Summary
To study the contribution of T-cell receptors (TCR) to resulting T cell responses, we studied three different human α TCRs, reactive to the same gp100-derived peptide presented in the context of HLA-A*0201. When expressed in primary CD8 T cells, all receptors elicited classic antigen-induced IFN-γ responses, which correlated with TCR affinity for peptide-MHC in the order T4H2>R6C12>SILv44. However, SILv44 elicited superior IL-17A release. Importantly, in vivo, SILv44-transgenic T cells mediated superior anti-tumor responses to 888-A2+ human melanoma tumor cells upon adoptive transfer into tumor-challenged mice while maintaining IL-17 expression. Modeling of the TCR ternary complexes suggested architectural differences between SILv44 and the other complexes, providing a potential structural basis for the observed differences. Overall, the data reveal a more prominent role for the T-cell receptor in defining host T-cell physiology than traditionally assumed, while parameters beyond IFN-γ secretion and TCR affinity ultimately determine the reactivity of tumor-reactive T cells.
Keywords: T cell receptor, melanoma, transgenic T cells, IL-17A, molecular modeling
Introduction
The T cell receptor (TCR) plays a decisive role in host T cell responses to cognate antigen through its variable regions and the properties they confer on each individual receptor (Smith et al., 2013). The TCR also contributes to T cell function through its membrane proximal regions that interact with other components of the TCR/CD3 complex to drive signaling through Zap-70 and downstream Raf, MEKK1 and MAP kinase pathways (van Panhuys, 2016). As a readout of T cell activation, investigators have generally turned to IFN-γ secretion (Meng et al., 2016). However, a recent body of work supports a critical role for IL-17 secreting T cells in defining the longevity and versatility of cytotoxic T cell populations responding to cognate antigen, suggesting that IFN-γ secretion alone will not adequately define superior T cell activation (Nelson et al., 2015).
One area where superior TCR activation is of particular importance is in tumor immunotherapy. T cell receptors specific for tumor antigens offer therapeutic potential (Golubovskaya, Berahovich, Xu, Harto, & Wu, 2017). By cloning such receptors, we can generate reagents to redirect T cells toward the tumor and activate immune responses in patients who do not develop adequate reactivity to keep the tumors in check. For example, TCR transgenic T cells targeting melanosomal differentiation antigens are currently in clinical trials for the treatment of malignant melanoma (Chodon et al., 2014). The first published trial of its kind was reported in 2006, using the F5 receptor against human MART-1 (Duval et al., 2006). The results were encouraging, and several investigators are currently pursuing T cell receptors for tumor-directed therapy, which appears to be associated with fewer severe side effects than T cells bearing chimeric antigen receptor transgenes (Ataca & Arslan, 2015).
In a general sense, understanding the role of TCRs in T cell activation is central to their biology. For therapeutic applications, TCRs with high affinity for relevant antigens can be identified through measurements that quantify binding affinities, or through functional parameters such as cytotoxicity towards target cells, proliferation of the T cells, or expression of representative cytokines (Hebeisen et al., 2015). As T cells are front and center in type 1 responses, IFN-γ has taken a prominent role as the prime cytokine to identify T cells and TCRs with optimal avidity or affinity towards their cognate antigen.
To challenge the hypothesis that TCR affinity and subsequent IFN-γ release define the optimal T-cells and TCRs, we studied three receptors that mediate reactivity towards a single antigenic peptide presented in the context of the same HLA-A*0201 molecule. TCR constructs were generated that defined expression of each TCR in an identical fashion, cloning each TCR into the same retroviral vectors with subunits separated by the same P2A viral slippage sequence, and introduced them into CD8 T cells from the same donors. Once each TCR was transduced in primary human T cells from these donors, we originally compared each TCR transgenic T cell population based on the amount of IFN-γ secreted in response to the gp100 antigen (Moore et al., 2009) and in response to naturally processed peptide. As differences in TCR stability may affect the level of TCR expression at the cell surface, which in turn affects T cell avidity, we measured the amount of TCR expression using antibodies to the TCR-β subunit. We next measured the classic IFN-γ secretion patterns in response to a range of peptide concentrations and related these to binding affinities determined using biochemical assays with recombinant molecules. We included analysis of IL-17A, a cytokine that may define the longevity of responses in vivo supported by signaling via the inducible costimulatory molecule called ICOS (Nelson et al., 2015). The phenotype and function of cytokine secreting cells was further defined by flow cytometry and cytokine array. To understand which TCR would define optimal responses toward tumor cells in vivo, we performed efficacy studies in SCID/beige immunodeficient mice challenged with 888-A2+ T cells, comparing tumor growth among groups of mice treated with transgenic CD8 T cells. The link between cytokine expression patterns and in vivo responsiveness to our TCR constructs was then further examined by modeling the position of each TCR in complex with the gp100/HLA-A2 complex. The striking differences in responsiveness of T cells from the same donors that express TCRs that respond to the same peptide-HLA complexes help us define the role of the TCR in T responsiveness and suggest parameters that will better predict the reactivity of host T cells when selecting T cells to be used in therapeutic settings.
Materials and methods
Cell lines
T2 cells were obtained from the American Type Culture Collection (VA, USA), and maintained in RPMI 1640 medium (Mediatech, VA, USA) with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, GA, USA), 250 ng/mL Amphotericin B, 100U/mL Penicillin, 100 μg/mL Streptomycin (Invitrogen, CA, USA), and 2 mM L-glutamine (Invitrogen, CA, USA). Cells were grown at 37° C and 5% CO2.
GP2–293 viral producing cells, retroviral vector pQCXIN, and pVSV-g were obtained from Clontech Laboratories, Inc (CA, USA), and cells were maintained in Dulbecco’s Modified Eagle Medium (Mediatech, VA, USA) with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, GA, USA), 250 ng/mL Amphotericin B, 100U/mL Penicillin, 100 μg/mL Streptomycin (Invitrogen), 4.5 g/L L-glutamine, 4.5 g/L glucose, and 4.61 g/L sodium pyruvate (Mediatech, VA, USA). Transfected GP2–293 were maintained in presence of 2 mg/mL G418 sulfate (Mediatech, VA, USA).
T cells purified from PBMCs were maintained in AIMV medium (Invitrogen, CA, USA) supplemented with 5% heat-inactivated pooled human AB serum (Sigma, VA, USA), 250 ng/mL Amphotericin B, 100U/mL Penicillin, 100 μg/mL Streptomycin (Invitrogen, CA, USA), 300 IU/mL recombinant human IL-2, 100ng/mL recombinant IL-15 (both R&D Systems, MN, USA), and 50 ng/mL OKT3 (eBioscience, CA, USA). PBMCs from healthy apheresis patients were isolated by Ficoll gradient centrifugation unless otherwise noted.
