Abstract
The advancement of glycoscience is critically dependent on the access to a large number of glycans for their functional study. Naturally occurring glycans are considered a viable source for diverse and biologically relevant glycan libraries. A mixture of free reducing glycans released from natural sources can be fluorescently tagged and separated by chromatography to produce a natural glycan library. Anthranilic acid (AA) has been widely used to fluorescently tag reducing glycans for HPLC or LC/MS analysis. However, AA conjugated glycans are not efficiently immobilized on microarray slides due to the lack of a primary alkylamine functional group. In this study, we have developed simple and efficient chemistry for bioconjugation and further functionalization of glycan–AA conjugates. This new approach enables quick preparation of glycan microarrays and neoglycoproteins from glycan–AA conjugates, which can be separated by weak anion exchange (WAX) and C18 reversed-phase HPLC.
INTRODUCTION
Glycans have crucial biological functions in many aspects of biology, including protein folding, cell–cell adhesion and trafficking, cell signaling, host–pathogen recognition and immune response, etc.1 With growing interest in glycans and their functions in modern biomedical research, functional glycomics, a systematic study of the glycome, has witnessed significant development in the past decades.2 High performance liquid chromatography (HPLC), mass spectrometry (MS), and online LC-MS have greatly advanced our ability to determine glycan structures. For functional studies, glycan microarray has become an indispensable technology for screening protein-glycan interactions.3–9 In a glycan microarray experiment, multiple glycan structures are immobilized on solid surfaces such as microscope slides for interrogation with glycan binding proteins (GBPs), providing a high throughput method to probe specific protein-glycan interactions. The value of a glycan microarray is dictated by the number, diversity, and biological relevance of the immobilized glycans, and the lack of these resources is currently a major obstacle in glycoscience. Unlike DNA, RNA, or proteins, biosynthesis of glycans is not template-driven but is an orchestrated process involving multiple glycosyltransferases with distinct substrate and linkage specificities. Despite recent advancements in chemoenzymatic synthesis of highly complex glycan structures,10–12 the access to biomedically relevant glycans is inadequate and limited to a number of very specialized laboratories, setting a high hurdle to detail their biological roles.13
An alternative route to access complex glycan structures is to isolate them from natural sources. Diverse and biologically relevant natural glycans, including N- and O-glycans linked to glycoproteins, glycans linked to glycosphingolipids, and others, can be released using chemical and/or enzymatic approaches.14–17 Because most glycans lack chromophores and are poorly ionized, the released glycans are often derivatized to facilitate HPLC, MS, or online LC-MS analysis. Fluorescent tagging at the reducing end through quantitative reductive amination has been commonly used to increase their hydrophobicity and amplify the detection limit in both normal-phase and reversed-phase HPLC. Among various fluorescent tags used in reductive amination of glycans,18 anthranilic acid or 2-aminobenzoic acid (AA) is widely used, and many glycan–AA conjugates are commercially available as HPLC and MS standards, albeit at high prices. Meanwhile, anthranilic acid has received much attention in biochemical analyses as a fluorophore in FRET systems.19,20 That the AA fluorescent tag holds one negative charge makes it highly flexible for HPLC separations such as hydrophilic interaction liquid chromatography (HILIC), mixed-mode HILIC/anion exchange, and weak anion exchange (WAX) chromatography separations and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis in both positive and negative modes.18 A comprehensive review discussed the pros and cons among commonly used florescent tags on glycans.18 One major drawback of glycan–AA conjugates is the low immobilization efficiency on amine-reactive surfaces due to the lack of a primary alkylamine functional group, limiting their use in glycan microarray preparation. To address this problem, we have developed bifunctional fluorescent tag 2-amino-N-(2-aminoethyl)-benzamide (AEAB),21,22 which can specifically conjugate with free reducing glycans via the aromatic amine, retaining a primary alkylamine for efficient immobilization on activated solid phases including microarray slides. AEAB and AA conjugations provide similar UV range fluorescence with similar intensity and detectability. The installation of an AEAB tag, however, reduces the negative charges on glycans and affects their separation based on anion exchange chromatography.
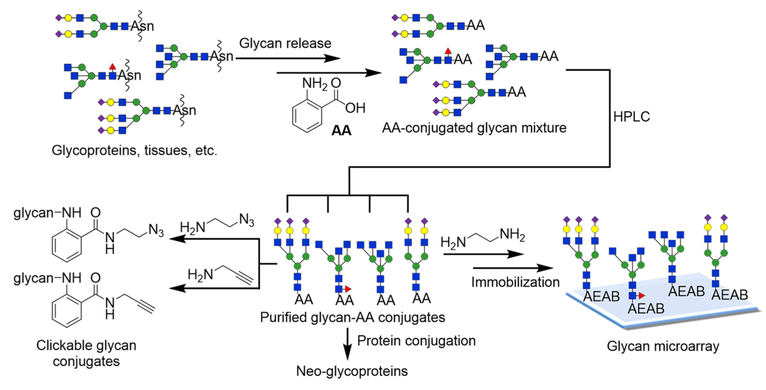
For preparative scale isolations of glycans, anion exchange chromatography is an important separation mechanism. While HILIC provides the best separation of glycans at analytical scale (μg), the high organic content of the mobile phase poses a significant challenge due to the low solubility of glycans. This problem has been largely overlooked since large quantities of complex glycans are generally not available. With more effort directed toward generating larger amounts of natural glycans, the solubility of glycans becomes an issue. For example, gram-quantity isolation of sialylglycopeptide from hen egg yolk and milligram-level separation of asparagine-linked Glc1Man9GlcNAc2 have been reported.23 We recently developed a novel method called oxidative release of natural glycans (ORNG) for large scale release of glycans linked to proteins and glycosphingolipids directly from kilograms of natural materials using household bleach.24,25 To separate grams of natural glycans, anion exchange and C18 reversed-phase chromatography are more desired due to the high water content of the mobile phases. In searching more compatible fluorescent tags, we re-explored the widely used AA tag. We envisioned that besides being a fluorescent tag for HPLC and MS, AA can serve as a versatile linker if the reactivity of the carboxylic acid functional group can be efficiently harnessed (Figure 1). Here we report simple chemistry for selective amidation of the AA tag, enabling facile immobilization of glycans to solid phases for preparation of glycan microarray and for synthesis of neoglycoproteins from glycan–AA conjugates.
