wells and positive peptide binding was assessed with an alkaline phosphatase conjugated secondary antibody and 4-Nitrophenyl phosphate disodium salt hexahydrate (PNPP). The development of the colorimetric signal was determined with a plate reader OD 405 450.
Intact, Denaturing and Sandwich ELISA
Intact binding ELISAs were performed by binding 500ng PsV to the wells of microtiter plates in PBS. The wells were blocked with blocking buffer (5% fat free dry milk in PBS/T). Hybridoma supernatants were added to the wells and binding was detected by an AP-conjugated secondary antibody (Southern Biotech). AP signal was developed with PNPP and the OD 405 450 was determined with a plate reader. For the denaturing ELISAs, PsV was added to the wells of the microtiter plate in 25 μl denaturing buffer (0.2M Na2CO3, 10mM DTT pH10.7). Plates were incubated overnight at 37°C or at 55°C for 1 hour allowing the protein to dry to the plate. Proteins were rehydrated for 15 minutes with 25μl PBS and further processed in the same manner as the intact ELISA. For sandwich ELISAs 10μg/ml purified mAb was bound to the microtiter plate in 25μl 50mM Na2CO3, pH 9.6 binding buffer overnight at 4°C. Wells were washed, blocked with blocking buffer and then 0.5μg PsV was added to the wells in blocking buffer for 1 hour. Wells were washed again and the detection mAb H16.V5 IgA was added followed by an IgA specific secondary antibody. Plates were further processed in the same manner as the intact ELISA.
Pseudovirus Neutralization
Neutralization of recombinant PsV particles was assessed by pre- or post-attachment neutralization assays in 293TT cells as previously described (45). For the pre-attachment assay PsV was incubated together with hybridoma supernatants or purified mAbs at 37°C for one hour and then added to 293TT cells plated in a 96-well plate. For the post-attachment assay PsV was incubated with 293TT cells at 4°C for 1 hour to allow for virus attachment to the cell surface. Cells were carefully washed with media and antibody dilutions were added at 4°C for 1 hour to allow for antibody attachment before viral entry into cells. Both pre and post-attachment assays were then incubated at 37°C for 72 hours. 30μl of the spent medium was removed and assayed with PNPP and optical density was determined by absorbance spectrometry with a plate reader at OD 405 450.
Fab Production and Fab Purity ELISA
The Pierce Fab production kit was used to digest mAbs into Fabs according to the manufacturers’ protocol (Thermo Scientific Pierce). 7I and 20C were processed with papain while 9E and 21T were processed with ficin to obtain Fab fragments. Fab purity and retention of the antigen binding site was assessed by ELISA. An intact ELISA was performed using PsV16 as the antigen and 100μg of purified mAb or Fab were added to the wells of the microtiter plate. mAbs or Fabs were detected with anti-Fc and/or anti-Fab AP-conjugated secondary antibodies (Southern Biotech).
Results
Epitope Mapping of Anti-L2 mAbs Using HPV16 L2 Peptides
Previous studies have produced and characterized anti-L2 mAbs, both neutralizing and non-neutralizing, and their epitopes are presented in (Table S1). Our panel of 59 HPV16 L2 reactive antibodies were produced using the HPV16 L2 peptide AA11-200 as antigen. Hybridoma supernatants were initially mapped by scanning for reactivity against AA1-88 or AA11-200 and selected for cloning based upon positivity in immunofluorescence (IF) assays and ELISA PsV binding studies (data not shown). Anti-L2 mAbs 1A and 2E were previously published (43) and included in these studies for comparison. From the panel of 59 hybridoma supernatants, 30 were selected and cloned.
Clones were isotyped and minimal amino acid epitopes of the cloned mAbs were mapped using overlapping peptides HPV16 L2 peptides AA13-90 and 76-200 (Table S2). The majority of the mAbs were mapped by this method, however; three mAbs (11D,13A, and 30A) failed to have any reactivity against the peptides. Six mAbs (1A, 2E, 9E, 32B, 46G, and 47I) were mapped using the first panel of peptides AA13-90, while the remaining mAbs were mapped further from the N-terminus using the second peptide panel AA76-200. The minimal epitopes are presented in Table 1 and epitopes of historical anti-L2 mAbs and novel epitopes of anti-L2 mAbs characterized in this study are diagramed in Figure 1 along with functional domains of the L2 protein. Figure 1 depicts the novelty of the mAbs that we have developed. With these probes, we are now able to target portions of the L2 protein that were not previously targeted by mAbs.
Table 1.
Cloned Anti-L2 mAbs were isotyped and minimal epitopes were mapped by HPV16 L2 and HPV18 peptide ELISA. No reactivity of the clone to peptide is designated as NR and NT means that the clones were not tested.