Melanocytes isolated from foreskin were maintained in Ham’s F-12 medium (Mediatech, VA, USA) with 2 mmol/L glutamine, 100 IU/ml penicillin,100 mg/ml streptomycin,100 mg/ml amphotericin, 0.1 mmol/l isobutylmethylxanthine (Sigma, MO, USA), 10 ng/ml 12-O-tetradecanoyl phorbol-13-acetate (OCT; Sigma, MO, USA), and 1% Ultroser G (PALL Life Sciences, NY, USA). HLA-A2 expression was validated by flow cytometry using FITC-conjugated antibody BB7.2 (BD Biosciences, CA, USA).
Human melanoma cell lines 624.38 (gp100+, HLA-A2+), 624.28 (gp100+, HLA-A2−), A375 (gp100−, HLA-A2+) and 888-A2+ (gp100+, HLA-A2+) (Klarquist et al., 2016; Rivoltini et al., 1995; Zhai et al., 1996) were maintained as described for GP293 cells above.
Retroviral vector construction and transduction
Retroviral vector pQCXIN (Clontech, CA, USA) served as a backbone for transductions. TCRα and β chains for T cell clones SILv44, T2H4 or R6C12 (Klarquist et al., 2016; Moore et al., 2009) were joined via a viral 2A self-cleavage sequence and inserted at NotI and PacI restriction sites on pQCXIN. Resulting plasmids were transfected into amphotropic viral producer cells GP2–293 using Lipofectamine 2000 (Invitrogen, CA, USA) and stable transfectants were selected using G-418 sulfate (Life Technologies, CA, USA). Infectious particles were generated following transfection with envelope gene from vesicular stomatitis virus. Virus production was confirmed by viral producing cell death through toxicity of secreted VSV-G.
For T cell transduction, PBMCs were activated in culture medium for 3 days. Non-adherent cells were negatively sorted for CD8 T cells (Stem Cell Technologies, BC, Canada) and transduced twice, one day apart using the retronectin-retrovirus method before selection in 1 mg/mL G-418 sulfate for 5 days. Transduced T cells were rapidly expanded using a 200-fold excess of 5000 rad irradiated feeders for 5 days.
Flow cytometry
Cells were stained with fluorochrome conjugated anti-CD3-FITC, anti-CD4-V450, anti-CD8-PE-Cy7 (all BD Biosciences, CA, USA), anti-Vβ8-PE, anti-Vβ17-PE (both Beckman Coulter, CA, USA). Data were collected on a Fortessa LSR II instrument (BD Biosciences, CA, USA).
Cytokine release assays
In cytokine release assays, 1×105 effector T cells were combined with T2 target cells at 1:1 in a 96 well U-bottom plates for 20 hours. T2 target cells were either unpulsed or peptide-pulsed for two hours prior to incubation with effectors. Cytokine release in the supernatant was measured using an ELISAPRO kit for INF-γ or IL-17A ELISA (Mabtech, OH, USA).
Anti-tumor efficacy of TCR transduced T cells in vivo
Therapeutic responses were measured in vivo, following subcutaneous injection of 106 human 888-A2+ into SCID/beige mice (n=4). Tumors were measured daily and grew for 35 days prior to retro-orbital injection of 2×106 TCR transduced human T cells expressing the SILv44, T4H2 or R6C12 TCR receptor, or untransduced T cells. A second injection was provided on day 58. Mice were sacrificed on day 63. Tumors were harvested and stored in OTC at −80°C for immunohistochemistry. Tissue staining was performed on tumor cryosections using anti-human CD3-FITC (BD Biosciences, CA, USA), Vβ8-PE, Vβ17-PE (both Beckman Coulter, CA, USA), and IL-17 and to IFN-γ (both BioLegend, CA, USA). The authors adhered to institutional guidelines and the NIH Guide for the Care and Use of Animals, under animal assurance number A3117–0 assigned to the Institution where said studies were performed.
Binding affinity measurements
Soluble constructs of SILv44, R6C12, T4H2, and HLA-A2 were expressed and refolded as described (Davis-Harrison, Armstrong, & Baker, 2005). Briefly, the TCR α and β chains, the HLA-A2 heavy chain and β2-microglobulin (β2m) were expressed separately in Escherichia coli. Isolated inclusion bodies were dissolved in 8 M urea. For TCR refolding, TCR α and β chains with α chain in 20% excess were rapidly diluted into 50 mM Tris-HCl (pH 8.3), 2.5 M urea, 6.3 mM cysteamine, 3.7 mM cystamine, 2 mM EDTA, and 0.2 mM PMSF with the addition of 400 mM ʟ-arginine for SILv44 and R6C12. HLA-A2 heavy chain and β2m were rapidly diluted into 100 mM Tris-HCl (pH 8.3), 400 mM ʟ-arginine, 6.3 mM cysteamine, 3.7 mM cystamine, 2 mM EDTA, and 0.2 mM PMSF in a 1:3 molar ratio with excess peptide. TCR and Peptide/MHC complexes were incubated for 12 hrs at 4˚C. TCR and Peptide/MHC complexes were dialyzed against 10 mM Tris-HCl (pH 8.3) for 48 hrs. Folded complexes were purified via anion exchange followed by size-exclusion chromatography. Steady-state binding experiments were performed with a Biacore T200 as described (Davis-Harrison et al., 2005). Briefly, experiments were performed at 25˚C in HBS-EP buffer containing 10 mM HEPES, 3 mM EDTA, 150 mM NaCl, and 0.0005% surfactant P20 at pH 7.4. TCR was immobilized on a CM5 sensor chip using standard amine coupling to final response units between 700–3000. Peptide/MHC complexes were injected at a flow rate of 10 μL/min until steady-state was reached. The concentration range of Peptide/MHC complexes spanned from 13 nM to 300 μM. The signal over the final 10 seconds of injection were averaged and subtracted from identical injections over a mock surface. Each injection was performed in duplicate and fit simultaneously, using global analysis to enhance accuracy and precision (Blevins & Baker, 2017). Data were processed using BiaEvaluation 4.0 and fit with Origin 2017 using a 1:1 binding model. Six experiments were performed for SILv44 and R6C12 and five experiments for T4H2.