Figure 1.
Strategy of the preparation of glycan–AA conjugates and versatile derivatizations of glycan–AA conjugates for further functionalization and microarray preparation.
RESULTS AND DISCUSSION
Amidation of Carboxylic Acid Group on Glycan–AA Conjugates.
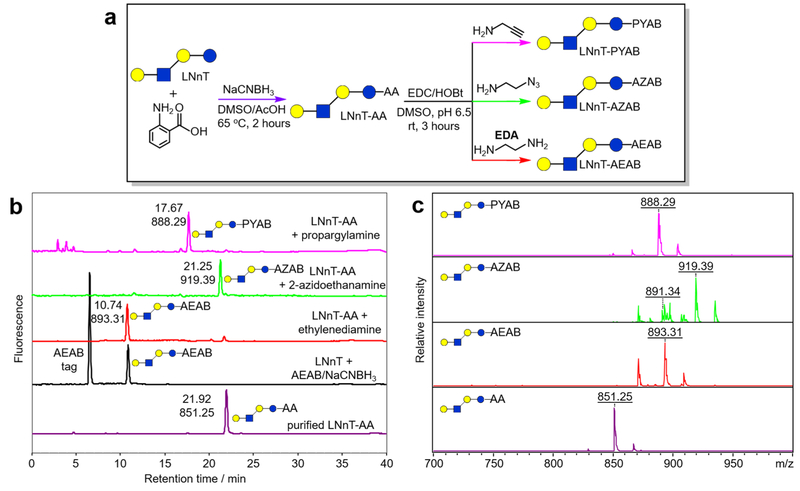
The condensation reaction between carboxylic acid and amine to form an amide bond is widely used in bioconjugation. Many reagents have been developed to activate carboxylic acid including carbodiimides, and additives such as N-hydroxysuccinimde (NHS) and hydroxybenzotriazole (HOBt) can reduce epimerization of activated amino acids.26,27 Our study suggests that the combination of 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC) and HOBt can efficiently convert glycan–AA conjugates to aromatic amide tagged glycans. As shown in Figure 2a, lacto-N-neotetraose (LNnT, Galβ1–4GlcNAcβ1–3Galβ1–4Glc) as the model compound reacted with AA to generate LNnT–AA via reductive amination. The purified LNnT–AA conjugate then reacted with excess amines (5% w/v amines in 0.5 M 2-(N-morpholino)ethanesulfonic acid (MES) aqueous buffer, pH = 6.5) in the presence of EDC and HOBt in DMSO solution for 3 h at room temperature. The reaction mixture was precipitated in 10 volumes of acetonitrile, and the precipitates were analyzed using C18 reversed-phase HPLC (Figure 2b, color: magenta, green, and red). HPLC profiles show that LNnT–AA is efficiently amidated with three selected amines, propargylamine, 2-azidoethylamine, and ethylenediamine (EDA), forming LNnT–PYAB, LNnT–AZAB, and LNnT–AEAB, respectively. All reactions yields are greater than 95% based on HPLC analysis. Since amidation of LNnT–AA with ethylenediamine produces LNnT–AEAB (overall yield greater than 90%), direct reductive amination of LNnT with AEAB· 2HCl was used to confirm conversion product identity (Figure 2b, color: black, yield21 greater than 98%). The amidation products were purified by HPLC and analyzed by MALDI-TOF-MS in positive ionization mode (Figure 2c). As expected, corresponding masses were observed for LNnT–AEAB (M + Na+, m/z = 893.31), LNnT–AZAB (M + Na+, m/z = 919.39), and LNnT–PYAB (M + Na+, m/z = 888.29). Interestingly, the azido product LNnT–AZAB showed more complex MALDI-MS including a minor peak at m/z = 891.34 (M + Na+). This Δ28 Da shift is likely due to fragmentation by N2 expulsion, which has been reported previously on MS analysis of azido compounds.28–30 This facile amidation of glycan–AA conjugates can quickly introduce new functionalities to glycan structures for further bioconjugation using either amine-reactive or clickable biomolecules or solid surfaces, making AA a versatile linker for glycan derivatizations (Figure S3).
Figure 2.
2-AA tagged-LNnT conversion with different amines. a) Scheme of LNnT reacting with 2-AA tag and then amidating with three different amines; b) reversed-phase HPLC profile (150 × 4.6 mm C18 column) of LNnT–AA conjugated with different amines; c) MALDI-TOF-MS of purified fluorophore tagged conjugates of LNnT. Only [M + Na]+ ions are labeled although [M + H]+ and [M + K]+ ions are also present.
Selective Amidation of Carboxylic Acid Group on Aromatic Ring of Sialylated Glycan–AA Conjugates.
