Abstract
Osteoporosis is a condition characterized by low bone mineral density and an increased risk of fracture. Traits contributing to osteoporotic fracture are highly heritable, indicating that a comprehensive understanding of bone requires a thorough understanding of the genetic basis of bone traits. Towards this goal, genome-wide association studies (GWASs) have identified over 500 loci associated with bone traits. However, few of the responsible genes have been identified, and little is known of how these genes work together to influence systems-level bone function. In this review, we describe how systems genetics approaches can be used to fill these knowledge gaps.
Keywords: Systems genetics, osteoporosis, genome-wide association study (GWAS), co-expression networks, bone mineral density (BMD), Bayesian networks
The Skeleton as a Dynamic System
The human skeleton is a dynamic, adaptive, and complex system impacting a wide array of physiological processes. It provides support and protection, enables locomotion, maintains hematopoiesis, serves as a reservoir for calcium and phosphorus, and has important endocrine functions [1–3]. Diseases of bone, however, inhibit the ability of the skeleton to carry out these functions. The most common disease of bone is osteoporosis (see Glossary), a condition of low bone mineral density (BMD) and an increased risk of fracture [4]. Osteoporosis affects over 12 million individuals in the U.S. and over 200 million worldwide [5]. Osteoporotic fractures are a serious clinical outcome associated with increased morbidity and mortality, particularly in the elderly. In fact, of the ~300,000 people in the U.S. over the age of 50 that suffer a hip fracture annually, 1 in 5 will die in the subsequent 12 months, and half of the survivors will not return to their prior independent living status [6]. Alarmingly, the incidence of fractures is expected to rise by 50% over the next decade, as the number of individuals over the age of 50 increases [7].
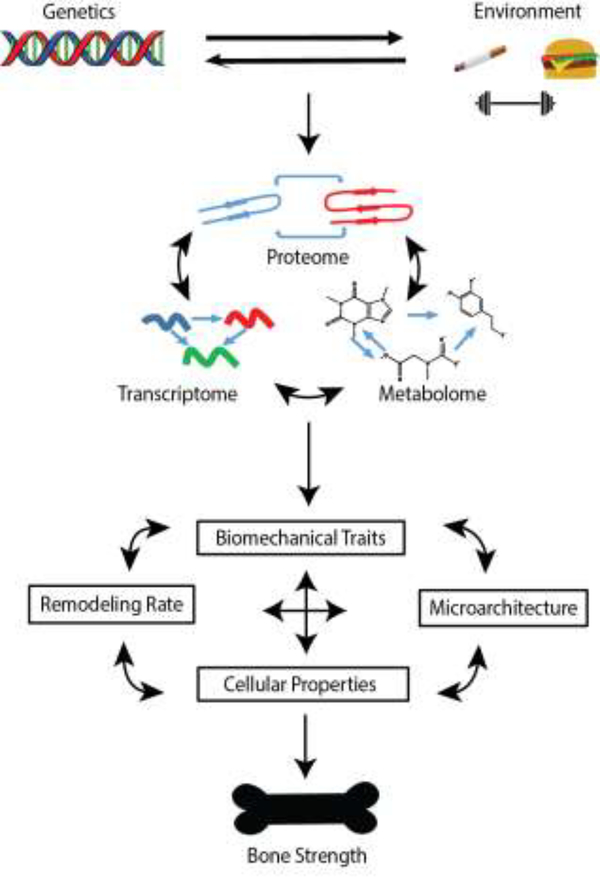
One of the hallmarks of quantitative traits related to osteoporosis (BMD, bone size, etc.) is their high heritability (h2=0.5 to 0.8) [8]. As a result, the development of a comprehensive understanding of bone biology necessitates a thorough understanding of the genetic factors underlying variation in bone traits. This not only includes defining the individual variants and genes contributing to osteoporosis, but also how they interact to impact molecular networks and systems-level function. Here, we discuss how systems genetics approaches are being used to accomplish these goals (Figure 1, Key Figure).
Figure 1, Key Figure. Overview of Systems Genetics.
Genetic and environmental factors, as well as their interactions, influence complex bone traits, such as bone strength. The influence of these factors is mediated through impacts on molecular intermediates (transcriptomes, proteomes, metabolomes, etc.) and can be assayed via appropriate “omics” techniques. Biological information is propagated through complex molecular networks to affect bones traits and ultimately the risk of fracture.
Current State of Osteoporosis Genetics
The genetic analysis of osteoporosis began in the early 1990s with candidate gene studies describing associations between polymorphisms in bone-relevant genes (e.g. vitamin D receptor and type I collagen) and BMD [9]. This was followed by a plethora of additional candidate gene investigations and linkage scans in families [10]. In retrospect, little information was gained from either approach [11,12]. In 2007, the tide began to turn with the first of many genome-wide association studies (GWASs) of BMD[13]. In a BMD GWAS, the genotypes of millions of single nucleotide polymorphisms (SNPs) across the genome are tested for an association with BMD in thousands, now often hundreds of thousands, of individuals [14]. To date, over 20 primary GWAS and GWAS meta-analyses have identified hundreds of associations for BMD [15–18]. The largest GWAS to date analyzed estimated BMD at the heel in 426,824 individuals and identified 1,103 independent genome-wide significant associations in 518 loci (Table 1) [19]. BMD has been the primary target of GWASs, mainly because of its strong association with fracture, high heritability, and relative ease of assessment in very large cohorts [20]. However, other traits such as bone size, bone geometry, and serum bone remodeling markers have been interrogated by GWAS [21–24]. Together, these studies have reinforced the importance of known genes and pathways (RANK-RANKL, WNT signaling, etc.) in human bone biology. More importantly, GWAS has provided a “treasure trove” of loci containing only novel genes with the potential to revolutionize our understanding of the genetics, and more importantly, the biology of bone.