Modeling of TCR-peptide/MHC complexes
TCR-peptide/MHC structural models were constructed using a template-based approach described recently (Riley et al., 2016). Briefly, sequences for R6C12, SILv44, and T4H2 were aligned and compared to a panel of HLA-A2 restricted TCRs with known TCR-peptide/MHC structures to serve as model templates. A template TCR was selected if the TCR alignment indicated strong sequence similarity and/or minor loop length changes. The DMF5-MART-1/HLA-A2 TCR-peptide/MHC complex (Borbulevych, Santhanagopolan, Hossain, & Baker, 2011) was selected as the template for the R6C12 and T4H2 models and the B7-Tax/HLA-A2 complex (Ding et al., 1998) was chosen for SILv44. Using PyRosetta, a python toolkit for the Rosetta protein design suite (Chaudhury, Lyskov, & Gray, 2010; Kaufmann, Lemmon, Deluca, Sheehan, & Meiler, 2010), the given TCR sequences and peptides were mapped onto the three-dimensional coordinates of the template TCRs and peptides in the TCR-peptide/MHC complexes. Repacking the amino acid sidechains and an energetic minimization of the CDR loops/peptides generated initial models of the target TCRs. Further design work performed in Rosetta followed a steepest descent design where many independent decoy structures were generated for each modeling stage. Each model underwent one stage for low resolution docking, one stage for high resolution docking, and multiple stages for CDR loop modeling. Using a previously described energy scoring function (Leaver-Fay et al., 2013), the lowest scoring decoys from each stage were chosen for the next step. Following generation of an initial TCR-peptide/MHC model, 10,000 decoys were generated with fully randomized peptide/MHC and TCR docking orientations coupled with a low resolution rigid body energy minimization move. Since many decoys generated in this stage were low scoring, preference was given to structures with crossing angles similar to the template. After the low resolution docking stage, loop randomization and modeling was performed as previously described with generation of 100 decoys for each CDR loop (Mandell, Coutsias, & Kortemme, 2009). The first round of loop modeling was followed by generation of 10,000 decoys with 3 Å, 8° rigid body perturbations and docked in high resolution. The final stages consisted of sequentially modeling each modified CDR loop until Rosetta scores were no longer decreasing between stages. The final model of R6C12 required 20 stages, SILv44 required 18, and T4H2 required only 13 stages due to a high template similarity. Structural analysis was performed with PyMol and Discovery Studio.
Statistics
Student’s t-test was used to compare relevant datasets, using two-sided comparisons for normally distributed data unless otherwise indicated. For in vivo experiments, a generalized estimating equation approach was used for making comparisons of growth rates across groups in a linear regression model with an assumed exchangeable correlation structure to account for repeated measures over time. Due to the non-linear patterns of tumor size by day, we used the square-root of volume as our outcome and day and indicators of group, and interactions between group and the day as our covariates. Robust Z-scores for the interaction terms were used to calculate p-values for comparisons of slopes across groups.
Results
TCR SILv44 preferentially imparts IL-17 responses on host T cells
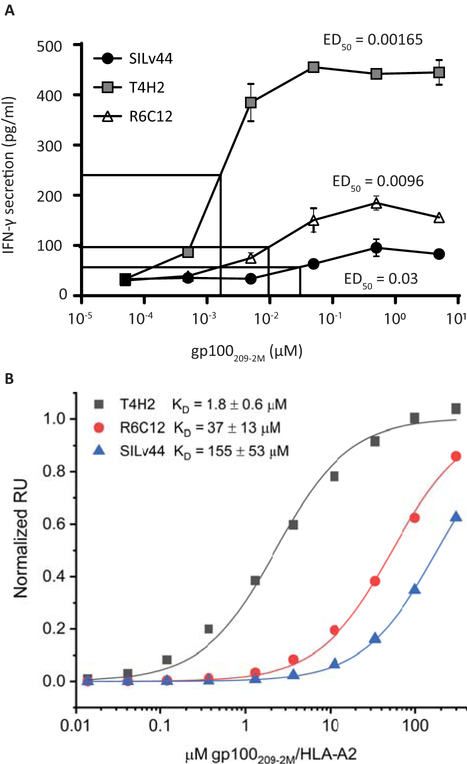
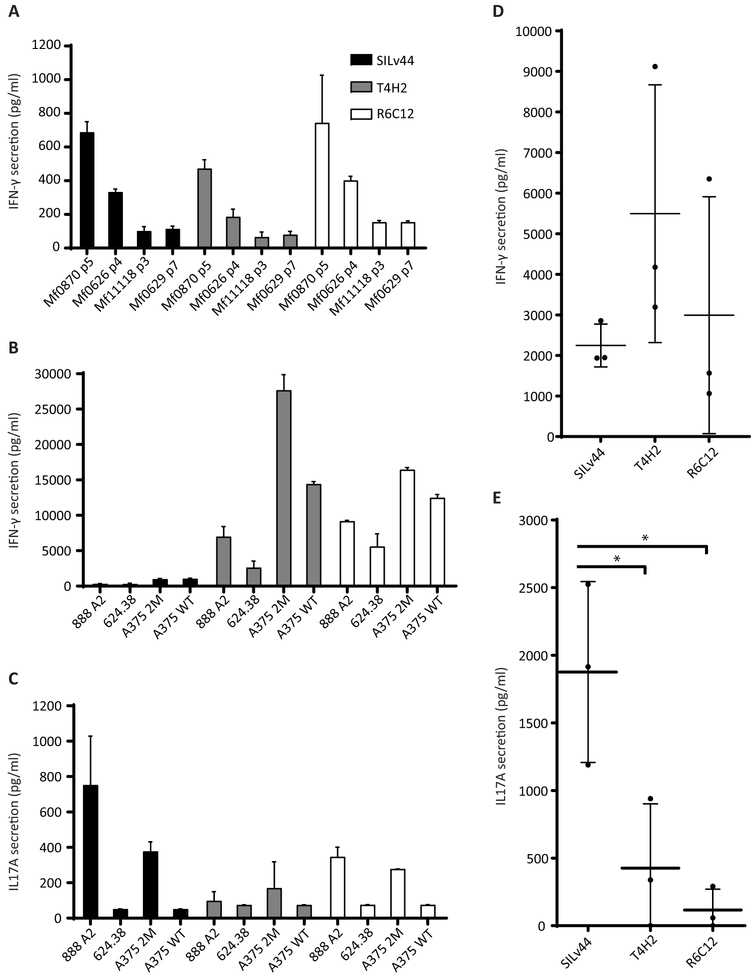
Given the role for IFN-γ in anti-tumor responses (Candeias & Gaipl, 2016), we initially examined secretion by CD8 T cells in response to increasing concentrations of the 2M-modified gp100 peptide with enhanced affinity for HLA-A2. In Figure 1A, the T4H2 TCR clearly displays superior reactivity towards peptide-pulsed T2 cells, with half-maximum IFN-γ secretion detected in response to 1.65 nM peptide as compared to 9.6 nM for R6C12 and 30 nM for SILv44. The ranking of IFN-γ release was generally congruent with TCR binding affinity measured with recombinant protein using surface plasmon resonance (SPF), which yielded Kd values of 1.8 ± 0.6 µM, 37 ± 13 µM and 155 ± 53 µM respectively (Figure 1B). Though the 2M-modified peptide used for the secretion assay matches the treatment provided to patients who gave rise to the T4H2 and R6C12 TCRs, the host rendering the SILv44 receptor had not been exposed to the 2M-modified peptide. The next step thus entailed a comparison of responses to natural peptide as processed and presented by melanocytes and melanoma cells. In response to melanocytes, we observed similar levels of secretion for the three TCRs (Figure 2A). We then included 624.38, 888-A2+, and A375 melanoma target cells pulsed with either the wild type or 2M-modified peptide (Figure 2B). Though no significant differences in IFN-γ were observed in response to the natural peptide, both T4H2 and R6C12 did trend towards a greater response to the 2M-modified peptide, consistent with the more potent immunogenicity of the 2M peptide owing to its stronger HLA-A2 binding affinity (Borbulevych, Baxter, Yu, Restifo, & Baker, 2005; Z. Yu et al., 2004). As IL-17 has also been implicated in anti-melanoma responses (Majchrzak et al., 2016), we next assessed IL-17 secretion. A TCR-dependent increase in IL-17 secretion was also observed in response to 888-A2+ melanoma cells (Figure 2C). To ensure that enhanced IL-17 secretion was not dependent on the donor T cells, the experiment was repeated with T cells from 3 different donors, showing that a trend toward preferential IFN-γ secretion by T4H2 transduced T cells (Figure 2D) was paralleled by a significantly stronger preference for IL-17 secretion among SILv44 transduced T cells (Figure 2E). Importantly, these findings clearly suggest that the cytokine profiles displayed in response to naturally processed antigen are defined by the T cell receptor, with enhanced IL-17 secretion resulting from the TCR with the weakest binding affinity to the 2M peptide.