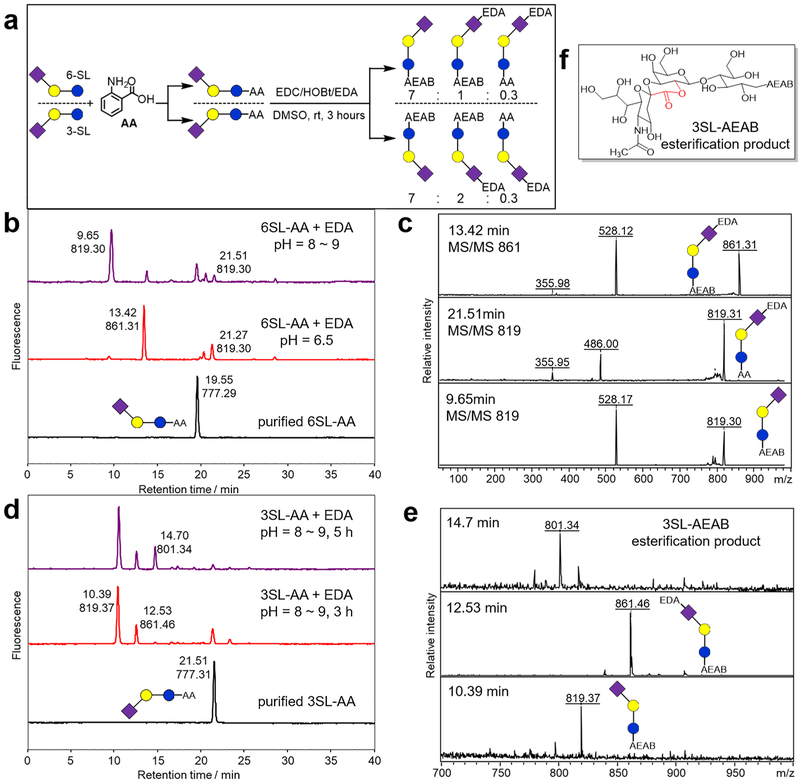
Natural glycans occurring in animal tissues often contain terminal sialic acids such as N-acetylneuraminic acid (Neu5Ac), which play important roles in biological and immunological as well as pathological processes.31,32 Chemical derivatizations on sialic acids such as permethylation, amidation, and esterification33–37 have been developed to facilitate MS analysis, but these derivatizations would likely abolish their biological activities. In our effort to install functional groups to glycan–AA conjugates, such as the conversion of glycan–AA to glycan–AEAB, it is vital to selectively amidate the benzoic acid but maintain the structural integrity of sialic acid (Figure 3). Many examples show evidence that conjugated carboxylic groups with a π bond or an aromatic ring diminish the rate of the reaction.38 However, using the same reaction condition described for LNnT–AA amidation, Neu5Acα2,6Galβ1,4Glc (6-SL)-AA with ethyl-enediamine afforded a diamidation product (Figure 3b, red, retention time = 13.42 min) showing a MALDI-MS peak at m/z = 861.31 (Figure 3c) along with a sialic acid monoamidation product (Figure 3b, red, retention time = 21.27 min) showing a MALDI-MS peak at m/z = 819.30 (Figure 3c). After screening the reaction condition, using 5% v/v EDA in 1.0 M MES aqueous solution at pH 8–9, the reaction generated the favored benzoic acid monoamidation product, i.e. 6-SL–AEAB (Figure 3b, purple, retention time = 9.65 min). The ratio of benzoic acid monoamidation:sialic acid monoamidation:diamindation is ~7:1:0.3, with conversion of 6SL–AA being greater than 90%. The two different monoamidation products can be easily separated by C18 RP-HPLC with a retention time of 9.65 and 21.51 min, respectively, and can be distinguished using MS/MS analysis (Figure 3c). The desired benzoic acid monoamidation product (6SL–AEAB) produced a characteristic fragment ion at m/z 528 confirming the conversion of AA to AEAB (Figure 3c, bottom). The fragment ions at m/z 486 indicate the underivatized AA, and the fragment ions at m/z 356 indicate monoamidated Neu5Ac (Figure 3c, middle). The selective AA amidation of Neu5Acα2,3Galβ1,4Glc (3-SL) was achieved similarly to produce 3-SL–AEAB (Figure 3d,e). However, for α2,3-linked sialic acid, an intraesterification byproduct of 3-SL–AEAB (Figure 3f) with an m/z peak at 801.34, can be observed (Figure 3d, top, retention time 14.70 min) upon extension of the reaction time. The typical reaction time was then set to 3 h to avoid this further esterification product. Similar intraesterification of 3-linked sialic acids was also reported by other groups.36
Figure 3.
AA tagged-3SL and −6SL conversions under different reaction conditions. (HPLC peaks are labeled with retention time, min, and base peak ion [M + Na]+ (m/z).) a) Scheme of two SL glycans reacting with an AA tag and then amidating with EDA; b) reversed-phase HPLC profile (150 × 4.6 mm C18 column) of a 6SL–AA tagged glycan conversion, bottom (black): purified 6SL–AA HPLC profile, middle (red:) conversion using EDA solution at pH = 6.5, upper (purple): conversion using EDA solution at pH = 8–9; c) structural analysis of peaks collected from (b) by MALDI-TOF MS/MS; d) reversed-phase HPLC profile of a 3SL–AA tagged glycan conversion, bottom (black): purified 3SL–AA HPLC profile, middle (red): conversion using EDA solution for 3 h, upper (purple): conversion using EDA solution for 5 h; e) MALDI-TOF-MS analysis of peaks collected (d); f) chemical structure of intraesterification product from 3-SL–AA when treated with EDA for an extended time.
Preparative 2D-HPLC Purification of AA-Tagged N-Glycans Released from Chicken Eggs by ORNG.