Table 1.
Examples of large-scale GWAS and GWAS meta-analyses for BMD illustrates the increase in identified loci as a function of sample size.
| Study | Phenotype | Sample size | Association count |
|---|---|---|---|
| Morris et al. (2018)[19] | Estimated heel BMD | 426,824 | 1103 independent associations (518 loci, 301 novel) |
| Kemp et al. (2017)[17] | Estimated heel BMD | 142,487 | 307 independent associations (203 loci, 153 novel) |
| Estrada et al. (2012) (meta-analysis) [16] | Lumbar spine and femoral neck BMD | 83,894 (32,961 discovery, 50,933 replication) | 64 independent associations (56 loci, 32 novel) |
| Rivadeneira et al. (2009) [40] | Lumbar spine and femoral neck BMD | 19,195 | 20 independent associations (20 loci, 13 novel) |
Note that the most recent discovery of 518 loci encompassed nearly all of the previously discovered loci identified in prior studies.
A limitation of current GWASs is that they have yet to fully uncover the genetic architecture of BMD [25]. In the heel eBMD study referenced above, the 518 independent associations explained only 20% of the phenotypic variance in eBMD [17]. These data suggest that BMD is highly polygenic, or even omnigenic (Box 1), and that much of the genetic basis of BMD remains to be discovered. GWASs are ideally suited to identify associations with common variants (minor allele frequencies (MAF) > 1%). Therefore, it is possible that rare variants (MAF < 1%) may explain part of the “missing heritability” [26]. In support of this hypothesis, recent whole genome-sequencing projects have identified rare variants with large effects on BMD [17,27–29]. It will likely require much larger GWASs and rare variant studies to fully dissect the genetic architecture of BMD and other bone traits.
BOX 1. Polygenicity of complex traits.
Recently, an omnigenic model was proposed to explain the genetic architecture of complex traits [81]. This model is essentially a modernization of Fisher’s infinitesimal model [82]. The omnigenic model states that any gene expressed in a disease-relevant cell type is likely genetically associated with the disease. The authors supported this model with data from large-scale GWASs for multiple diseases/traits suggesting that hundreds of thousands of variants spread uniformly across the genome have non-zero genetic effects. They also provide data suggesting that disease heritability is not concentrated in biologically-relevant processes. They postulate that this is due to the fact that genes expressed within a cell are members of a highly interconnected network made up of “core” (genes participating in a biological process directly influencing a disease) and “peripheral” genes (genes connected to core genes to varying extents). In the omnigenic model, peripheral genes influence disease indirectly through subtle network interactions with core genes.
An opposing viewpoint to the classification of genes as “core” or “peripheral” argues that assuming only core genes directly affect disease is underestimating biological complexity [83]. Alternatively, it is possible that highly polygenic, complex diseases are affected by common variants not through their indirect effects on core genes, but by altering complex biological relationships across many genes. Thus, without the distinction between core and peripheral genes, the omnigenic model is no different than a polygenic model and it is evident that complex disease genes work together in complex networks. Thus, the polygenicity of complex disease reinforces the importance of systems genetics approaches.
By any standard, GWASs have been wildly successful at identifying new loci; however, to date this information has done little to increase our understanding of bone biology or disease. Of the hundreds of loci impacting BMD and other bone traits, the genes responsible for nearly all of the associations are unknown. There are many reasons for this knowledge gap, including the fact that most associations are due to non-coding variation, the lack of bone-specific “-omic” resources, and the inherent difficulties in experimentally establishing causality between variants, genes, and traits.
Using Systems Genetics to Inform Bone GWAS
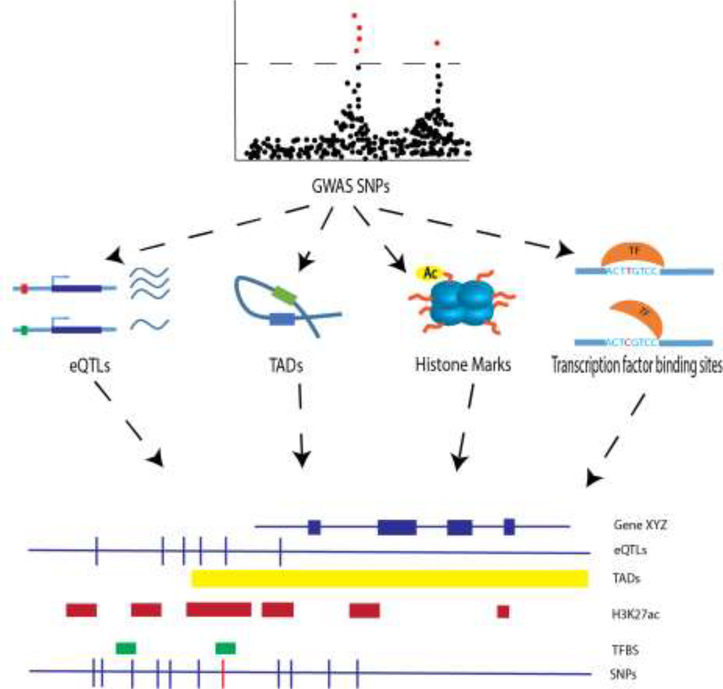
One approach that has the potential to increase our understanding of bone genetics is the emerging field of systems genetics [30,31]. Systems genetics integrates the principles of systems biology with genetics to determine how genetic variation affects molecular phenotypes and cellular networks [30]. In the context of GWAS, systems genetics approaches have proven extremely useful for connecting associated variants with molecular functions (e.g. transcription). The “layering” of different “-omics” datasets (transcriptomics, metabolomics, proteomics, etc.) onto a set of GWAS loci is the most direct way to begin to identify the molecular consequences of disease-associated variants (Figure 2). Most importantly, it also serves to connect disease-associated variants to the genes they regulate.