| Anti-L2 Clone | Ig Isotype | Minimal Amino Acid HPV16 | Minimal Amino Acid HPV18 |
|---|---|---|---|
| Epitope | Epitope | ||
| 16.L2.1A | IgG1 | 18-32 | 17-31 |
| 16.L2.2E | IgG1/IgG2b | 18-32 | NR |
| 16.L2.1C | IgG1 | 121-135 | NR |
| 16.L2.2R | IgG1 | 161-170 | NR |
| 16.L2.3G | IgG1 | 176-185 | NR |
| 16.L2.7I | IgG2a | 141-150 | 136-145 |
| 16.L2.9E | IgG1 | 38-52 | NR |
| 16.L2.11D | IgG1 | NR | NR |
| 16.L2.13A | IgG1 | NR | NR |
| 16.L2.14L | IgM | 101-110 | 161-175 |
| 16.L2.17O | IgG2b | 161-170 | NR |
| 16.L2.19M | IgG2a | 86-95 | NR |
| 16.L2.20C | IgG2a | 121-135 | NR |
| 16.L2.21T | IgG1 | 121-135 | NR |
| 16.L2.24F | IgG2a | 126-135 | NR |
| 16.L2.30A | IgG1 | NR | NR |
| 16.L2.31B | IgG1 | 176-185 | NR |
| 16.L2.32B | IgG1 | 68-87 | NR |
| 16.L2.35X | IgG2b | 86-95 | NR |
| 16.L2.36C | IgG1 | 161-170 | NR |
| 16.L2.37A | IgG1 | 71-95 | NR |
| 16.L2.38A | IgG1 | 176-185 | NR |
| 16.L2.42L | IgG1 | 86-95 | NR |
| 16.L2.43J | IgG1 | 161-170 | NR |
| 16.L2.46G | IgG1 | 18-32 | 17-36 |
| 16.L2.47I | IgM | 33-52 | NT |
| 16.L2.48A | IgG1 | 86-95 | NR |
| 16.L2.50F | IgG1 | 86-95 | NR |
| 16.L2.51A | IgG1 | 176-185 | NR |
| 16.L2.52E | IgG2a | 161-175 | NT |
| 16.L2.54C | IgM | 120-125 | NR |
| 16.L2.58E | IgG1 | 86-95 | NR |
Figure 1. Mapped L2 interaction sites.
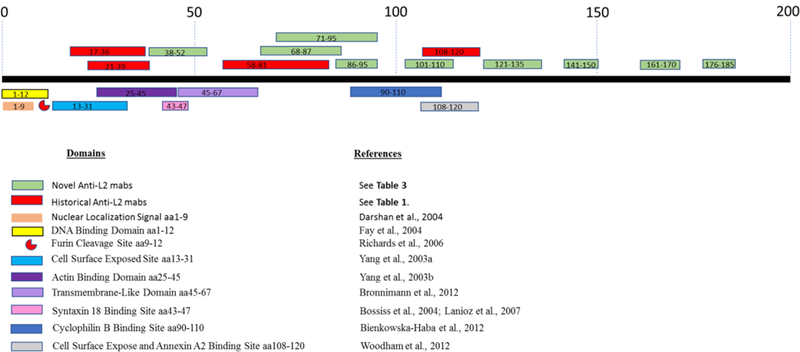
Diagram of known L2 protein interaction domains, epitopes of historical anti-L2 mAbs, and novel epitopes covered by anti-L2 mAbs presented in this manuscript. Diagram was adapted and updated from (46): Wang etal. 2013.
Anti-L2 mAbs Differentially Target Linear and Conformational Epitopes
To better qualify the nature of the epitopes recognized by the mAb panel, we assessed the ability of the mAbs to bind intact or denatured PsV16 by ELISA. We defined the ability to bind intact particles as the recognition of a conformational epitope. Binding to denatured antigen signifies the recognition of linear epitopes. The cloned mAb panel in addition to 1A and 2E was assessed in a conformational ELISA and the optical density (OD) was graphed (Figure 2A). Detection of linear epitopes was analyzed in parallel with the previously defined linear detecting anti-L1 mAb H16.D9 (44) (Figure 2B). Additional mAbs H16.J4 and H16.V5 were also used as controls. H16.J4 recognizes linear epitopes and served as an additional positive denaturing control and H16.V5 was a negative control as it only recognizes conformational sites (44). OD’s were determined to be positive if the averaged signal with the background and standard deviation subtracted was greater than 3 standard deviations of the background. Nine of the mAbs, including 1A and 2E (1A, 2E, 9E, 13A, 19M, 32B, 36C, 46G, 58E), only recognized PsV in an intact ELISA and thus bind conformationally-dependent epitopes (Table 2). The majority of the mAb panel was able to bind both intact and denatured antigen. Three mAbs (38A, 51A, 54C) detected strictly denatured PsV. Despite being screened by peptide ELISA and IF, mAb 47I failed to detect PsV in either the intact or the denatured ELISA.
Figure 2. Anti-L2 mAb recognition of intact or denatured capsids.
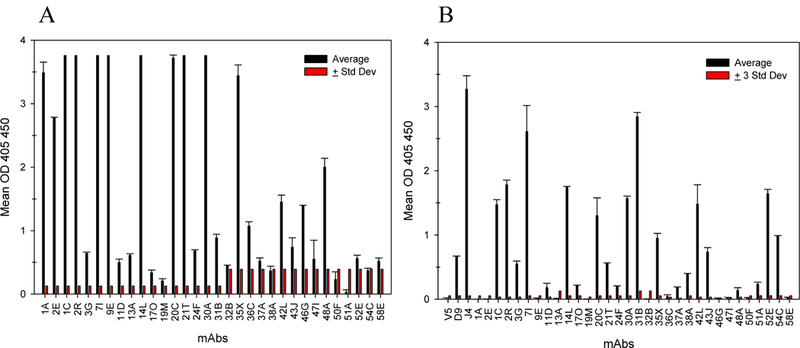
Binding of cloned anti-L2 mAbs to PsV16 was assessed by ELISA under native (A) and denaturing conditions (B). (A) Optical densities for the intact ELISA and (B) denaturing ELISA are displayed with +1 standard deviation of the duplicate sample (black) and +3 standard deviations of the blank value (red).
Table 2.