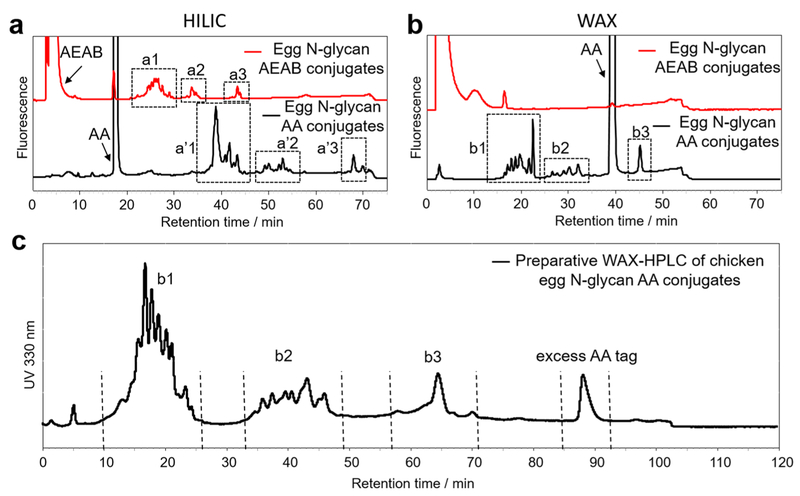
Lack of a large scale method to release N- and O-glycans from glycoproteins is still a roadblock for functional glycomics as traditional enzymatic and chemical methods are only practical in analytical scale.13 We recently reported the oxidative release of natural glycans (ORNG) using sodium hypochlorite (NaOCl) to release free reducing N-glycans from different natural sources including animal/plant tissues or commercial protein isolates at kilogram scale to acquire gram scale or larger amounts of glycans.24,25 The released glycan mixture, however, needs to be tagged and separated by HPLC to yield purified, individual glycans. While HILIC normally provides the best separation at analytical scale, its use of high organic content mobile phases creates a serious solubility issue when dealing with a gram or higher scale glycan mixture. Anion exchange and C18 chromatography, on the other hand, use an aqueous mobile phase and can be scaled up relatively easily. Using N-glycans released from chicken eggs by ORNG, we compared the separation of glycan–AEAB and glycan–AA conjugates on an amine column using both HILIC and WAX modes at analytical scale. As shown in Figure 4a,b, while both chicken egg N-glycan–AA and –AEAB conjugates showed good separation using the HILIC mode, only glycan–AA conjugates can be well resolved using the WAX mode. This is not surprising since the installation of AEAB on glycans significantly reduces the overall negative charge of sialylated glycans. Therefore, we employed AA as a fluorescent tag for chicken egg N-glycans and performed preparative scale WAX-HPLC separation of the AA conjugates (Figure 4c). Neutral, monosialylated and disialylated glycans are well separated into three groups. The preparative WAX-HPLC profile of chicken egg N-glycan AA conjugates (Figure 4c) showed a very similar resolution compared to the analytical profile (Figure 4b, bottom), proving that WAX-HPLC on an amino column can be used for large scale glycan separation. Interestingly, the excess AA tag was eluted later than disialylated glycans during a preparative run, while it was eluted before the disialylated glycans during the analytical run. It is interesting to note that although three groups of glycans are well separated in the WAX mode (Figure 4c), suggesting their different charge statuses, neutral glycans were also observed in the b2 group. This is apparently due to the installation of the AA tag, which may dramatically change the charge density of glycans.
Figure 4.
HPLC separation of chicken egg N-glycan AEAB and AA conjugates. a) Analytical HILIC-HPLC profiles of chicken egg N-glycan–AA conjugates (bottom, black) and –AEAB conjugates (upper, red) on an amino column, a1–3 and a′1–3 are three well separated groups of glycans; b) analytical WAX-HPLC profiles of chicken egg N-glycan–AA conjugates (bottom, black) and –AEAB conjugates (upper, red) on an amino column, b1–3: three well separated groups of glycans; c) the elution profile (UV 330 nm) of crude chicken egg N-glycan–AA conjugates on preparative WAX-HPLC (50 × 250 mm amino column). Fractions were collected based on the UV profile (flow rate: 50 mL/min).
Fractions were collected from preparative WAX-HPLC and subjected to preparative C18 reversed-phase HPLC for second dimensional purification (Figure S1 and Table S1). After obtaining purified AA tagged N-glycans, a selection of individual glycan–AA conjugates, including both neutral and sialylated glycans, was converted to glycan–AEAB conjugates under the optimized reaction condition described above for LNnT–AA (neutral glycans) or 3-SL–AA (sialylated glycans). A total of 30 glycan–AEAB conjugates were purified for microarray preparation.
Microarray Printing and Binding Assay.
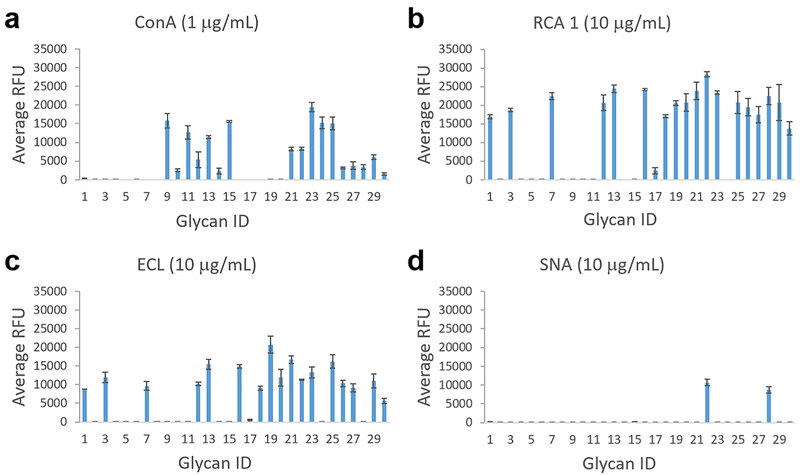
To compare the immobilization efficiency between AA tagged glycans and AEAB tagged glycans, LNnT–AA and its conversion product LNnT–AEAB and 2′-fucosyllactose–AA (2′-FL–AA) and its conversion product 2′-FL–AEAB were printed on an NHS-activated slide and assayed with two lectins (Figure S2). The relative binding of the lectins AAL and ECL to glycans 2′-FL and LNnT, respectively, is in agreement with the known binding specificities, and binding decreases with decreasing concentration of printed glycan. The converted AEAB tagged glycans clearly showed stronger binding signals than AA tagged glycans, indicating a much better printing efficiency, which is consistent with the higher reactivity of primary alkylamine on AEAB and is consistent with our previous study.21 Using this facile chemistry to