Figure 2. Systems genetics approaches for prioritizing GWAS data.
By integrating biological data with data from GWAS, SNPs affecting traits can be prioritized for functional follow-up. For example, transcriptomic, biophysical and epigenetic data pertaining to lists of GWAS SNPs can be leveraged in order to prioritize the most likely causal SNPs (red bar, bottom row). TFBS = transcription factor binding sites, H3K27ac = histone H3 lysine 27 acetylation.
One of the most widely used systems genetics approaches for informing GWAS is the identification of expression quantitative trait loci (eQTL) [32]. Just like a clinical trait, GWAS can be used to identify associations for the expression of a gene [33]. These analyses identify sets of genetics variants, or eQTL, that influence transcript levels of any gene expressed in a given cell-type or tissue. There are two types of eQTL, local and distant [34]. Local eQTL influence the transcript levels of genes in close proximity; whereas distant eQTL influence gene expression over a long genomic distance. The identification of eQTL is a logical follow-up to a GWAS, given that the vast majority of GWAS loci are due to non-coding variants that presumably have a role in gene regulation. A typical analysis consists of identifying local eQTL for genes located within a GWAS locus and then determining if the two signals are due to the same sets of variants (referred to as colocalizing eQTL) [35,36].
One of the major considerations for eQTL studies (and for that matter the generation of any other -omics dataset) is the cell-type or tissue used for the generation of gene expression profiles. Recently, the Genotype-Tissue Expression (GTEx) project demonstrated that many eQTL are tissue specific, thus ideally the transcriptomics data would be from a disease relevant source [37]. In the context of bone GWAS, this would be either bone tissue or bone cells, i.e. osteoblasts and osteoclasts [37,38]. However, to date only three relatively small studies have generated bone relevant eQTL data. One such study generated microarray profiles on trans-iliacal bone biopsies from 84 postmenopausal women [39]. These data were used to identify colocalizing eQTL for a GWAS of vertebral volumetric BMD [21]. Microarray profiles of undifferentiated osteoblasts from 95 individuals have also been used to identify eQTL and inform several bone GWASs [16,17,40]. More recently, eQTL were identified in cultured primary osteoclasts using RNA-seq profiles in 158 individuals. These data were used to identify colocalizing osteoclast eQTL for genes in eight loci [41]. Integrating transcriptomics data from non-disease relevant sources has also been shown to provide insight given the expectation that some eQTL will be active across many different tissues [37]. For example, peripheral blood eQTL were recently used to identify ASB16 Antisense RNA 1( ASB16-AS1) and Synapsin II (SYN2) as potentially causal BMD GWAS genes [42]. Furthermore, GTEx expression data from thyroid tissue was used in a recent study to link the expression of Microtubule Affinity Regulating Kinase 3 (MARK3) to BMD-associated variants on Chr. 14q32.32 [43].
Another systems genetics approach that has proven useful for informing GWAS is the integration of epigenetics data [44]. As mentioned above, the vast majority of BMD GWAS loci implicate only non-coding variation that presumably impacts gene regulation. Thus, it is likely that most causal GWAS variants reside in regulatory elements, such as promoters and enhancers, which can be identified as regions of open chromatin marked by histone modifications, including H3K4me1, H3K4me2, H3K4me3, and H3K27ac. In support of this hypothesis, studies have demonstrated an enrichment for GWAS variants overlapping enhancers in disease-relevant tissues [45–48].
Epigenetic data has recently been used to inform BMD GWAS [49]. Using publicly available data, an eQTL in blood cells was identified for Long Intergenic Non-Protein Coding RNA 339 (LINC00339) that colocalized with a BMD GWAS association on Chr. 1p36.12. It was then found, using chromosome conformation capture (HI-C) data, that one of the eQTL SNPs (rs6426749) was located in a genomic region interacting with the promoter of LINC00339. Using epigenetics data from the ENCODE project, this SNP was found to overlap and influence the activity of an enhancer element in osteoblasts by altering a binding site for Transcription Factor AP-2 Alpha (TFAP2A) [50]. Furthermore, alteration of LINC00339 expression influenced the transcript levels of a nearby gene, Cell Division Control Protein 42 Homolog (CDC42), which plays a key role in bone modeling and remodeling [51]. Using a similar approach, another recent study determined that the BMD GWAS SNP rs9533090 affects the expression of Receptor Activator of Nuclear factor Kappa-Β Ligand (RANKL), which plays a central role in osteoclastogenesis, by disrupting a Nuclear Factor 1 C-type (NFIC) binding site and enhancer activity [52,53]. These studies demonstrate the power of systems genetics approaches that combine multiple data types to unravel the molecular consequences of BMD-associated variants.