Fig. 1.
T4H2 transduced T cells produce superior IFNγ responses to gp100209–2M modified peptide in concert with TCR binding affinity. (A) 50,000 CD8 T cells transduced to express the SILv44, T4H2 or R6C12 TCRs were combined 1:1 with peptide-pulsed T2 target cells for 20 hours. IFN-γ levels were measured in the supernatant, with ED50 T4H2<R6C12<SILv44-transduced cells. (B) Binding of T4H2, R6C12, and SILv44 TCRs to gp100209–2M/HLA-A2 measured by SPR. Affinities in terms of KD values are shown in the inset; values are averages and standard deviations of six independent measurements for SILv44 and R6C12 and five independent measurements for T4H2.
Fig. 2.
Naturally processed peptide elicits an IL17A response from SILv44-tranduced T cells. (A) IFN-γ secretion in response to HLA-A2+ melanocytes Mf0870 and Mf0626 does not differ significantly among PBMCs transduced to express the 3 TCRs. (B) Expression of T4H2 and R6C12 receptors trend towards greater IFN-γ secretion in response to melanoma cells. (C) IL-17A secretion, however, may be greater by SILv44-transduced T cells. (D) The trend observed in B is confirmed when comparing transduced T cells from 3 different donors responding to 888-A2+ cells. (E) Significantly greater IL-17A production observed among TCR transduced T cells from another 3 different donors responding to 888-A2+ cells.
TCR dependent differences in cytokine profiles are fine-tuned by TCR stimulation
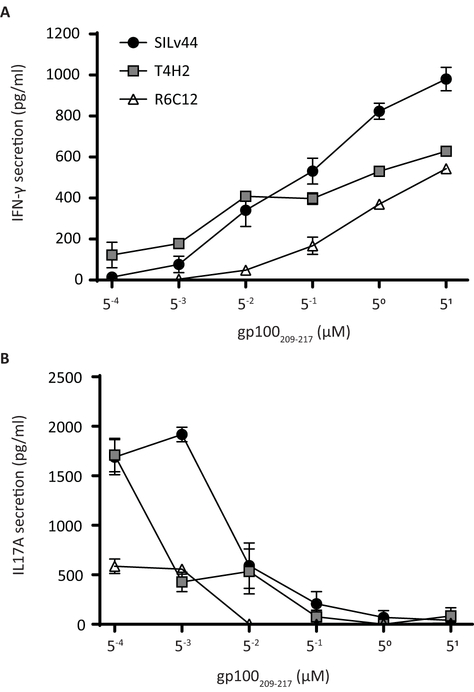
We next considered whether differences in T cell avidity driven either by TCR cell surface levels or by TCR affinity for the peptide-MHC complex could contribute to the cytokine production differences. When TCR Vβ expression of transduced T cells was measured for each TCR in a representative experiment and compared to expression by untransduced T cells, transductions efficiencies of 60.2% (SILv44), 74.9% (T4H2) and 69% (R6C12) were observed, though percentages varied by T cell donor. Importantly, the amount of TCR Vβ staining on successfully transduced cells did not differ significantly for the three TCRs (supplemental Fig. 1). If stronger TCR binding to the peptide/MHC complex dictates enhanced signaling and preferential secretion of IFN-γ, the data from Figures 1 and 2 would suggest that IL-17 secretion is differently governed in this interaction and therefore that TCR affinity may not fully reflect resulting T cell responses. Interestingly, the only difference between the 2M-modified and wild type peptides observed thus far is that the modified peptide binds HLA-A2 approximately 10 times more strongly than the wild type peptide (Borbulevych et al., 2005; Z. Yu et al., 2004). In order to assess how changes in peptide levels driven by lower MHC-binding affinities may impact the responses of TCR-transduced or untransduced T cells, we exposed both T cell populations to T2 cells pulsed with the wild type peptide at a wide range of concentrations. While we saw increases in IFN-γ production with increased peptide concentration among TCRs, with SILv44 the most responsive at higher peptides concentrations (Figure 3A), we did not observe any plateauing of the IFN-γ responses. This is in contrast to the response of the TCRs to the modified peptide (Figure 1) and is consistent with the weaker binding of the wild type versus modified peptide in the binding groove (Borbulevych et al., 2005; Z. Yu et al., 2004). In contrast to IFN-γ production, we observed decreasing IL-17A production with increasing peptide concentration among all TCRs (Figure 3B). Furthermore, subtle differences among the TCRs appear to show that in response to low concentrations comparable to those of naturally processed antigen, specifically at the 0.5nM range, the SILv44 TCR drives relatively more IL-17 secretion than the other TCRs whereas T4H2 preferentially drives IFN-γ secretion. Moreover, a broader cytokine profile by SILv44 transduced T cells was observed in response to 888-A2+ target cells when compared to T4H2 and R6C12-transduced T cells in a cytokine bead array (Supplemental figure 2). Taken together, these data suggest that the functional response of the T cells may be dictated by features beyond both antigen specificity and affinity of the peptide in the binding groove, such as differences in the TCR variable region.