convert glycan–AA to glycan–AEAB, we printed a microarray of 30 chicken egg N-glycans. This microarray was interrogated with four plant lectins: ConA, RCA-1, ECL, and SNA (Figure 5 and Table S2). As expected, ConA showed binding to many N-glycans. However, there are a number of glycans that showed no binding, presumably tri- and tetraantennary ones. Interestingly, 4 N-glycans with composition of H3N7 were printed. While glycan ID #2, 5, 6 showed no binding to ConA, glycan ID #10 showed clear binding, suggesting the occurrence of isomeric structures. Both with H5N7 composition, glycan ID #8 showed no binding, while glycan ID #26 showed clear binding to ConA. SNA showed bindings to two sialylated glycans. Both ECL and RCA-1 are known to bind terminal Galβ4GlcNAc. One major difference is that terminal 6-sialylation on the Galβ4GlcNAc motif abolishes binding to ECL but does not affect RCA-1 binding. As expected, all ECL bound glycans also showed binding to RCA-1. Glycan ID 28 showed binding to RCA-1 but not ECL (chart ID 28) and also showed clear binding to SNA, consistent with a disialylated structure. On the other hand, glycan ID 22 showed bindings to RCA I, ECL, and SNA, consistent with a monosialylated structure. Glycan ID #1, 7, and 16, with the same H4N8 composition, all showed clear binding to ECL and RCA-I, suggesting a terminal Galβ4Glc-NAc. On the other hand, while glycan ID #8, 17, and 20 all have the same composition H5N5, they show none, weak, and strong binding to ECL and RCA-1, respectively, suggesting distinct structures. The binding results provide important structural information to the printed glycans. While more study is required to further elucidate the glycan structures and is not the focus of this study, the results clearly demonstrated that glycan–AA conjugates can be easily converted to glycan–AEAB conjugates for microarray preparation.
Figure 5.
Plant lectin binding of a) Concanavalin A (Con A), b) Ricinus communis agglutinin I (RCA 1), c) Erythrina cristagalli (ECL), and d) Sambucus nigra (SNA) on the glycan microarray of chicken egg N-glycan–AEAB conjugates converted from –AA conjugates. The X-axis represents glycan ID, and the Y-axis represents the relative fluorescence unit of bound proteins. Error bars represent standard deviation of 6 replicates.
Preparation of Neoglycoproteins from Glycan–AA Conjugates.
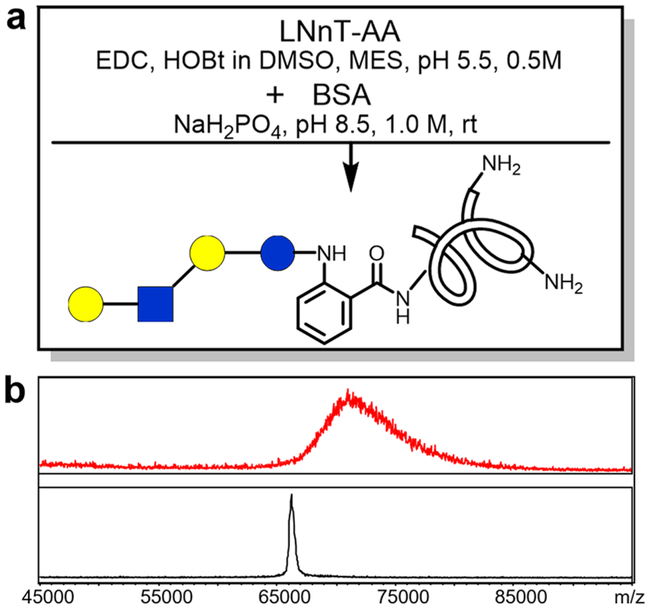
We predicted that glycan–AA conjugates can also be linked to proteins to produce neoglycoproteins using similar chemistry. Neoglycoproteins prepared by linking glycans to proteins have many potential applications, including microarray preparation and carbohydrate-based vaccine development.39–41 Previously we reported using p-nitrophenyl anthranilate (PNPA) as a heterobifunctional linker to prepare neoglycoproteins.39 The glycan–PNPA conjugate, as an activated benzoic ester, is only stable for short-term storage. Since glycan–AA conjugates can be converted to amides using various amines, they can potentially be conjugated directly with amino groups on proteins (Figure 6a) to form neoglycoproteins. To explore this possibility, LNnT–AA was activated by EDC/HOBt followed by addition of bovine serum albumin (BSA). When the product was analyzed by MALDI-TOF-MS, the attachment of glycans onto proteins is evident (Figure 6b). The resulting neoglycoprotein BSA-LNnT showed increased mass and wider molecular weight distribution. By estimation based on the mass shift from unconjugated BSA, more than 5 molecules of LNnT per BSA molecule are attached on average. This procedure provides a straightforward way to prepare neoglycoproteins.
Figure 6.
BSA conjugation with LNnT–AA: a) scheme and reaction condition of BSA conjugation with LNnT–AA and b) MALDI-TOF profiles of BSA (bottom) and BSA–LNnT conjugation product.
CONCLUSION
In this study, we have developed facile chemical methods to greatly expand the use of AA as a fluorescent tag and bifunctional linker. Glycan–AA conjugates prepared from natural glycans can be separated using WAX-HPLC and C18-HPLC at large scale. Besides being a sensitive fluorescent tag for glycan analysis and preparation, the AA tag can be easily and selectively amidated with various amines to convert glycan–AA conjugates into various forms of functionalized glycans. These functionalized glycans can be adopted for further conjugation by click chemistry, microarray printing, and neoglycoprotein preparation. Because of its simplicity, versatility, and the current wide use of AA tags for glycan derivatization, we believe that this method can contribute greatly to functional glycomics.