There are limitations to using eQTL data to inform GWAS. As described above, the most powerful sets of eQTL data (e.g. GTEx) are from non-bone tissue. While such data have been informative for identifying colocalizing eQTL, it is likely that well powered eQTL studies in bone tissue and bone cells will provide more insight. It has also become evident that tissue and cell-type specificity is a critical factor when trying to dissect how GWAS loci influence BMD. As a result, not only do we need efforts focused on generating data in bone tissue and bone cells, but also specific bone cell populations at different stages of their lifecycle exposed to varying stimuli. It should also be noted that differences in the genetic backgrounds (with differences in linkage-disequilibrium structure) of GWASs and eQTL studies impact the interpretability of results. This can be solved by efforts to generate both types of data from racially diverse populations.
Network-based Approaches
As described above, systems genetics approaches are critical for identifying individual genes contributing to BMD. However, gene discovery is just the beginning. The next step is to understand how variants, genes and their products work together to enable proper bone function. GWASs for BMD have identified many genetic loci implicating disparate biological processes and mechanisms, suggesting a complex web of networks operating within and between various bone cell-types. Identifying these interactions is important as they can inform our understanding of “emergent properties” of bone that are not evident from the function of individual genes in isolation. This is analogous to identifying a car battery and alternator as elements involved in starting an engine. However, it would be impossible to understand their true function without knowing that they worked together in a car’s electrical system. It is also likely that genetic variation is a major perturbation that shapes underlying biological networks. As a result, systems, rather than reductionist, approaches to bone genetics are critical to understand the role of genetics in systems-level function. Understanding bone molecular networks and how they are influenced by genetic variation is also important in the context of discovering and evaluating potential anti-osteoporotic therapeutic targets [54–56].
Biological networks
Networks are prevalent in all aspects of our lives. The internet, social media, and economic markets are all examples of networks that impact us daily. In biology, many types of networks exist including protein-protein interaction, transcriptions factor binding, metabolic, and gene regulatory networks. Mathematically, a network (or graph) is a set of nodes (elements) connected by edges, which represent relationships between nodes [57]. Edges can be directed or undirected and either weighted or unweighted. An undirected gene co-expression network represents the relationships in co-expression between genes without an indication of which node is upstream of the other, while a directed network models the information flow between nodes (e.g. increased expression of gene A causes increased expression of gene B). Weights can represent the strength of evidence for the edge or the strength of the relationship between nodes. Methods used to generate and analyze networks are indispensable to systems genetics, as they allow for a shift of focus from reductionist methods, like GWAS, to more holistic, systems-level approaches. Mostly due to the scarcity of bone-relevant data, and the relative paucity of investigators applying such approaches, the use of network biology in the bone field has lagged behind others. However, there are emerging use cases. For example, by combining BMD GWAS data with functional genomic analysis, a PU.1-dependent transcription factor network essential for osteoclast differentiation has been identified [58].
Co-expression networks
The most popular types of biological networks used in systems genetics applications are based on co-expression. There are many methods for generating co-expression networks and one of the most widely used is weighted gene co-expression network analysis (WGCNA) [59]. WGCNA organizes transcriptomics data into modules, or clusters, of co-expressed genes. It does this by analyzing co-expression (i.e. correlation in expression) across a set of perturbations, such as genetic background in mice or environmental exposures in a human population. Modules have been found to have a number of important features, such as containing functionally related genes that may be subject to co-regulation by similar factors [59,60]. As a result, one can think of co-expression network analysis as a way to organize biology in a relatively unbiased way, similar to the way that file folders are used to organize documents by topic.
There are two aspects of co-expression networks that make them particularly useful for systems genetics studies. First, unlike many other popular biological networks they retain tissue or cell-type specific information. While recent advances in proteomic technology have facilitated the study of protein-protein interactions in vivo, the vast majority of extant data is generated through in vitro methods, which may not accurately reflect physiological interactions [61]. Second, unlike other biological networks, co-expression modules can be related to phenotypes from the individuals used to generate the transcriptomic profiles. For example, a WGCNA network was recently generated from blood cells in individuals with BMD measurements [62]. These data were then used to identify a module whose behavior (as summarized by its first principal component) was correlated with BMD. Once trait-correlated modules are identified they can be further analyzed to identify key genes and relationships. For example, highly connected “hub” genes have been shown to drive modular associations with a trait [63]. A recent study generated a WGCNA network using bone transcriptomic data on 96 strains from the Hybrid Mouse Diversity Panel (HMDP) [64,65]. An osteoblast-lineage specific module was identified (module 9) and shown to be highly correlated with femoral BMD in the same HMDP strains. The study showed that knockdown of the top two module 9 hub genes (Melanoma Antigen Family D1 (Maged1) and Par-6 Family Cell Polarity Regulator Gamma (Pard6g)) altered osteoblast proliferation, differentiation and mineralization in vitro and knockout of Maged1 decreased BMD in mice [63,65]. The authors mapped the first principal component of module 9 and demonstrated that the overall expression levels of module 9 genes were influenced by a local eQTL for Secreted Frizzled-related Protein 1 (Sfrp1), a key regulator of osteoblastogenesis [66]. This demonstrates how co-expression network analysis in a genetics population can be used to understand the systems-level organization of genes. Similarly, another study generated a WGCNA network using gene expression data from female transiliac bone biopsies in humans. Through the integration of BMD GWAS data, this study identified a gene module and several candidate genes (Homer Protein Homolog 1 (HOMER1) and Spectrin Beta, Non-Erythrocytic 1 (SPTBN1)), with putatively important roles in bone mass regulation [67].