Anti-L2 mAbs were categorized by their ability to bind native, denatured, or both epitopes as assayed by ELISA. Signals greater than 3 standard deviations of the negative control were assumed to be positive.
| Native | 1A, 2E, 1C, 2R, 3G, 7I, 9E, 11D, 13A, 14L, 17O, 19M, 20C, 21T, 24F, 30A, 31B, 32B, 35X, 36C, 37A, 42L, 43J, 46G, 48A, 52E, 58E |
| Denatured | 1C, 2R, 3G, 7I, 11D, 14L, 17O, 20C, 21T, 24F, 30A, 31B, 35X, 37A, 38A, 42L, 43J, 48A, 51A, 52E, 54C |
| Native/Denatured | 1C, 2R, 3G, 7I, 11D, 14L, 17O, 20C, 21T, 24F, 30A, 31B, 35X, 37A, 42L, 43J, 48A, 52E |
| Native Only | 1A, 2E, 9E, 13A, 19M, 32B, 36C, 46G, 58E |
| Denatured Only | 38A, 51A, 54C |
The mAbs 7I, 9E, 20C and 21T were purified using affinity columns and were also tested in ELISA binding studies to approximate binding titers. The half maximum binding titer for 7I is 66.7 pM, 20C is 3.3 nM, 9E is 6.7 nM and 21 T is 1.2 nM. Our unpublished studies showed that the half maximum binding titer of 16.V5 is 233.3 pM.
Anti-L2 mAbs are Neutralizing
We screened the large panel of cloned antibody supernatants for their ability to neutralize infection using the PsV16 pre-attachment neutralization assay (Figure 3) and preliminarily in a QV16 post-attachment neutralization assay (Figure S1). These assays demonstrate the neutralizing capacity of mAbs 7I and 9E both pre- and post-attachment, as well as 20C and 21T post-attachment. We then assessed the ability of high-titer supernatants 7I, 9E and 21T to neutralize NV HPV16. The dogma in the field suggests that L2 requires association with components of the cell-surface for the exposure of certain neutralizing epitopes (37) (47) and therefore we assessed neutralization of the NV in a post-attachment assay (Figure S2). H16.V5 served as an anti-L1 control and both 1A and 2E were used as anti-L2 neutralization controls. All four antibodies neutralize NV HPV16 by 80% or more.
Figure 3. Neutralization by anti-L2 mAb panel.
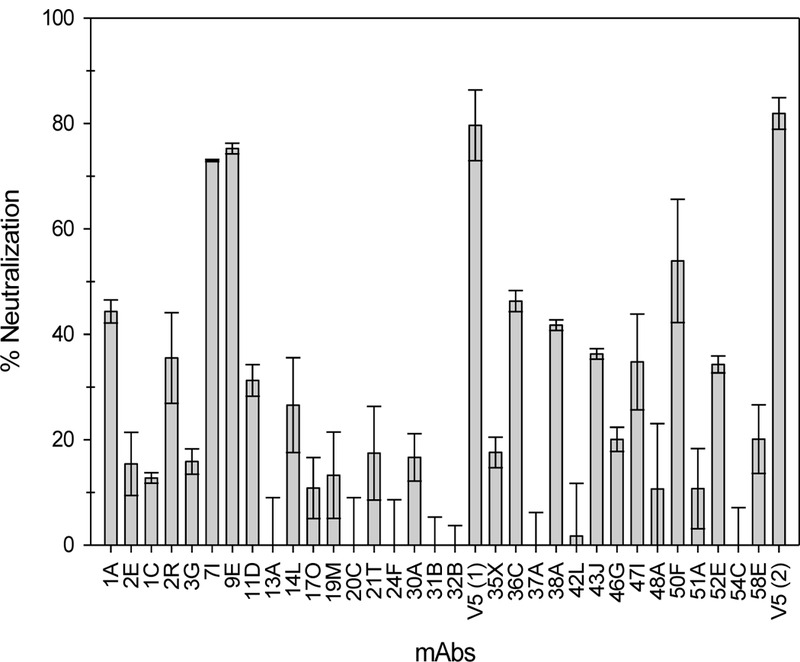
The anti-L2 mAb panel was tested for the ability to neutralize PsV16 in a pre-attachment neutralization assay. Neutralization is displayed as a percentage + SEM.
Purified mAbs were also used for neutralization to determine approximate neutralization titers in the PsV pre-attachment assay. The half-maximum neutralization titer for 7I is 46.7 pM and 9E is 2.7 nM. 20C and 21T did not neutralize above background levels in the pre-attachment neutralization assay even with purified mAb. 20C and 21T were tested in a post-attachment neutralization assay with PsV16 in 293TT cells. The half maximum neutralization titer for 20C is 467 nM. 21T did not reach 50% neutralization in this assay and only achieved about 46% neutralization.
Anti-L2 mAbs bind intact virions
Previous reports showed that anti-L2 mAb RG-1 failed to bind PsV in IP but was able to bind when PsV was bound to an ELISA well indicating conformational changes were required for RG-1 to bind (15), (37). Our previous studies showed that 1A and 2E L2 mAbs are able to bind PsV in a sandwich ELISA. Sandwich ELISAs are not thought to induce conformational changes of the viral capsids, contrary to ELISA methods where viral capsids are adsorbed directly to the wells of the microtiter plate (43). We tested a subpanel of anti-L2 mAbs for their ability to bind PsV in a sandwich ELISA. Figure 4 shows that the previously reported 1A and 2E mAbs, as well as 7I, 20C, 21T and 9E are able to capture PsV16. 16.V5 was used as a positive capture control and 11.B2 (HPV11 specific) and 58.J6 (HPV58 specific) were negative controls and subtracted from the final OD before graphing.
Figure 4. L2 mAbs capture PsV16 in a sandwich ELISA.