Fig. 3.
IL-17A secretion increases in response to decreasing wild type peptide concentrations. Target cells, pulsed with increasing concentrations of wild type gp100 peptide, were incubated at a 1:1 ratio with peripheral blood mononuclear cells transduced with one of the SILv44, T4H2 or R6C12 TCRs. At concentrations lower than 5 nM gp100209–217 peptide, (A) IFN-γ concentrations are gradually outperformed by (B) IL-17A secretion, revealing a lower optimum peptide concentration for the inflammatory cytokine.
IL-17A-producing SILv44-tranduced T cells demonstrate superior in vivo tumor control
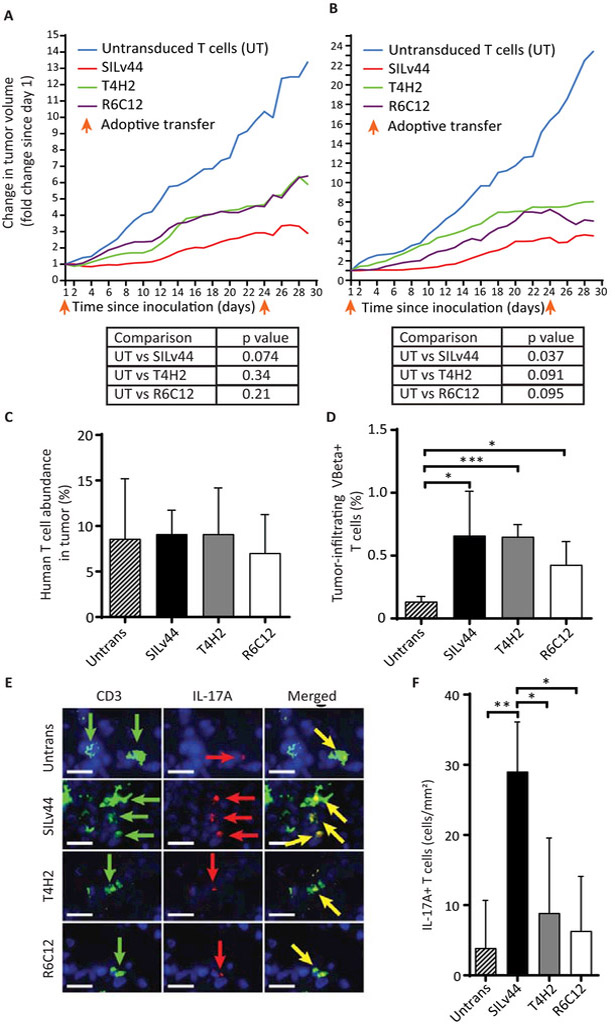
We next asked whether the distinct cytokine responses observed among the TCRs would translate to differential tumor control in vivo. We therefore performed adoptive transfer experiments in SCID/beige mice challenged with human 888-A2+ tumor cells. Here, transduced human lymphocytes were selected for SILv44, T4H2 or R6C12 expression based on gentamycin resistance and rapidly expanded. TCR expression was confirmed by flow cytometry before injecting equal numbers of TCR-transduced human T cells, or a corresponding number of untransduced T cells, into each mouse at days 35 and 58 post tumor challenge. To account for irrelevant pre-treatment differences in tumor size, the average fold change in tumor size relative to treatment day 1 is depicted; results are shown for two separate experiments to account for inter-experimental variation (Figure 4A, B). Very similar growth curves were observed in both experiments, and significant inhibition of tumor growth was observed only for SILv44 transduced T cells compared to mice receiving untransduced T cells as listed underneath the growth curves. Moreover, tumors treated with SILv44 expressing T cells revealed marked necrosis (data not shown), suggesting that this TCR affected tumor cell viability to a greater extent than is reflected by tumor sizes alone. However, when we compared T cell infiltration and transgenic TCR Vβ expression (Figure 4C, D), we observed no differences among groups, suggesting that the quality of the cellular response rather than the quantity of intratumoral antigen-specific T cells may be responsible for differences in anti-tumor activity. Immunofluorescent staining subsequently revealed higher levels of IL-17A+ T cells in the tumors of mice that received SILv44-transduced T cells compared to those that received either T4H2, R6C12 or untransduced T cells demonstrating that IL-17A+ T cell density in tumors tracked with greater anti-tumor responses in vivo (Figure 4E) and that IFN-γ expression alone may not correspond to superior tumor control.
Fig. 4.
TCR-transduced T cells differentially contain melanoma tumors. (A) SCID/beige mice were subcutaneously injected with 106 888-A2+ human melanoma tumor cells at three mice per group and allowed to firmly establish tumors over three weeks, before 2×106 human T cells were adoptively transferred twice with a 23 day interval. Tumor growth was significantly contained only by SILv44 transduced T cells. (B) The experiment was repeated at five mice per group and again, only the SILv44 TCR transduced T cells significantly contained tumor growth. (C) Intratumoral T cells were equally abundant at euthanasia regardless of the expressed TCR, showing that it was T cell function rather than T cell number holding tumors in check. (D) Transgenic TCR expression (as % of tumor infiltrating T cells) was limited to transduced T cell groups as determined using antibodies to the TCRβ subunit. (E) An increased frequency of IL-17A+ T cells was found in tumors infiltrated by SILv44 expressing T cells. (F) Immunohistology was performed and quantified on multiple sections of two tissue samples per group. Scale bars represent 17µm. *P<0.05, ** P<0.1, *** P<0.001.