Another use of co-expression networks is to inform GWAS. A number of studies have demonstrated that network information is a useful prioritization strategy for predicting causal genes for sets of GWAS associations [68]. As an illustration, a recent study mapped genes located in 64 BMD GWAS associations onto the HMDP bone network described above [43,68]. This led to the identification of two modules that were enriched for genes implicated by GWAS. Using information on module genes with known roles in bone, it was predicted that novel module genes located in GWAS loci were causal and likely altered BMD via a role in osteoblasts. Two of the module genes, Microtubule Affinity Regulating Kinase 3 (MARK3) and Spectrin Beta, Non-Erythrocytic 1 (SPTBN1), were experimentally confirmed to influence BMD when perturbed in mice. This study indicates that viewing GWAS data through the lens of a disease-relevant co-expression network can begin to highlight how key GWAS genes function together to regulate BMD.
Bayesian Networks
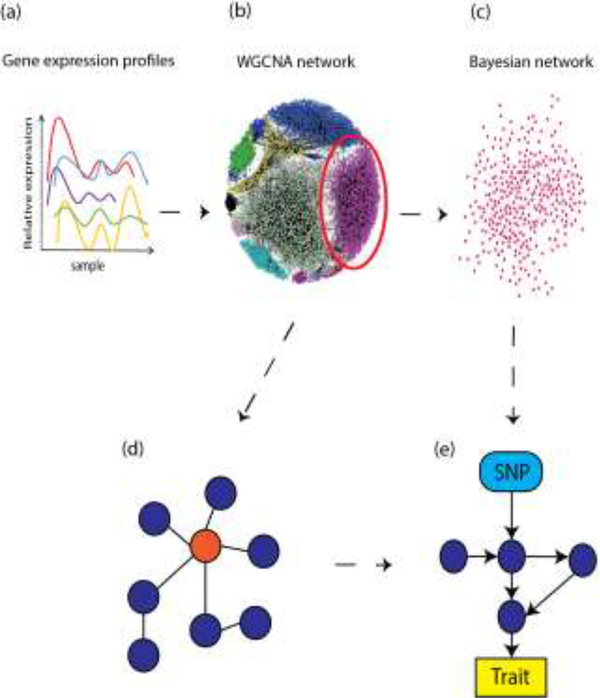
Though initially described in the mid 1980’s, Bayesian networks (BNs) have only recently begun to gain traction in biological research [69]. BNs are directed, acyclic graph representations of conditional dependencies between random variables [57]. The directed, acyclic nature of the graphs is informative for reconstructing systems-level relationships between genes. For example, in a systems genetics context it is possible to apply a BN structure learning algorithm to a WGCNA module, as the dependence of gene expression on other genes can be observed in a hierarchical manner, which allows for an elucidation of the direction of the flow of molecular information. One scenario is where BN analysis methods are applied to trait-relevant WGCNA modules, in order to direct relationships between genes and identify key regulatory elements (Figure 3). This strategy was employed in a recent study, where an undirected co-expression network was constructed. Directional relationships between nodes were then established using Bayesian network analysis. This led to the identification of causal network structures relevant to late-onset Alzheimer’s disease (LOAD) pathology as well as the identification of TYRO Protein Tyrosine Kinase Binding Protein (TYROBP) as a key regulator [70]. In another study, a BN generated from co-expression modules was used to reveal regulatory driver genes affecting coronary artery disease [71]. To our knowledge, BNs have not yet been applied in a systems genetics context in the bone field, and therefore provide an exciting avenue for future research.
Figure 3. Network analysis can reveal systems-level information.
In a typical network analysis workflow, gene expression profiles (a) (typically RNA-seq) can be analyzed from a network perspective. In this example, a co-expression network (b) is generated using WGCNA. In a WGCNA network, modules (colored clusters, b) consist of gene expression profiles connected by undirected edges, which signify the strength of the connection between two genes. A trait-correlated WGCNA module (within red circle, b) can be further dissected through Bayesian network analysis (c). Highly-connected hub genes (orange node, d) can signify functionally important genes. Bayesian networks differ from WGCNA networks in that they contain directed edges, and are acyclic (e). Furthermore, diverse biological information, such as SNP and trait data (blue and yellow boxes respectively, e) can be incorporated into Bayesian networks as prior information to improve network reconstruction. An advantage of Bayesian network analysis is the generation of more mechanistic hypotheses.
A great advantage of BNs is that they allow for the incorporation of prior knowledge, which allows for more informative modeling of gene relationships within modules. For example, network structure learning can be biased by “whitelisting” high-confidence edges (such as well-known gene-gene relationships or protein-protein interactions) a priori, or “blacklisting” improbable edges. Disparate data sources can be easily incorporated into BNs as well. For example, a BN from a WGCNA module can also include SNP nodes and trait nodes, in order to model information transfer from genetic element to gene expression and phenotypic outcomes [72].
Network-based approaches are not without limitations. One limitation involves the quality and type of the investigated bone phenotype. For example, BMD can be assayed in different anatomical locations by several different methods, which can lead to heterogeneity in the data that can obfuscate meaningful network relationships, or lead to network connections that are artificial and not mechanistically viable. Furthermore, a phenotype such as BMD is actually a composite of many different aspects of bone, which can also exacerbate the problems of interpretability. Therefore, careful selection of phenotypes should be performed a priori. Furthermore, biological networks often encompass multiple cell types, tissues and physiological microenvironments. In silico analyses based on data from in vitro sources, such as cultured osteoblasts, will not uncover many physiological relationships that exist in vivo. This drawback is not unique to network analyses and pervades biological science, but should be carefully considered when designing experiments and drawing conclusions. Methodological drawbacks exist as well. A significant drawback of using BNs to model biological relationships is in their acyclic nature. In biological processes, structures like feedback and feed-forward loops are prevalent. As BNs are acyclic, these network structures will be missed. Furthermore, depending on the algorithm used, it can be computationally impractical to learn the network structure of large sets of genes. These shortcomings make the aforementioned strategy of using BNs to dissect WGCNA modules an attractive one [73].