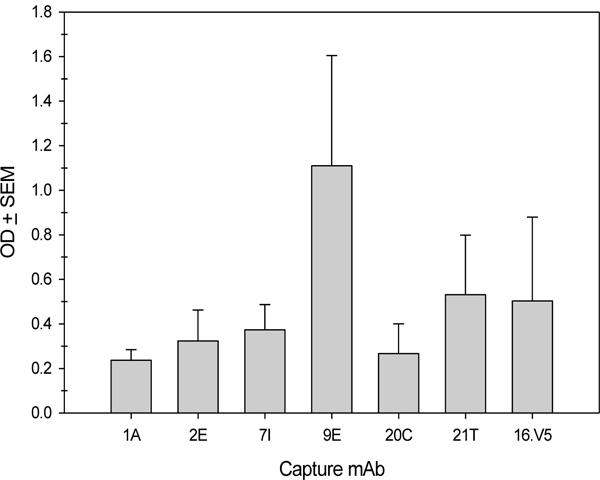
10μg/ml mAbs were bound to an ELISA plate in protein binding buffer, wells were blocked with milk and then PsV16 was added in milk. Captured PsV16 was detected by 10μg/ml 16.V5-IgA mAb followed by an anti-mouse IgA-AP secondary antibody and reading at OD 405 450. 3 individual experiments were conducted and the averages of the negative control mAbs (11.B2 and 58.J6) ODs were subtracted from the average of the L2 mAbs before graphing.
Selected anti-L2 mAbs 7I, 9E, 20C and 21T Exhibit Cross-Reactive Binding
The four mAbs for which high-titer supernatants were made (7I, 9E, 20C and 21T) all bind epitopes not previously recognized by historical anti-L2 mAbs and they all are capable of neutralizing HPV16 capsids. Therefore, we assessed the ability of these selected mAbs to cross- react with different HPV types. The ability of these mAbs to cross-react with divergent L2 proteins HPV5,6,11,16,18,31,33,35,39,45,52,58, and 59 as well as BPV, MmuPVl, and CRPV was assessed by IF. Aside from HPV16 L2, 7I and 9E mAbs recognized several additional L2 proteins (images in Figure S3 and summarized in Table 3). A positive control anti-L1 antibody is included in Figure S3 and is listed as presented in the figure: H16.V5, H5.A6, H6M.48, H11.B2, H16.V5, H18.J4, H31.A6, H33.B6, H35.Q8, H39.B6, H45.N5, H52.D11, H58.J63, H59.G1, B1.A1, MPV.A4, and CRPV. 2C. Both 7I and 9E mAbs recognized HPVs 11, 16, 31, and 35, but as individual mAbs they also uniquely recognize other L2 proteins. 7I detected the L2 protein of HPV6, 18, 39, 45 and 59 while 9E identified HPV5, 33, 52, 58 and the L2 protein of CRPV. The two mAbs 20C and 21T are significantly less cross-reactive by IF. While both recognize HPV16, the only cross-reactivity observed was between 20C and HPV11 L2.
Table 3.
Cross-reactivity of anti-L2 mAbs 7I, 9E, 20C and 21T identified by Immunofluorescence.
| L2 | 16.L2.7I mAb | 16.L2.9E mAb | 16.L2.20C | 16.L2.21T |
|---|---|---|---|---|
| HPV5 | - | + | - | - |
| HPV6 | + | - | - | - |
| HPV11 | + | + | + | - |
| HPV16 | + | + | + | + |
| HPV18 | + | - | - | - |
| HPV31 | + | + | - | - |
| HPV33 | - | + | - | - |
| HPV35 | + | + | - | - |
| HPV39 | + | - | - | - |
| HPV45 | + | - | - | - |
| HPV52 | - | + | - | - |
| HPV58 | - | + | - | - |
| HPV59 | + | - | - | - |
| BPV | - | - | - | - |
| MmuPV1 | - | - | - | - |
| CRPV | - | + | - | - |
Since both 7I and 9E appear to have great cross-reactivity, we examined the L2 amino acid sequences to see if the 7I or 9E epitopes share common sequences between the different HPV types. Sequences were aligned in Figure S4 but no obvious commonality was observed at these epitopes. The 7I epitope is fairly conserved across the different HPV types. The 9E epitope is conserved to a degree, but the CRPV epitope stands out as being drastically different. Inexplicably, 9E is still able to recognize the CRPV L2 protein by IF.
Cross-reactive Anti-L2 mAb Binding of Conformational PsV Predicts CrossReactive Neutralization
We are aware that positive binding of mAb in IF does not equate to binding in ELISA or neutralization of virus. In order to better examine the ability of these mAbs to cross-reactively bind the virion surface, we performed intact binding ELISAs. We examined the ability of 7I and 9E to bind PsVs of the following PV types: CRPV, HPV16, HPV18, HPV39, HPV45 and HPV59 (Figure 5A). Both mAbs bound HPV16 and the reactivity to CRPV was predictable as 7I failed to bind but 9E was successful. Binding by 7I to HPV39, 45, and 59 was above 3 standard deviations of the blank value, but the signal was still extremely weak. There was no binding activity observed for HPV18 despite positive 7I binding in IF. We also investigated the ability of 7I and 9E to neutralize the same divergent HPV types in a pre-attachment neutralization assay (Figure 5B). Both 7I and 9E exhibited neutralizing activity in a pre-attachment neutralization assay with HPV16. As expected by the IF and ELISA assay, 9E also neutralized CRPV. Despite positive binding as assayed by IF, none of the other HPV types were neutralized. It was surprising that the positive binding observed by IF did not translate better into neutralization. We speculated that even by IF, some of the protein may not be completely folded and assembled within cells thereby enabling binding to linear epitopes that may not be exposed on the surface of an assembled virion. Although cross-reactive binding was prevalent by IF, these assays indicate that binding ELISAs are better tools for predicting in vitro neutralization.
Figure 5. Correlation between cross-reactive 7I/9E binding and neutralization.