EXPERIMENTAL SECTION
Materials.
All chemicals and HPLC solvents were purchased from Sigma-Aldrich, St. Louis, MO and Fisher Scientific, Pittsburgh, PA. Milli-Q water was used to prepare all aqueous buffers. All solutions were prepared freshly to use.
Mass Spectrometry (MS).
A Bruker Daltonics UltraflexII MALDI-TOF/TOF system and an anchorchip target plate were used for MS analysis. A reflective positive mode was used for glycans. 2,5-Dihydroxybenzoic acid (DHB) (10 mg/mL in 50% acetonitrile with 0.1% trifluoroacetic acid) was used as matrix.
Synthesis of 2-Azidoethanamine.
2-Chloroethylamine hydrochloride (2.32 g, 20 mmol) and sodium azide (NaN3, 2.6 g, 40 mmol) were dissolved in 40 mL of Milli-Q water. After stirring for 12 h at 80 °C, 10 mL of 1 M sodium hydroxide solution was added to adjust the reaction to alkaline condition. After stirring for another 10 min at room temperature, the reaction was extracted with dichloromethane (3 × 10 mL). The organic layer was dried with magnesium sulfate and then concentrated in vacno at 0 °C to give product (yield: 60%).
Synthesis of Glycan–AA.
In a 1.5 mL snap-cap microcentrifuge tube, 100 μg of free reducing glycan was mixed with 50 μL of 60 mg/mL 2-AA in DMSO/AcOH (7:3) solution and 50 μL of 64 mg/mL NaCNBH3 in DMSO/AcOH (7:3) solution and vortexed for 1 min. After 2 h at 65 °C, the reaction solution was quenched with 10X volume of acetonitrile. The cloudy reaction mixture was centrifuged for 2 min at 10000 × g. The supernatant was decanted, and the pellet was dried under vacuum and dissolved into 100 μL of Milli-Q water for further analysis and purification.
The synthesis of glycan–AEAB by reductive amination is essentially the same as that for glycan–AA conjugates except that 84 mg/mL AEAB in DMSO/AcOH (7:3) solution was used instead.
LNnT–AA Conversion via Amidation Reaction.
In a 1.5 mL snap-cap microcentrifuge tube, 20 μg of LNnT–AA was mixed with 50 μL of 10 mg/mL EDC in DMSO solution and 50 μL of 10 mg/mL HOBt in DMSO solution. Then 10 μL of 5% (v/v) amine (EDA, propargylamine, or 2-azidoethanamine) and 0.5 M MES buffer (pH = 6.5) were added to the mixture and vortexed for 1 min. After 3 h at room temperature, the reaction solution was quenched by 10X volume acetonitrile. The cloudy reaction mixture was centrifuged for 10 min at 10000 × g. The supernatant was decanted, and white precipitate was dried under vacuum and then dissolved into 100 μL of Milli-Q water for further analysis and purification.
α-2,6-Linked Sialylated glycan–AA Conversion to AEAB Tagged Glycan.
In a microcentrifuge tube, 10 μg of α-2,6-linked sialylated glycan–AA was mixed with 50 μL of 10 mg/mL EDC in DMSO solution and 50 μL of 10 mg/mL HOBt in DMSO solution. Then 10 μL of 5% (v/v) EDA and 1.0 M MES aqueous solution (pH 8.2) was added to the DMSO mixture solution and vortexed for 1 min. After 5 h at room temperature, the reaction solution was dissolved with an equal volume of Milli-Q water for further analysis and purification.
α-2,3-Linked Sialylated glycan–AA Conversion to AEAB Tagged Glycan.
In a microcentrifuge tube, 10 μg of α-2,3-linked sialylated glycan–AA was mixed with 50 μL of 10 mg/mL EDC in DMSO solution and 50 μL of 10 mg/mL HOBt in DMSO solution. Then 10 μL of 5% (v/v) EDA and 1.0 M MES aqueous solution (pH 8.2) was added to the DMSO mixture solution and vortexed for 1 min. After 3 h at room temperature, the reaction solution was dissolved with an equal volume of Milli-Q water for further analysis and purification.
Release of N-Glycans from Chicken Eggs by ORNG.
Forty-eight chicken eggs (contain about 310 g proteins) were mixed with 7.1 L of H2O and 3.6 L of 6% NaOCl under open air and stirred for 30 min. Concentrated HCl was added to quench the reaction and adjust the pH to 4.8. The final reaction mixture, ~10.7 L, was then centrifuged to remove undissolved material. The supernatant was then concentrated to ~500 mL in vacno. The further process followed the procedure described in our previous publication.25
High Performance Liquid Chromatography (HPLC) Analyses.
A Shimadzu HPLC CBM-20A system with UV detector SPD-20A and fluorescence detector RF-10Axl was used for analytical HPLC profiles. A Shimadzu HPLC LC-8a preparative system with UV detector SPD-10Av was used for preparative HPLC separation. UV absorption at 330 nm or fluorescence at 330 nm excitation and 420 nm emission was used for detection of anthranilic acid (AA) and 2-amino-N-(2-aminoethyl)benzamide (AEAB). Phenomenex amino columns were used for both HILIC and WAX HPLC. Phenomenex C18 columns were used for reversed-phase HPLC separation. For WAX HPLC, the mobile phase gradient was from 0 to 200 mM ammonium acetate (pH 4.5) containing 5% acetonitrile over 50 min (analytical run) or 120 min (preparative run). For reversed-phase HPLC, the mobile phase gradient was from 2% to 15% acetonitrile containing 0.1% trifluoroacetic acid over 30 or 45 min.
2D-HPLC Separation.