Concluding Remarks and Future Perspectives
In the past decade or so, advances in sequencing technology have completely revolutionized biological science. In practically every biological field, a wealth of “-omics” data is being generated. However, our understanding of the underpinnings of biological processes and diseases is still far from complete. This is evident in the bone field, as many genetic associations with BMD have been described, yet we still know few of the responsible genes. These limitations reinforce the need for complementary strategies, such as systems genetics, to further advance our understanding of bone genetics.
One of the major limitations of genetic studies of bone is the primary focus on BMD. Although BMD is the single strongest predictor of osteoporotic fracture, there are many individuals with normal BMD that fracture [74,75]. The use of BMD has been necessitated by the difficulty, or impossibility, of measuring other aspects of bone fragility in humans. For example, biomechanical properties of bone strength, the single most important fracture-related trait, can only be measured in cadavers. Due to these limitations, a possible alternative is to use GWAS and systems genetics in mice and rats as a way of developing a more complete understanding of osteoporosis [76–78].
The adaptive nature of bone creates an additional layer of complexity in understanding osteoporosis. Studies have demonstrated that recombinant inbred mice with different genetic backgrounds build functional bones in different ways. For example, mice with genetically slender bones will compensate for this deficiency by increasing cortical thickness and mineralization, whereas mice with mineralization defects will increase bone size [79,80]. This genetically-based covariation in traits serves as an example of a system adapting to perturbations. It also illustrates the importance of understanding not only how genetic variation impacts individual traits, but also the relationships between traits. A more encompassing approach to systems genetics has the potential to begin to understand how genetic variation contributes to these relationships and overall system function.
In the field of systems genetics it is of the utmost importance to develop approaches for the effective understanding and utilization of available data (Box 2). As biology is inherently complex, it is unreasonable to believe that a single, or few, types of genetic analyses will be sufficient to gain a thorough understanding of the genetics of complex bone traits. We argue that more realistic models of biological processes can be generated and analyzed by synthesizing and incorporating seemingly disparate data sources. While barely scratching the surface, the systems genetics approaches described herein provide an avenue for such an endeavor. Of course, our understanding of many systems-level principles is still evolving (see Outstanding Questions). With increasingly accessible computational resources and by training researchers adept in the computational sciences, the transition to understanding bone biology and the impacts of genetic variation from a holistic, systems perspective will be within our reach.
BOX 2. Nascent approaches in systems genetics.
In order to capitalize on advances in biological knowledge, such as the recent abundance of “–omics” datasets, continuous development of analytical approaches that can utilize large-scale, varied data is necessary. Machine learning is an example of such approaches, and has generated a high level of interest in recent years. Machine learning is a broad term that encompasses many widely used algorithms and techniques; for example, hierarchical clustering, a main component of WGCNA network construction, and linear regression are considered machine learning algorithms. In the bone field, machine learning approaches have been utilized to predict bone loss rates, osteoporosis and bone fracture risk [84–87]
In the context of systems genetics, machine learning approaches are of high interest due to their utility in analyzing large, highly-dimensional data, such as multi-omic data sets [88]. Within machine learning, “deep learning” approaches (such as artificial neural networks) represent the state-of-the-art, and refer to a class of approaches that “learn” patterns in data through structuring algorithms in hierarchical layers. Deep learning approaches can be utilized to learn patterns in data for the prediction of traits and for the identification of important data features. For example, patterns that classify samples as healthy or diseased can be discerned in multi-omic datasets, facilitating the prediction of diseased individuals from input data, as well as identifying causal determinants of disease, leading to a deeper understanding of the biological networks underlying phenotypes [89]. Similarly, machine learning approaches can also be used to learn patterns in data that lead to the identification of more accurate phenotypes and biomarkers [90]. Deep learning approaches can even be used to predict genetic elements, such as enhancers, from genomic data [91]. To our knowledge, such approaches have not yet been implemented in in the context of the systems genetics of bone, but have begun to be utilized in other biological fields [92–95].
In addition to being limited by their relatively recent genesis, the use of such approaches has been limited in the bone field due to the same limitation that affects network analyses; the sparsity of large, high-quality data sets encompassing multiple omics sources. In order for the bone field to ascend to the forefront of systems genetics, it is imperative to generate such data sets from relevant samples.
Outstanding Questions Box.
To date, the genetic analysis of osteoporosis has focused on connecting variants to genes to disease. Equally important will be understanding how genetic variation impacts gene networks and how network perturbations lead to disease.
The bone field has generated several large-scale GWAS of BMD, should we now focus on the generation of other “omics” data to facilitate systems genetics approaches? If so, what data types are lacking, which tissue/cell-types should be the focus and what are the most pressing questions to examine?
Does a “best” approach or algorithm for network-based analyses exist? What are the best practices for validation and interpretation of the methodology and results of network analyses?
How can systems genetics approaches be utilized to understand genetically-influenced relationships between traits, which ultimately underlie bone strength?
Highlights.
Osteoporosis is a common, complex disease characterized by low bone mineral density (BMD). Quantitative bone traits influencing osteoporotic fracture, such as BMD, are highly heritable (h2>0.5).