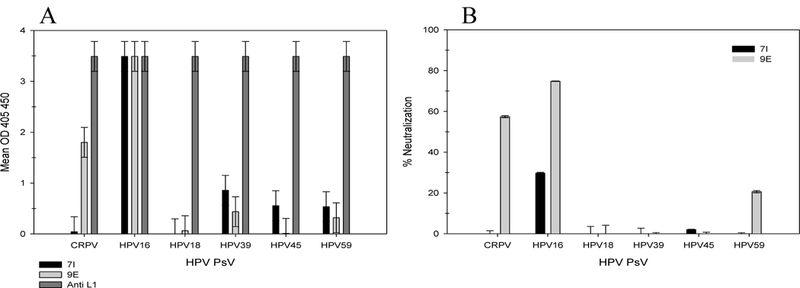
The cross-reactivity of anti-L2 mAbs 7I and 9E were tested by intact binding ELISA (A) and a pre-attachment neutralization assay (B) for the following PsVs: CRPV, HPV16, HPV18, HPV39, HPV45 and HPV59. (A) Binding is shown as the OD + 3 standard deviations of the negative control. (B) Percent neutralization of each PsV is shown relative to infection in the absence of antibody + 3 standard deviations of the blank value.
In addition to 7I and 9E, we tested the ability of 20C and 21T to neutralize HPV18 NV (data not shown). The only mAb with any neutralizing activity was 20C but the infectivity in the presence of 20C was still greater than 50% of the infection in the controls (no mAbs). We also tested the ability of 7I, 9E, 20C and 21T to neutralize PsV31, 33, 35, 52 and 58 in a pre-attachment neutralization assay (as previously described (17)) and these mAbs were not able to neutralize the assayed PsV types (data not shown).
Functional Antibody Fragments Generated from Anti-L2 mAbs
Enzymes are frequently used to alter the structure of mAbs. Select enzymes cleave the mAb separating both antigen-binding arms from each other and from the Fc domain of the antibody. These fragments are useful for numerous studies including but not limited to (1) the assessment of antibody neutralization mechanisms, (2) study of epitopes and structure using techniques such as Cryo-EM or crystallography, (3) eliminate Fc-mediated effector cells and non-specific binding in immunological studies, (4) increase sensitivity of binding by reducing potential steric hindrance. While several other groups have successfully produced anti-L2 mAbs, there has been difficulty producing Fab fragments. We attempted Fab production with anti-L2 mAbs 1A and 2E, but we were unable to detect Ig binding following processing with the enzyme (43). Other researchers have shared their difficulties in anti-L2 Fab production (personal correspondence and (48)). Our next attempt at anti-L2 Fab production was with 7I, 9E, 20C and 21T. Purified mAbs 7I and 20C were treated with papain for the production of Fab fragments and 9E and 21T were digested with the enzyme ficin. Following processing, we verified by ELISA the purity of the Fab fragments and the ability to bind PsV16 (Figure 6).
Figure 6. Assessment of Fab fragment purity.
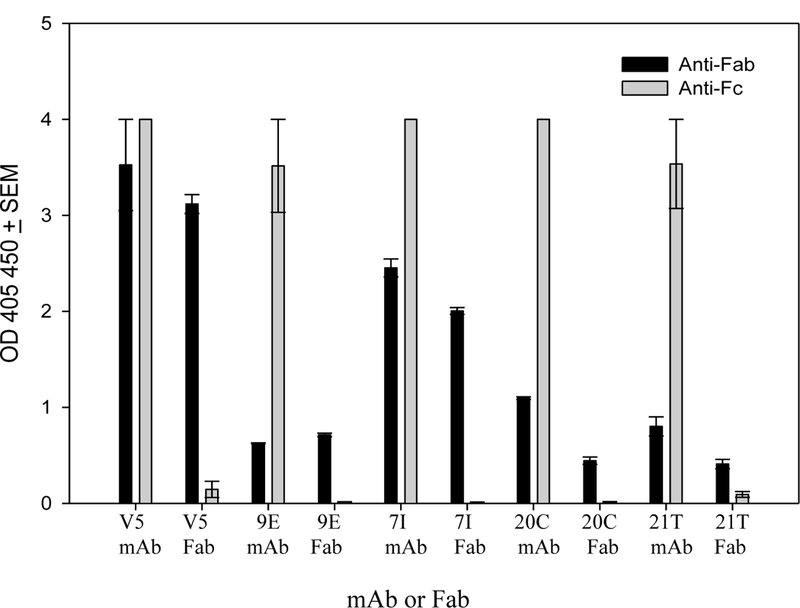
The completion of Fab digestion and the functionality of Fab fragments was assessed by ELISA. Anti-Fab and anti-Fc signals are displayed as the optical density (OD) + SEM. Purity of Fab was based upon a positive anti-Fab signal and a negative anti-Fc signal.
Discussion
In this work, a panel of 30 mAbs reactive to the L2 protein of HPV16 was developed and characterized. Many mAbs in the panel recognize epitopes previously defined by other anti-L2 mAbs, but several also bound unique epitopes (Figure 1). Novel regions of the HPV16 L2 protein recognized by our panel of mAbs include AA39-52, 82-85, 121-135, 141-150, 161-170 and 176-185. We assessed the epitopes recognized by our panel and characterized them as buried or conformational (Figure 2A, B and Table 2). Buried epitopes are defined by the ability of the antibody to recognize denatured PsV capsids but not intact PsV. Conformational epitopes are defined by the ability of the antibody to bind intact PsV capsids but the loss of binding capacity with denatured PsV. Nearly all of the mAbs have the ability to detect native (conformational) structure and many are able to detect both native and denatured PsVs. Only three mAbs (38A, 51A, 54C) strictly require denaturation of the PsV to bind a buried epitope, albeit weakly.