Preparative WAX HPLC separation: 1.8 g of a white solid powder containing AA conjugated glycans was dissolved into 10 mL of H2O. Then the 10 mL solution was injected into a preparative amino HPLC column (column size: 250 × 50 mm). Fractions (0.4 min) were collected, and selected fractions were analyzed using MALDI-TOF-MS. C18 reversed-phase HPLC separation: fractions collected from preparative WAX-HPLC separation were individually lyophilized, redissolved in water, and then applied to preparative C18 reversed-phase HPLC (column size: 250 × 21.2 mm). Peaks were collected and characterized using MALDI-TOF-MS.
LNnT-BSA Conjugation.
In a 1.5 mL snap-cap microcentrifuge tube, 50 μg of LNnT–AA was mixed with 5 μL of 100 mg/mL EDC in DMSO solution, 5 μL of 100 mg/mL HOBt in DMSO solution, and 5 μL of 0.5 M MES buffer (pH = 5.5). After 15 min at room temperature, 5 μL of BSA (1 mg/mL in water) and 5 μL of phosphate buffer (1.0 M, pH = 8.5) were added to the mixed solution and reacted for 1 h under room temperature. The reaction solution was then subjected to a Sephadex G-25 column to remove salts and other impurities before MALDI-TOF-MS analysis.
Microarray Printing, Binding Assay, and Scanning.
Contact printing on NHS-activated slides was carried out using an Aushon’s 2470 microarrayer. N-Glycans were printed at 5 μM to 100 μM in 100 mM sodium phosphate (pH = 8.5) in replicates of 6. Biotinylated AAL, ECL, SNA, ConA, and RCA were assayed on the microarray slides at concentrations described in the figures. Five μg/mL cyanine 5-streptavidin (Invitrogen) was used for the detection of lectin binding using a fluorescent scanner (Innopsys).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Common Fund Glyco-science (U01GM116254) and partially by a STTR grant R41GM122139. We thank Dr. David F. Smith for discussions and comments on the manuscript. We also acknowledge Emory Comprehensive Glycomics Core (ECGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acs.bioconj-chem.8b00678.
Figures of HPLC profiles of chicken egg glycan–AA purification, lectin binding to a microarray of LNnT–AA and LNnT–AEAB at multiple concentrations, and click chemistry validation of LNnT–AZAB with propargylamine; tables of summary of chicken egg glycan compositions included in microarray printing and lectin binding data to the glycan microarray (PDF)
Notes
The authors declare the following competing financial interest(s): X.S. is a cofounder of NatGlycan LLC, which is commercializing the ORNG technology.
REFERENCES
- (1).Varki A (2017) Biological roles of glycans. Glycobiology 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gaunitz S, Nagy G, Pohl NLB, and Novotny MV (2017) Recent Advances in the Analysis of Complex Glycoproteins. Anal. Chem 89, 389–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong C-H, and Paulson JC (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci U. S. A 101, 17033–17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).de Paz JL, and Seeberger PH (2008) Deciphering the Glycosaminoglycan Code with the Help of Microarrays. Mol. BioSyst 4, 707–711. [DOI] [PubMed] [Google Scholar]
- (5).Feizi T, Fazio F, Chai W, and Wong C-H (2003) Carbohydrate Microarrays-a New Set of Technologies at the Frontiers of Glycomics. Curr. Opin. Struct. Biol 13, 637–645. [DOI] [PubMed] [Google Scholar]
- (6).Oyelaran O, and Gildersleeve JC (2009) Glycan Arrays: Recent Advances and Future Challenges. Curr. Opin. Chem. Biol 13, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Paulson JC, Blixt O, and Collins BE (2006) Sweet spots in functional glycomics. Nat. Chem. Biol 2, 238. [DOI] [PubMed] [Google Scholar]
- (8).Rillahan CD, and Paulson JC (2011) Glycan Microarrays for Decoding the Glycome. Annu. Rev. Biochem 80, 797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Stevens J, Blixt O, Paulson JC, and Wilson IA (2006) Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol 4, 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wen L, Edmunds G, Gibbons C, Zhang J, Gadi MR, Zhu H, Fang J, Liu X, Kong Y, and Wang PG (2018) Toward Automated Enzymatic Synthesis of Oligosaccharides. Chem. Rev 118, 8151–8187. [DOI] [PubMed] [Google Scholar]
- (11).Kulkarni SS, Wang C-C, Sabbavarapu NM, Podilapu AR, Liao P-H, and Hung S-C (2018) One-Pot” Protection, Glycosylation, and Protection–Glycosylation Strategies of Carbohydrates. Chem. Rev 118, 8025–8104. [DOI] [PubMed] [Google Scholar]
- (12).Nielsen MM, and Pedersen CM (2018) Catalytic Glycosylations in Oligosaccharide Synthesis. Chem. Rev 118, 8285–8358. [DOI] [PubMed] [Google Scholar]
- (13).Seeberger PH (2015) The Logic of Automated Glycan Assembly. Acc. Chem. Res 48, 1450–1463. [DOI] [PubMed] [Google Scholar]
- (14).Plummer TH Jr., Elder JH, Alexander S, Phelan AW, and Tarentino AL (1984) Demonstration of peptide:N-glycosidase F activity in endo-β-N-acetylglucosaminidase F preparations. J. Biol. Chem 259, 10700–10704. [PubMed] [Google Scholar]
- (15).Tarentino AL, Gomez CM, and Plummer TH Jr. (1985) Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry 24, 4665–4671. [DOI] [PubMed] [Google Scholar]
- (16).Mulagapati S, Koppolu V, and Raju TS (2017) Decoding of O-Linked Glycosylation by Mass Spectrometry. Biochemistry 56, 1218–1226. [DOI] [PubMed] [Google Scholar]
- (17).Li Y-T, Chou C-W, Li S-C, Kobayashi U, Ishibashi Y. h., and Ito M (2009) Preparation of Homogenous Oligosaccharide Chains from Glycosphingolipids. Glycoconjugate J 26, 929–933. [DOI] [PubMed] [Google Scholar]