Genome-wide association studies (GWASs) have identified >300 loci for BMD. However, most causal genes, their mechanisms of action, and how they interact are unknown.
Systems genetics approaches can be used to integrate bone GWAS with other “-omics” data to identify causal variants and genes.
Network-based analysis approaches can provide insight into how genetic variation impacts molecular networks and how network dysfunction leads to osteoporosis.
Acknowledgements
CRF is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R01AR064790, R01AR068345 and R01AR071657. BMA is supported by a National Institutes of Health, Biomedical Data Sciences Training Grant (5T32LM012416).
Glossary:
- Acyclic graph
A graph that does not contain cycles. A cycle is a set of nodes and edges, where a node can be reachable from itself. For example, the graph with nodes A, B and C with edges A -> B, B -> C, C -> A contains a cycle
- BMD
Bone mineral density is the amount of mineral (hydroxyapatite) per volume of bone. BMD is the main diagnostic measure for osteoporosis and is one of the strongest predictors of fracture
- Edge
An edge is a connection between two nodes in a network
- Epigenetics
The study of processes that affect gene expression or function but do not involve changes in DNA sequence. These include processes such as histone modifications (acetylation, phosphorylation, etc.) and DNA methylation
- eQTL
expression quantitative trait loci are genomic regions harboring genetic variation influencing RNA levels (via transcription, splicing, stability, etc.)
- GWAS
Genome-Wide Association Studies identify genomic regions harboring genetic variation influencing a disease or quantitative trait
- Genetic architecture
For a given trait it refers to the number, mode of action, effect size and frequency of genetic variants that contribute to that trait in the population and their interactions with each other and the environment
- Heritability
The fraction of variance in a trait that is attributable to genetic variance
- Machine learning
A branch of artificial intelligence concerned with creating algorithms that can analyze data, and improve analytical performance, with minimal instruction. For example, machine learning algorithms can analyze data to uncover patterns that predict certain outcomes, while progressively improving metrics like the selectivity and sensitivity of predictions
- Node
Along with edges, nodes are the basic units of a network. In a co-expression networks, nodes are genes or transcripts
- Osteoblast
Specialized bone-forming cells of mesenchymal origin
- Osteoclast
Specialized bone-resorbing cells of hematopoietic origin
- Osteoporosis
A metabolic bone disease characterized by decreased bone mass and increased risk of fracture
- Single Nucleotide Polymorphisms
SNPs are single base pair substitutions. SNPs are the most common type of variation in the human genome, occurring on average every 300 bases. SNPs can have many molecular consequences such as altering protein structure, gene regulation or splicing
- Structure learning
In Bayesian networks, this refers to the process of learning the structure of the directed acyclic graph. That is to say, learning the connections and directionality between nodes. Two main classes of algorithms are used for structure learning: score-based and constraint-based algorithms
- Systems genetics
An integrative field that aims to understand how genetic information is transmitted and integrated through biological systems and networks
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Grabowski P (2015) Physiology of Bone. Endocr. Dev 28, 33–55 [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G and Oury F (2012) Biology without walls: the novel endocrinology of bone. Annu. Rev. Physiol 74, 87–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riddle RC and Clemens TL (2017) Bone Cell Bioenergetics and Skeletal Energy Homeostasis. Physiol. Rev 97, 667–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black DM and Rosen CJ (2016) Clinical Practice. Postmenopausal Osteoporosis. N. Engl. J. Med 374, 254–262 [DOI] [PubMed] [Google Scholar]
- 5.Cummings SR and Melton LJ (2002) Epidemiology and outcomes of osteoporotic fractures. Lancet 359, 1761–1767 [DOI] [PubMed] [Google Scholar]
- 6.Harvey N et al. (2010) Osteoporosis: impact on health and economics. Nat. Rev. Rheumatol 6, 99–105 [DOI] [PubMed] [Google Scholar]
- 7.Burge R et al. (2006) Incidence and Economic Burden of Osteoporosis-Related Fractures in the United States, 2005–2025. J. Bone Miner. Res 22, 465–475 [DOI] [PubMed] [Google Scholar]
- 8.Peacock M et al. (2002) Genetics of osteoporosis. Endocr. Rev 23, 303–326 [DOI] [PubMed] [Google Scholar]
- 9.Morrison NA et al. (1994) Prediction of bone density from vitamin D receptor alleles. Nature 367, 284–287 [DOI] [PubMed] [Google Scholar]
- 10.Ralston SH and Uitterlinden AG (2010) Genetics of osteoporosis. Endocr. Rev 31, 629–662 [DOI] [PubMed] [Google Scholar]
- 11.Richards JB et al. (2009) Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann. Intern. Med 151, 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ioannidis JP et al. (2007) Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J. Bone Miner. Res 22, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiel DP et al. (2007) Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med. Genet 8 Suppl 1, S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush WS and Moore JH (2012) Chapter 11: Genome-wide association studies. PLoS Comput. Biol 8, e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards JB et al. (2012) Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat. Rev. Genet 13, 576–588 [DOI] [PubMed] [Google Scholar]
- 16.Estrada K et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet 44, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp JP et al. (2017) Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet 49, 1468–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabik OL and Farber CR (2017) Using GWAS to identify novel therapeutic targets for osteoporosis. Transl. Res 181, 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris JA et al. 11-Jun-(2018), An Atlas of Human and Murine Genetic Influences on Osteoporosis., bioRxiv, 338863
- 20.Cummings SR et al. (1990) Appendicular bone density and age predict hip fracture in women. The Study of Osteoporotic Fractures Research Group. JAMA 263, 665–668 [PubMed] [Google Scholar]