Despite not localizing to epitopes previously defined to be surface exposed, many mAbs also demonstrated neutralizing potential (Figure 3). One of these epitopes (AA38-52) was defined by mAb 9E. There are several reported domains of L2 that overlap with the 9E epitope. These domains may or may not be targeted by the antibody and contribute to neutralization by interfering with trafficking (49-52). Two additional sites further away from the extreme N- terminus AA121-135 and AA141-150 were uniquely identified by mAbs 20C/21T and 7I respectively. 21T and 7I both neutralize NV in a post-attachment assay, but neutralization of PsV16 is limited (Figure S2 and Figure 3). We are unable to speculate at this time as to their mechanism of neutralization and acknowledge that the different particle types utilize different genomes and the production differs by cell line and cell stratification. Any or all of these factors could be the reason for the different results in neutralizing capacity. Despite these differences, the PsV16 neutralization titer of 7I (46.7 pM) is comparable to the L1 mAb, 16.V5 as we showed previously that the 16.V5 half maximum neutralization titer is approximately 20 pM (45).
Epitopes of mAbs 11D, 13A, and 30A were not able to be finely mapped using overlapping peptides (Table 1). A common explanation for failure to map linear epitopes using peptides is that the epitope lies between two peptides; however, we used highly overlapping peptides that overlap by fifteen AAs and are offset by five. Therefore, such difficulty in epitope mapping is unexpected. Failure to map an mAb epitope may be an expected result in the case of an antibody which strictly recognizes conformational epitopes. Although peptides do have the potential for some secondary structure or even tertiary structure, it is generally limited since it is only a small piece of protein. If an antibody requires a quaternary structure for recognition, it will not be able to bind a single peptide because by definition, quaternary structures result from the interactions of two or more polypeptide chains. While this rationale could explain the difficulty to map 13A using peptides, the argument for the requirement of a structural epitope is challenged by 11D and 30A recognition of denatured PsV in a denaturing ELISA.
A parallel argument is that denaturation in a denaturing ELISA is not complete and therefore there remains a degree of structure which could be sufficient for antibodies requiring conformational epitopes. However, we were able to map the epitopes of 1A and 2E using overlapping peptides and both mAbs bound virions in intact ELISAs, yet these epitopes were lost in a denaturing ELISA. Therefore, if the denaturation conditions of the denaturing ELISA are not harsh enough to completely denature the virion proteins, they are at least enough to eliminate the detection of 1A and 2E binding by ELISA. Perhaps immunoblotting is more sensitive than ELISA as contradictory results were observed when 1A and 2E positively detected HPV16 and HPV18 L2 by a denaturing and reducing SDS-PAGE/immunoblot (data not shown).
An important aspect of mAbs generated against the L2 protein is the cross-reactivity of the mAb to a variety of HPV types. RG-1 targets a highly conserved portion of the L2 N- terminus AA17-36 and antiserum to the L2 peptide has been demonstrated to neutralize HPV types 5,6,16,18,31,45,52,58, BPV1, and CRPV (15). We assessed the ability of four of our high-titer mAbs 7I, 9E, 20C and 21T to detect several PV types by IF (Table 3 and Figure S3). We knew from our epitope mapping that 7I, 9E and 20C/21T bind L2 at unique sites. 7I and 9E were the most cross-reactive and while there were some PV types recognized by both mAbs, differential recognition was also observed. Another interesting result includes the finding that 7I mAb bound HPV18 in IF assay and the fine epitope was eventually mapped to AA136-145 in subsequent studies by peptide ELISA. Despite recognition of HPV18 L2 in those studies, 7I mAb failed to detect HPV18 PsV in an intact ELISA and also did not exhibit any neutralizing capacity against HPV18 PsV (Figure 5A and B). The lack of cross-neutralization observed in vitro may be a limitation of current L1- based in vitro neutralization assays given how L2 epitopes are often not exposed on the surface of HPV PsVs until post-attachment (47). Indeed, it was shown previously that passive transfer of mAbs into mice followed by challenge by HPV PsVs offered greater protection because of an additional protective mechanism of Fc-mediated phagocytosis of L2 antibody-virion complexes (48).
We are not the first group of investigators to develop antibodies targeting the minor capsid protein, but we are unaware of any investigators that have successfully generated functional anti-L2 Fabs until those reported here. The generation of anti-L2 Fabs creates opportunities for structural analyses by Cryo-EM or crystallography. These types of studies require a Fab because the Fc portion of the mAb is too flexible. Fabs are also desirable for competition studies because they create less potential for steric hindrance. The ability to generate Fabs in addition to mAbs will also aid in future studies to examine neutralization mechanisms of these antibodies.
The generation of this diverse panel of mAbs creates a novel set of biological probes for the assessment of HPV structure. Several mAbs targeting L1 have been available for years and these have aided the community of HPV researchers in many great discoveries. The probes developed for L1 have been superior to those available for L2 and perhaps L2 is simply more complex. The development of these well-characterized mAb probes targeting L2 should enable many more great discoveries, helping to better define the role of L2 in the HPV lifecycle as well as the placement of the protein within the context of the overall virion structure.
Supplementary Material
Acknowledgments:
Research was funded by the NCI Training Grant “Viruses and Cancer” 5 T32 CA60395 and the Jake Gittlen Laboratories for Cancer Research.