Structural models of the TCR-peptide/MHC complexes predict different TCR binding architectures
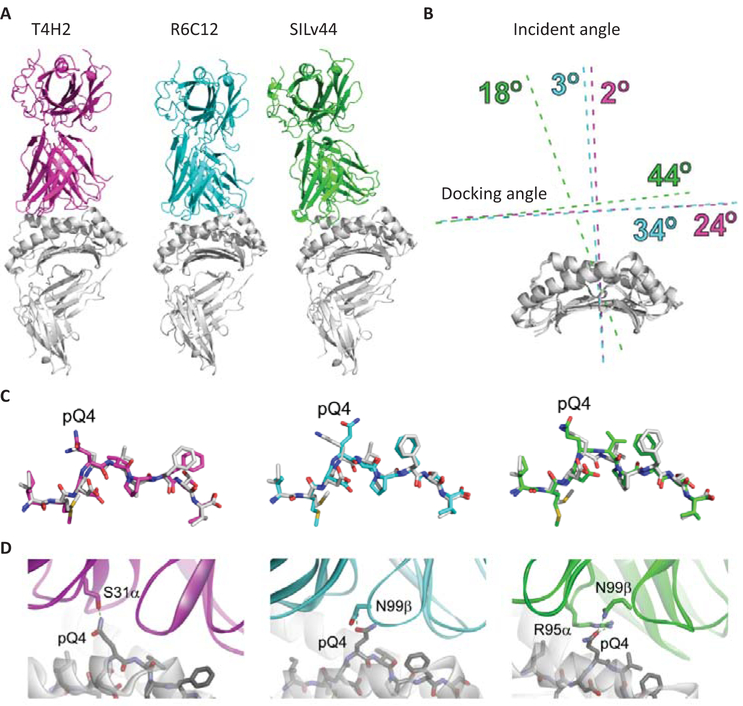
To explore possible underpinnings of the differential cytokine expression and associated outcomes with the three TCRs, and SILv44 in particular, we generated models of the three TCR-peptide/MHC complexes using a template-based approach that combines loop and peptide modeling with low and high resolution docking procedures. We previously demonstrated this modeling procedure can reproduce key structural features such as interface contacts and TCR docking modes (Riley et al., 2016). After identifying appropriate template structures for T4H2, R6C12, and SILv44, the TCRs and peptides were computationally morphed and several rounds of structural and energetic refinement were applied. The complexes were subsequently subjected to multiple rounds of randomized low resolution docking, high resolution interface refinement, and restrained high resolution docking. An overview of the resulting structural models is shown in Figure 5A. The most striking observation is the altered mode of engagement predicted for SILv44 compared to T4H2 and R6C12. As summarized in Figure 5B, although all three TCRs have similar crossing angles over the gp100209–2M/HLA-A2 complex (24°−44°, within the range of TCR complexes with class I proteins) (Rudolph, Stanfield, & Wilson, 2006; Wucherpfennig, Call, Deng, & Mariuzza, 2009), the model of the SILv44 complex has a substantially different incident angle (18° vs. 2–3°), resulting in a more ‘tilted’ complex. Beyond the gross topological difference, this altered mode of engagement impacts overall interface features. We focused on the central p4 glutamine of the gp100209–2M peptide, which in the unbound gp100209–2M/HLA-A2 complex protrudes up from the peptide backbone (Borbulevych et al., 2005; Riley et al., 2016). Although the peptide conformation in each complex is predicted to be similar (Figure 5C), the three TCRs engage pGln4 differently (Figure 5D). Whereas SILv44 uses a positive charge from Arg95 in CDR3α, both T4H2 and R6C12 use only non-charged side chains to engage pGln4. Burial of positive charges is often unfavorable, even when hydrogen bonds or salt bridges are formed (Hendsch & Tidor, 1994), and this could contribute to the weaker affinity of SILv44 towards the 2M antigen. Lastly, we observed that in the R6C12 complex, pGln4 is engaged by Asn99 of CDR3β (Fig. 5D). This is consistent with alanine scanning mutagenesis of this TCR, which revealed a greater role for CDR3β than CDR3α in ligand binding, and a role for Asn99β in particular (Malecek et al., 2013). Thus, in silico modeling of the interaction of our TCRs with their cognate peptide predicted a different conformation for the combination that offered superior anti-tumor responses in vivo.
Fig. 5.
Structural models of the T4H2, R6C12, and SILv44 TCRs with gp100209–2M/HLA-A2. (A) Overview of the complexes. In the three models, the T4H2 complex is magenta, R6C12 is cyan, and SILv44 is green (this coloring is maintained for all panels). The orientation of the peptide/MHC is the same in all three images. The altered binding mode predicted for SILv44 is clearly apparent. (B) Quantification of TCR binding modes for the three complexes. The primary difference is in the incident angle, which is 18° off the vertical for SILv44, but 3° for R6C12 and 2° for T4H2. (C) Conformations of the modeled peptides in the three TCR-peptide/MHC models, compared to the conformation of the peptide in the unbound gp100209–2M/HLA-A2 crystal structure (white). The overall conformations are very similar, except for that with R6C12, in which pGln4 is predicted to be positioned more centrally in the TCR-peptide/MHC interface. (D) Engagement of pGln4 in the three TCR-peptide/MHC models.
Discussion
Using in vitro, in vivo and in silico approaches, we performed in-depth characterization of T cells, transduced to express TCRs that respond to the same peptide-MHC complex but display distinct variable regions. Strikingly, we observed markedly different functional profiles among the TCRs, with the most potent in vivo anti-tumor responses produced by the SILv44 TCR and associated with the elevated production of IL-17A rather than IFN-γ. Indeed, despite demonstrating the weakest binding affinity towards the 2M antigen among the three TCRs, SILv44 showed a remarkable ability to produce superior levels of IL-17A and a broader cytokine profile in response to sub nanomolar concentrations of peptide and appears to have a unique binding conformation as predicted by computational models. These data showed that beyond antigen specificity and affinity, other TCR molecular properties may be important in determining the functional response of the T cells, in this case promoting a potent IL-17A-producing phenotype that delivers superior tumor control in a model of melanoma.
As the main difference among the TCRs is the variable region and the antigen recognition properties it confers, it would appear that the interaction between the TCR and the peptide-MHC complex is a driver that defines the cytokine profile exhibited by each transgenic T cell population. This may follow similar principles as those involved in thymic selection of T cells, whereby the affinity of TCRs for peptide-MHC complexes presented by epithelial cells in the thymus provides a key component of positive and negative selection (Nitta, Murata, Ueno, Tanaka, & Takahama, 2008). In this setting, the recognition of weak TCR signals in the thymic cortex results in positive selection whereas strong TCR signals in the medulla lead to deletion to protect against self-reactive T cell responses. This elegant process delivers a pool of functional T cells that possess the ability to recognize a range of signals including those from weak, low affinity peptide-MHC complexes. Thus, the type of T cell that results from antigen recognition in the thymus is dependent on the strength of the TCR interaction with the peptide-MHC complex.