- (18).Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, and Wuhrer M (2010) Glycan Labeling Strategies and Their Use in Identification and Quantification. Anal. Bioanal Chem 397, 3457–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Narawane S, Budnjo A, Grauffel C, Haug BE, and Reuter N (2014) In Silico Design, Synthesis, and Assays of Specific Substrates for Proteinase 3: Influence of Fluorogenic and Charged Groups. J. Med. Chem 57, 1111–1115. [DOI] [PubMed] [Google Scholar]
- (20).Schulz-Fincke A-C, Tikhomirov SA, Braune A, Girbl T, Gilberg E, Bajorath J, Blaut M, Nourshargh S, and Gutschow M (2018) Design of an Activity-Based Probe for Human Neutrophil Elastase: Implementation of the Lossen Rearrangement To Induce Förster Resonance Energy Transfers. Biochemistry 57, 742–752. [DOI] [PubMed] [Google Scholar]
- (21).Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, and Cummings RD (2009) Novel Fluorescent Glycan Microarray Strategy Reveals Ligands for Galectins. Chem. Biol 16, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, and Smith DF (2011) Shotgun glycomics: a microarray strategy for functional glycomics. Nat. Methods 8, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang N, Seko A, Takeda Y, and Ito Y (2015) Preparation of asparagine-linked monoglucosylated high-mannose-type oligosaccharide from egg yolk. Carbohydr. Res 411, 37–41. [DOI] [PubMed] [Google Scholar]
- (24).Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, and Cummings RD (2016) Oxidative release of natural glycans for functional glycomics. Nat. Methods 13, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhu Y, Yan M, Lasanajak Y, Smith DF, and Song X (2018) Large scale preparation of high mannose and paucimannose N-glycans fromsoybean proteins by oxidative release of natural glycans (ORNG). Carbohydr. Res 464, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Montalbetti CAGN, and Falque V (2005) Amide bond formation and peptide coupling. Tetrahedron 61, 10827–10852. [Google Scholar]
- (27).Valeur E, and Bradley M (2009) Amide bond formation: beyond the myth of coupling reagents. Chem. Soc. Rev 38, 606–631. [DOI] [PubMed] [Google Scholar]
- (28).Raynaud J, Absalon C, Gnanou Y, and Taton D (2009) N-Heterocyclic Carbene-Induced Zwitterionic Ring-Opening Polymerization of Ethylene Oxide and Direct Synthesis of a,w-Difunctionalized Poly(ethylene oxide)s and Poly(ethylene oxide)-b-poly(e-caprolactone) Block Copolymers. J. Am. Chem. Soc 131, 3201–3209. [DOI] [PubMed] [Google Scholar]
- (29).Li Y, Hoskins JN, Sreerama S, and Grayson SM (2010) MALDI-TOF Mass Spectral Characterization of Polymers Containing an Azide Group: Evidence of Metastable Ions. Macromolecules 43, 6225–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Guillaneuf Y, Dufils P-E, Autissier L, Rollet M, Gigmes D, and Bertin D (2010) Radical Chain End Chemical Transformation of SG1-Based Polystyrenes. Macromolecules 43, 91–100. [Google Scholar]
- (31).Varki A (2008) Sialic Acids in Human Health and Disease. Trends Mol. Med 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chen X, and Varki A (2010) Advances in the Biology and Chemistry of Sialic Acids. ACS Chem. Biol 5, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sekiya S, Wada Y, and Tanaka K (2005) Derivatization for Stabilizing Sialic Acids in MALDI-MS. Anal. Chem 77, 4962–4968. [DOI] [PubMed] [Google Scholar]
- (34).Toyoda M, Ito H, Matsuno Y. k., Narimatsu H, and Kameyama A (2008) Quantitative Derivatization of Sialic Acids for the Detection of Sialoglycans by MALDI MS. Anal. Chem 80, 5211–5218. [DOI] [PubMed] [Google Scholar]
- (35).Reiding KR, Blank D, Kuijper DM, Deelder AM, and Wuhrer M (2014) High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification. Anal. Chem 86, 5784–5793. [DOI] [PubMed] [Google Scholar]
- (36).de Haan N, Reiding KR, Haberger M, Reusch D, Falck D, and Wuhrer M (2015) Linkage-Specific Sialic Acid Derivatization for MALDI-TOF-MS Profiling of IgG Glycopeptides. Anal. Chem 87, 8284–8291. [DOI] [PubMed] [Google Scholar]
- (37).Jiang K, Zhu H, Li L, Guo Y, Gashash E, Ma C, Sun X, Li J, Zhang L, and Wang PG (2017) Sialic acid linkage-specific permethylation for improved profiling of protein glycosylation by MALDI-TOF MS. Anal. Chim. Acta 981, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nahmany M, and Melman A (2004) Chemoselectivity in Reactions of Esterification. Org. Biomol Chem 2, 1563–1572. [DOI] [PubMed] [Google Scholar]
- (39).Luyai A, Lasanajak Y, Smith DF, Cummings R, and Song X (2009) Facile Preparation of Fluorescent Neoglycoproteins Using p-Nitrophenyl Anthranilate as a Heterobifunctional Linker. Bioconjugate Chem. 20, 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Prasanphanich NS, Song X, Heimburg-Molinaro J, Luyai AE, Lasanajak Y, Cutler CE, Smith DF, and Cummings RD (2015) Intact Reducing Glycan Promotes the Specific Immune Response to Lacto-N-neotetraose-BSA Neoglycoconjugates. Bioconjugate Chem. 26, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Toonstra C, Wu L, Li C, Wang D, and Wang L-X (2018) Top-Down Chemoenzymatic Approach to Synthesizing Diverse High-Mannose N-Glycans and Related Neoglycoproteins for Carbohydrate Microarray Analysis. Bioconjugate Chem. 29, 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.