Footnotes
Disclosure:
We have read the journal’s policy and have the following competing interests. Joshua W. Wang is a founder of PathoVax LLC. Richard Roden is a member of Papivax LLC, has Papivax Biotech Inc. stock options and is a member of Papivax Biotech Inc.’s Scientific Advisory Board. Under a license agreement between PathoVax LLC and the Johns Hopkins University, Richard Roden is entitled to distributions of payments associated with an invention described in this study. Richard Roden also owns equity in PathoVax LLC and is a member of its scientific advisory board. Under a license agreement between Bravovax and the Johns Hopkins University, Richard Roden is entitled to distributions of payments associated with an invention described in this study. Thesearrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. 2015. Human papillomavirus molecular biology and disease association. Rev Med Virol 25 Suppl 1:2–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreider JW, Howett MK, Leure-Dupree AE, Zaino RJ, Weber JA. 1987. Laboratory production in vivo of infectious human papillomavirus type 11. J Virol 61:590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen ND, Kreider JW, Cladel NM, Patrick SD, Welsh PA. 1990. Monoclonal antibody-mediated neutralization of infectious human papillomavirus type 11. J Virol 64:5678–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludmerer SW, Benincasa D, Mark GE 3rd. 1996. Two amino acid residues confer type specificity to a neutralizing, conformationally dependent epitope on human papillomavirus type 11. J Virol 70:4791–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White WI, Wilson SD, Palmer-Hill FJ, Woods RM, Ghim SJ, Hewitt LA, Goldman DM, Burke SJ, Jenson AB, Koenig S, Suzich JA. 1999. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J Virol 73:4882–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell 5:557–567. [DOI] [PubMed] [Google Scholar]
- 7.Christensen ND, Kirnbauer R, Schiller JT, Ghim SJ, Schlegel R, Jenson AB, Kreider JW. 1994. Human papillomavirus types 6 and 11 have antigenically distinct strongly immunogenic conformationally dependent neutralizing epitopes. Virology 205:329–335. [DOI] [PubMed] [Google Scholar]
- 8.Campo MS, O’Neil BW, Grindlay GJ, Curtis F, Knowles G, Chandrachud L. 1997. A peptide encoding a B-cell epitope from the N-terminus of the capsid protein L2 of bovine papillomavirus-4 prevents disease. Virology 234:261–266. [DOI] [PubMed] [Google Scholar]
- 9.Embers ME, Budgeon LR, Pickel M, Christensen ND. 2002. Protective immunity to rabbit oral and cutaneous papillomaviruses by immunization with short peptides of L2, the minor capsid protein. J Virol 76:9798–9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambhira R, Jagu S, Karanam B, Gravitt PE, Culp TD, Christensen ND, Roden RB. 2007. Protection of rabbits against challenge with rabbit papillomaviruses by immunization with the N terminus of human papillomavirus type 16 minor capsid antigen L2. J Virol 81:11585–11592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagu S, Karanam B, Gambhira R, Chivukula SV, Chaganti RJ, Lowy DR, Schiller JT, Roden RB. 2009. Concatenated multitype L2 fusion proteins as candidate prophylactic pan¬human papillomavirus vaccines. J Natl Cancer Inst 101:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawana K, Kawana Y, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. 2001. Nasal immunization of mice with peptide having a cross-neutralization epitope on minor capsid protein L2 of human papillomavirus type 16 elicit systemic and mucosal antibodies. Vaccine 19:1496–1502. [DOI] [PubMed] [Google Scholar]
- 13.Kawana Y, Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. 2001. Human papillomavirus type 16 minor capsid protein l2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J Virol 75:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YL, Borenstein LA, Selvakumar R, Ahmed R, Wettstein FO. 1992. Effective vaccination against papilloma development by immunization with L1 or L2 structural protein of cottontail rabbit papillomavirus. Virology 187:612–619. [DOI] [PubMed] [Google Scholar]
- 15.Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, Alphs H, Culp T, Christensen ND, Roden RB. 2007. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol 81:13927–13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo K, Ishii Y, Ochi H, Matsumoto T, Yoshikawa H, Kanda T. 2007. Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology 358:266–272. [DOI] [PubMed] [Google Scholar]
- 17.Pastrana DV, Gambhira R, Buck CB, Pang YY, Thompson CD, Culp TD, Christensen ND, Lowy DR, Schiller JT, Roden RB. 2005. Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology 337:365–372. [DOI] [PubMed] [Google Scholar]
- 18.Roden RB, Yutzy WHt, Fallon R, Inglis S, Lowy DR, Schiller JT. 2000. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology 270:254–257. [DOI] [PubMed] [Google Scholar]
- 19.Heino P, Skyldberg B, Lehtinen M, Rantala I, Hagmar B, Kreider JW, Kirnbauer R, Dillner J. 1995. Human papillomavirus type 16 capsids expose multiple type-restricted and type-common antigenic epitopes. J Gen Virol 76 ( Pt 5):1141–1153. [DOI] [PubMed] [Google Scholar]
- 20.Liu WJ, Gissmann L, Sun XY, Kanjanahaluethai A, Muller M, Doorbar J, Zhou J. 1997. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 227:474–483. [DOI] [PubMed] [Google Scholar]
- 21.Kawana K, Matsumoto K, Yoshikawa H, Taketani Y, Kawana T, Yoshiike K, Kanda T. 1998. A surface immunodeterminant of human papillomavirus type 16 minor capsid protein L2. Virology 245:353–359. [DOI] [PubMed] [Google Scholar]
- 22.Knowles G, Grindlay GJ, Campo MS, Chandrachud LM, O’Neil BW. 1997. Linear B-cell epitopes in the N-terminus of L2 of bovine papillomavirus type 4. Res Vet Sci 62:289–291. [DOI] [PubMed] [Google Scholar]
- 23.Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. 1999. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J Virol 73:6188–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen ND, Kreider JW, Kan NC, DiAngelo SL. 1991. The open reading frame L2 of cottontail rabbit papillomavirus contains antibody-inducing neutralizing epitopes. Virology 181:572–579. [DOI] [PubMed] [Google Scholar]