Interestingly, we found that host T cells responded with greater IL-17A secretion in response to lower peptide concentrations whereas IFN-γ secretion increased at higher peptide concentrations within the physiologic range (as deduced from the amounts generated in response to melanocytes and melanoma cells). The relative role of TCR strength and ligand availability for T cells responsiveness is an open question (Persaud, Parker, Lo, Weber, & Allen, 2014) and, based on the present data, we speculate that a greater number of TCR molecules engaged in the synapse preferentially drive Tc1 responses, whereas limited availability of the MHC-peptide complex would correspond to fewer TCR molecules participating, driving a Tc17 phenotype and allowing T cell responses in environments that display a range of antigen availability. A driving force behind T cell subset development is found in Notch stimulation, as we have reported for the role of Notch-1 in IL-17+ T cell development (Keerthivasan et al., 2011), while others view Notch as an amplifier of existing differentiation signals, including IFN-γ (Bailis et al., 2013). As Notch-1 will also drive a Th1 response, there is similarity between both T cell subsets, explaining in part how IL-17A and IFN-γ expression can co-exist in a plastic T cell population that can convert into single positive T cells dependent on the microenvironment (Geginat et al., 2016). Indeed, a distinct subset of Tc17 cells capable of producing IFN-γ has been demonstrated in humans (Dusseaux et al., 2011) consistent with the plasticity observed among the SILv44 Tc17 population here. Furthermore, the Tc17 population has been reported to display a long-lived effector phenotype compared to Tc1 (Y. Yu et al., 2013). It is therefore plausible that the presence of a long-lived population of Tc17 cells with the plasticity to convert to Tc1 cells as warranted may account for the superior protective ability of the SILv44 receptor-transduced T cell population in our studies. Moreover, our in vivo data suggest that the gp100 antigen is expressed in the tumor at a level that selects for a strong IL-17A response and weaker IFN-γ response by SILv44-transduced T cells. Indeed, at the wild type peptide concentration at which we observe this in vitro, IFN-γ fails to distinguish the TCRs, supporting the conclusions that IFN-γ may be insufficient to identify the most potent T cell responses and that other cytokines including, but not limited to IL-17A, may play a key role in tumor control.
Given the question of how differences in the TCR variable regions might influence downstream T cell signaling and cytokine responses, we turned to TCR in silico modeling, which predicted gross architectural differences between the complex formed by the SILv44 TCR and the R6C12/T4H2 TCRs with the 2M-modified peptide, whose conformation matches that of the wild type peptide in the binding groove of the HLA-A2 molecule (Borbulevych et al., 2005; Z. Yu et al., 2004). TCR binding geometries impact T cell function (Adams et al., 2011; Gras et al., 2016). Thus, differences in the binding modes of the TCRs might influence the assembly of a TCR signaling complex, including CD8 and/or CD3 geometries and/or their binding affinities. This should impact signaling and contribute differential cytokine profiles, potentially driven by different TCR binding affinities towards antigen. The crystal structures of each TCR-peptide/MHC complex could confirm the nature of differences predicted by in silico modeling, and further work will be required to assess actual roles of such differences in host T cell reactivity.
Overall we demonstrate that, inconsistent with traditional expectations from studies relating TCR binding to IFN-γ cytokine production, SILv44 elicited the most potent anti-tumor response, unexpectedly eliciting a Tc17 phenotype and capable of inducing broad cytokine responses, rather than mediating a classical anti-tumor IFN-γ-centric profile. Yet structural modeling of the TCR-peptide-MHC complex suggested a different conformational profile for SILv44 that could support a link between binding conformation and differential cytokine profiles. In conclusion, it is clear that properties of TCRs responsible for interacting with the peptide-MHC complex play a previously underappreciated role in defining the phenotype of host T cells, and may define the therapeutic outcome of adoptive T cell transfer beyond supporting the specific recognition of antigen alone.
Supplementary Material
Significance.
T cell receptor transgenic therapies are in clinical trials for malignant melanoma. Currently, TCRs for such therapies are selected based primarily on binding affinity to the peptide/MHC complex and the resultant production of IFN-γ by transduced T cells. Our data provide evidence that identification of the most potent TCRs in tumor control may lie beyond these features and that by widening the selection criteria to include non-conventional cytokines such as IL-17A, we may be able to more successfully identify the most potent TCRs for future immunotherapies.
Acknowledgments
These studies were supported by R01CA109536, R01CA191317 and a LAM Foundation Established Investigator Award to CLP, PO1CA154778 to MN, and NIGMS R35GM118166 and R01AI29543 to BMB. TPR was supported by a fellowship from the Indiana CTSI, funded in part by NIH grant UL1TR001108. ARS was supported by a fellowship from the Walther Cancer Foundation Interdisciplinary Interface Training Project. We acknowledge excellent help from the Loyola FACS core (Pat Simms), tissue donations from patients and helpful advice and materials from Dr. Shikhar Mehrotra, Gina Scurti and Aravind Seetharaman.
Footnotes
Conflict of interest
The authors have no conflict of interest to declare.
References
- Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, . . . Garcia KC (2011). T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity, 35(5), 681–693. doi: 10.1016/j.immuni.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataca P, & Arslan O(2015). Chimeric Antigen Receptor T Cell Therapy in Hematology. Turk J Haematol, 32(4), 285–294. doi: 10.4274/tjh.2015.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis W, Yashiro-Ohtani Y, Fang TC, Hatton RD, Weaver CT, Artis D, & Pear WS (2013). Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity, 39(1), 148–159. doi: 10.1016/j.immuni.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins SJ, & Baker BM (2017). Using Global Analysis to Extend the Accuracy and Precision of Binding Measurements with T cell Receptors and Their Peptide/MHC Ligands. Front Mol Biosci, 4, 2. doi: 10.3389/fmolb.2017.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbulevych OY, Baxter TK, Yu Z, Restifo NP, & Baker BM (2005). Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J Immunol, 174(8), 4812–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbulevych OY, Santhanagopolan SM, Hossain M, & Baker BM (2011). TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol, 187(5), 2453–2463. doi: 10.4049/jimmunol.1101268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias SM, & Gaipl US (2016). The Immune System in Cancer Prevention, Development and Therapy. Anticancer Agents Med Chem, 16(1), 101–107. [DOI] [PubMed] [Google Scholar]
- Chaudhury S, Lyskov S, & Gray JJ (2010). PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics, 26(5), 689–691. doi: 10.1093/bioinformatics/btq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodon T, Comin-Anduix B, Chmielowski B, Koya RC, Wu Z, Auerbach M, . . . Ribas A (2014). Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin Cancer Res, 20(9), 2457–2465. doi: 10.1158/1078-0432.Ccr-13-3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Harrison RL, Armstrong KM, & Baker BM (2005). Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol, 346(2), 533–550. doi: 10.1016/j.jmb.2004.11.063 [DOI] [PubMed] [Google Scholar]
- Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, & Wiley DC (1998). Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity, 8(4), 403–411. [DOI] [PubMed] [Google Scholar]
- Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, . . . Lantz O (2011). Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood, 117(4), 1250–1259. doi: 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Duval L, Schmidt H, Kaltoft K, Fode K, Jensen JJ, Sorensen SM, . . . von der Maase H (2006). Adoptive transfer of allogeneic cytotoxic T lymphocytes equipped with a HLA-A2 restricted MART-1 T-cell receptor: a phase I trial in metastatic melanoma. Clin Cancer Res, 12(4), 1229–1236. doi: 10.1158/1078-0432.Ccr-05-1485 [DOI] [PubMed] [Google Scholar]
- Geginat J, Paroni M, Kastirr I, Larghi P, Pagani M, & Abrignani S (2016). Reverse plasticity: TGF-beta and IL-6 induce Th1-to-Th17-cell transdifferentiation in the gut. Eur J Immunol, 46(10), 2306–2310. doi: 10.1002/eji.201646618 [DOI] [PubMed] [Google Scholar]
- Golubovskaya V, Berahovich R, Xu S, Harto H, & Wu L (2017). Major Highlights of the CAR-TCR Summit, Boston, 2016. Anticancer Agents Med Chem, 17(10), 1344–1350. doi: 10.2174/1871520617666170110151900 [DOI] [PubMed] [Google Scholar]
- Gras S, Chadderton J, Del Campo CM, Farenc C, Wiede F, Josephs TM, . . . La Gruta NL (2016). Reversed T Cell Receptor Docking on a Major Histocompatibility Class I Complex Limits Involvement in the Immune Response. Immunity, 45(4), 749–760. doi: 10.1016/j.immuni.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Hebeisen M, Allard M, Gannon PO, Schmidt J, Speiser DE, & Rufer N (2015). Identifying Individual T Cell Receptors of Optimal Avidity for Tumor Antigens. Front Immunol, 6, 582. doi: 10.3389/fimmu.2015.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendsch ZS, & Tidor B (1994). Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci, 3(2), 211–226. doi: 10.1002/pro.5560030206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, & Meiler J (2010). Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry, 49(14), 2987–2998. doi: 10.1021/bi902153g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthivasan S, Suleiman R, Lawlor R, Roderick J, Bates T, Minter L, . . . Osborne BA (2011). Notch signaling regulates mouse and human Th17 differentiation. J Immunol, 187(2), 692–701. doi: 10.4049/jimmunol.1003658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarquist J, Eby JM, Henning SW, Li M, Wainwright DA, Westerhof W, . . . Le Poole IC (2016). Functional cloning of a gp100-reactive T-cell receptor from vitiligo patient skin. Pigment Cell Melanoma Res, 29(3), 379–384. doi: 10.1111/pcmr.12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, O’Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, . . . Kuhlman B (2013). Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol, 523, 109–143. doi: 10.1016/b978-0-12-394292-0.00006-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majchrzak K, Nelson MH, Bailey SR, Bowers JS, Yu XZ, Rubinstein MP, . . . Paulos CM (2016). Exploiting IL-17-producing CD4+ and CD8+ T cells to improve cancer immunotherapy in the clinic. Cancer Immunol Immunother, 65(3), 247–259. doi: 10.1007/s00262-016-1797-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecek K, Zhong S, McGary K, Yu C, Huang K, Johnson LA, . . . Krogsgaard M (2013). Engineering improved T cell receptors using an alanine-scan guided T cell display selection system. J Immunol Methods, 392(1–2), 1–11. doi: 10.1016/j.jim.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Coutsias EA, & Kortemme T (2009). Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods, 6(8), 551–552. doi: 10.1038/nmeth0809-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Liu Z, Rangelova E, Poiret T, Ambati A, Rane L, . . . Maeurer MJ (2016). Expansion of Tumor-reactive T Cells From Patients With Pancreatic Cancer. J Immunother, 39(2), 81–89. doi: 10.1097/cji.0000000000000111 [DOI] [PubMed] [Google Scholar]
- Moore TV, Lyons GE, Brasic N, Roszkowski JJ, Voelkl S, Mackensen A, . . . Nishimura MI (2009). Relationship between CD8-dependent antigen recognition, T cell functional avidity, and tumor cell recognition. Cancer Immunol Immunother, 58(5), 719–728. doi: 10.1007/s00262-008-0594-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MH, Kundimi S, Bowers JS, Rogers CE, Huff LW, Schwartz KM, . . . Paulos CM (2015). The inducible costimulator augments Tc17 cell responses to self and tumor tissue. J Immunol, 194(4), 1737–1747. doi: 10.4049/jimmunol.1401082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Murata S, Ueno T, Tanaka K, & Takahama Y (2008). Thymic microenvironments for T-cell repertoire formation. Adv Immunol, 99, 59–94. doi: 10.1016/s0065-2776(08)00603-2 [DOI] [PubMed] [Google Scholar]
- Persaud SP, Parker CR, Lo WL, Weber KS, & Allen PM (2014). Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol, 15(3), 266–274. doi: 10.1038/ni.2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley TP, Ayres CM, Hellman LM, Singh NK, Cosiano M, Cimons JM, . . . Baker BM (2016). A generalized framework for computational design and mutational scanning of T-cell receptor binding interfaces. Protein Eng Des Sel, 29(12), 595–606. doi: 10.1093/protein/gzw050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoltini L, Barracchini KC, Viggiano V, Kawakami Y, Smith A, Mixon A, . . . et al. (1995). Quantitative correlation between HLA class I allele expression and recognition of melanoma cells by antigen-specific cytotoxic T lymphocytes. Cancer Res, 55(14), 3149–3157. [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, & Wilson IA (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol, 24, 419–466. doi: 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Smith SN, Sommermeyer D, Piepenbrink KH, Blevins SJ, Bernhard H, Uckert W, . . . Kranz DM (2013). Plasticity in the contribution of T cell receptor variable region residues to binding of peptide-HLA-A2 complexes. J Mol Biol, 425(22), 4496–4507. doi: 10.1016/j.jmb.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Panhuys N (2016). TCR Signal Strength Alters T-DC Activation and Interaction Times and Directs the Outcome of Differentiation. Front Immunol, 7, 6. doi: 10.3389/fimmu.2016.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Call MJ, Deng L, & Mariuzza R (2009). Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol, 21(6), 590–595. doi: 10.1016/j.coi.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Cho HI, Wang D, Kaosaard K, Anasetti C, Celis E, & Yu XZ (2013). Adoptive transfer of Tc1 or Tc17 cells elicits antitumor immunity against established melanoma through distinct mechanisms. J Immunol, 190(4), 1873–1881. doi: 10.4049/jimmunol.1201989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, . . . Restifo NP (2004). Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. J Clin Invest, 114(4), 551–559. doi: 10.1172/jci21695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Yang JC, Kawakami Y, Spiess P, Wadsworth SC, Cardoza LM, . . . Rosenberg SA (1996). Antigen-specific tumor vaccines. Development and characterization of recombinant adenoviruses encoding MART1 or gp100 for cancer therapy. J Immunol, 156(2), 700–710. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.