- 25.Volpers C, Sapp M, Snijders PJ, Walboomers JM, Streeck RE. 1995. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J Gen Virol 76 ( Pt 11):2661–2667. [DOI] [PubMed] [Google Scholar]
- 26.Roden RB, Weissinger EM, Henderson DW, Booy F, Kirnbauer R, Mushinski JF, Lowy DR, Schiller JT. 1994. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol 68:7570–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Embers ME, Budgeon LR, Culp TD, Reed CA, Pickel MD, Christensen ND. 2004. Differential antibody responses to a distinct region of human papillomavirus minor capsid proteins. Vaccine 22:670–680. [DOI] [PubMed] [Google Scholar]
- 28.Rubio I, Seitz H, Canali E, Sehr P, Bolchi A, Tommasino M, Ottonello S, Muller M. 2011. The N-terminal region of the human papillomavirus L2 protein contains overlapping binding sites for neutralizing, cross-neutralizing and non-neutralizing antibodies. Virology 409:348–359. [DOI] [PubMed] [Google Scholar]
- 29.Nakao S, Mori S, Kondo K, Matsumoto K, Yoshikawa H, Kanda T. 2012. Monoclonal antibodies recognizing cross-neutralization epitopes in human papillomavirus 16 minor capsid protein L2. Virology 434:110–117. [DOI] [PubMed] [Google Scholar]
- 30.Wang JW, Jagu S, Kwak K, Wang C, Peng S, Kirnbauer R, Roden RB. 2014. Preparation and properties of a papillomavirus infectious intermediate and its utility for neutralization studies. Virology 449:304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Li Z, Xiao J, Wang J, Zhang L, Liu Y, Fan F, Xin L, Wei M, Kong Z, Yu H, Gu Y, Zhang J, Li S, Xia N. 2015. Identification of Broad-Genotype HPV L2 Neutralization Site for Pan-HPV Vaccine Development by a Cross-Neutralizing Antibody. PLoS One 10:e0123944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang JW, Jagu S, Wu WH, Viscidi RP, Macgregor-Das A, Fogel JM, Kwak K, Daayana S, Kitchener H, Stern PL, Gravitt PE, Trimble CL, Roden RB. 2015. Seroepidemiology of Human Papillomavirus 16 (HPV16) L2 and Generation of L2-Specific Human Chimeric Monoclonal Antibodies. Clin Vaccine Immunol 22:806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaegashi N, Jenison SA, Valentine JM, Dunn M, Taichman LB, Baker DA, Galloway DA. 1991. Characterization of murine polyclonal antisera and monoclonal antibodies generated against intact and denatured human papillomavirus type 1 virions. J Virol 65:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubio I, Bolchi A, Moretto N, Canali E, Gissmann L, Tommasino M, Muller M, Ottonello S.2009. Potent anti-HPV immune responses induced by tandem repeats of the HPV16 L2 (20 -- 38) peptide displayed on bacterial thioredoxin. Vaccine 27:1949–1956. [DOI] [PubMed] [Google Scholar]
- 35.Jiang RT, Schellenbacher C, Chackerian B, Roden RB. 2016. Progress and prospects for L2-based human papillomavirus vaccines. Expert Rev Vaccines 15:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schellenbacher C, Roden RBS, Kirnbauer R. 2017. Developments in L2-based human papillomavirus (HPV) vaccines. Virus Res 231:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Day PM, Gambhira R, Roden RB, Lowy DR, Schiller JT. 2008. Mechanisms of human papillomavirus type 16 neutralization by l2 cross-neutralizing and l1 type-specific antibodies. J Virol 82:4638–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selinka HC, Giroglou T, Nowak T, Christensen ND, Sapp M. 2003. Further evidence that papillomavirus capsids exist in two distinct conformations. J Virol 77:12961–12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. 2007. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J Virol 81:8784–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards RM, Lowy DR, Schiller JT, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamper N, Day PM, Nowak T, Selinka HC, Florin L, Bolscher J, Hilbig L, Schiller JT, Sapp M. 2006. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol 80:759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bywaters SM, Brendle SA, Tossi KP, Biryukov J, Meyers C, Christensen ND. 2017. Antibody Competition Reveals Surface Location of HPV L2 Minor Capsid Protein Residues 17–36. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus¬like particles as defined by monoclonal antibodies. Virology 223:174–184. [DOI] [PubMed] [Google Scholar]
- 45.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 483:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JW, Roden RB. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day PM, Kines RC, Thompson CD, Jagu S, Roden RB, Lowy DR, Schiller JT. 2010. In vivo mechanisms of vaccine-induced protection against HPV infection. Cell Host Microbe 8:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JW, Wu WH, Huang TC, Wong M, Kwak K, Ozato K, Hung CF, Roden RBS. 2018. Roles of Fc Domain and Exudation in L2 Antibody-Mediated Protection against Human Papillomavirus. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bossis I, Roden RB, Gambhira R, Yang R, Tagaya M, Howley PM, Meneses PI. 2005. Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J Virol 79:6723–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laniosz V, Nguyen KC, Meneses PI. 2007. Bovine papillomavirus type 1 infection is mediated by SNARE syntaxin 18. J Virol 81:7435–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang R, Yutzy WHt, Viscidi RP, Roden RB. 2003. Interaction of L2 with beta-actin directs intracellular transport of papillomavirus and infection. J Biol Chem 278:12546–12553. [DOI] [PubMed] [Google Scholar]
- 52.Bronnimann MP, Chapman JA, Park CK, Campos SK. 2013. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J Virol 87:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyers C, Frattini MG, Hudson JB, Laimins LA. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971–973. [DOI] [PubMed] [Google Scholar]
- 54.Meyers C, Mayer TJ, Ozbun MA. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol 71:7381–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