- 21.Nielson CM et al. (2016) Novel Genetic Variants Associated With Increased Vertebral Volumetric BMD, Reduced Vertebral Fracture Risk, and Increased Expression of SLC1A3 and EPHB2. J. Bone Miner. Res 31, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu Y-H et al. (2010) An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 6, e1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prins BP et al. (2017) Genome-wide analysis of health-related biomarkers in the UK Household Longitudinal Study reveals novel associations. Sci. Rep 7, 11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L-J et al. (2010) Genome-wide association study for femoral neck bone geometry. J. Bone Miner. Res 25, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timpson NJ et al. (2017) Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat. Rev. Genet 19, 110–124 [DOI] [PubMed] [Google Scholar]
- 26.Manolio TA et al. (2009) Finding the missing heritability of complex diseases. Nature 461, 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Styrkarsdottir U et al. (2013) Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 [DOI] [PubMed] [Google Scholar]
- 28.Styrkarsdottir U et al. (2016) Two Rare Mutations in the COL1A2 Gene Associate With Low Bone Mineral Density and Fractures in Iceland. J. Bone Miner. Res 31, 173–179 [DOI] [PubMed] [Google Scholar]
- 29.Zheng H-F et al. (2015) Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 526, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadeau JH and Dudley AM (2011) Genetics. Systems genetics. Science 331, 1015–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civelek M and Lusis AJ (2014) Systems genetics approaches to understand complex traits. Nat. Rev. Genet 15, 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cookson W et al. (2009) Mapping complex disease traits with global gene expression. Nat. Rev. Genet 10, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farber CR and Lusis AJ (2008) Integrating global gene expression analysis and genetics. Adv. Genet 60, 571–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockman MV and Kruglyak L (2006) Genetics of global gene expression. Nat. Rev. Genet 7, 862–872 [DOI] [PubMed] [Google Scholar]
- 35.Hormozdiari F et al. (2016) Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet 99, 1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giambartolomei C et al. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium GTEx et al. (2017) Genetic effects on gene expression across human tissues. Nature 550, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonewald LF (2011) The amazing osteocyte. J. Bone Miner. Res 26, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reppe S et al. (2010) Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone 46, 604–612 [DOI] [PubMed] [Google Scholar]
- 40.Rivadeneira F et al. (2009) Twenty bone-mineral-density loci identified by largescale meta-analysis of genome-wide association studies. Nat. Genet 41, 1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullin BH et al. (2018) Expression quantitative trait locus study of bone mineral density GWAS variants in human osteoclasts. J. Bone Miner. Res DOI: 10.1002/jbmr.3412 [DOI] [PubMed] [Google Scholar]
- 42.Meng X-H et al. (2018) Integration of summary data from GWAS and eQTL studies identified novel causal BMD genes with functional predictions. Bone 113, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calabrese GM et al. (2017) Integrating GWAS and Co-expression Network Data Identifies Bone Mineral Density Genes SPTBN1 and MARK3 and an Osteoblast Functional Module. Cell Syst 4, 46–59.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tak YG and Farnham PJ (2015) Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in noncoding regions of the human genome. Epigenetics Chromatin 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onengut-Gumuscu S et al. (2015) Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet . 47, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farh KK-H et al. (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castaldi PJ et al. (2014) Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am. J. Respir. Crit. Care Med . 190, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hazelett DJ et al. (2014) Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 10, e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X-F et al. (2018) An Osteoporosis Risk SNP at 1p36.12 Acts as an AlleleSpecific Enhancer to Modulate LINC00339 Expression via Long-Range Loop Formation. Am. J. Hum. Genet . 102, 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito Y et al. (2010) Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J. Clin. Invest . 120, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teitelbaum SL and Ross FP (2003) Genetic regulation of osteoclast development and function. Nat. Rev. Genet . 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 53.Zhu D-L et al. (2018) Multiple Functional Variants at 13q14 Risk Locus for Osteoporosis Regulate RANKL Expression Through Long-Range Super-Enhancer. J. Bone Miner. Res . 33, 1335–1346 [DOI] [PubMed] [Google Scholar]
- 54.Nelson MR et al. (2015) The support of human genetic evidence for approved drug indications. Nat. Genet . 47, 856–860 [DOI] [PubMed] [Google Scholar]
- 55.Plenge RM et al. (2013) Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov . 12, 581–594 [DOI] [PubMed] [Google Scholar]
- 56.Barabási A-L et al. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genet . 12, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Needham CJ et al. (2007) A primer on learning in Bayesian networks for computational biology. PLoS Comput. Biol . 3, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carey HA et al. (2018) Enhancer variants reveal a conserved transcription factor network governed by PU.1 during osteoclast differentiation. Bone Res 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P and Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang R et al. (2006) Comprehensive analysis of pathway or functionally related gene expression in the National Cancer Institute’s anticancer screen. Genomics 87, 315–328 [DOI] [PubMed] [Google Scholar]
- 61.Kaake RM et al. (2014) A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell. Proteomics 13, 3533–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farber CR (2010) Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J. Bone Miner. Res . 25, 2359–2367 [DOI] [PubMed] [Google Scholar]
- 63.Langfelder P et al. (2013) When is hub gene selection better than standard meta-analysis? PLoS One 8, e61505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bennett BJ et al. (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calabrese G et al. (2012) Systems genetic analysis of osteoblast-lineage cells. PLoS Genet. 8, e1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao W et al. (2010) Overexpression of secreted frizzled-related protein 1 inhibits bone formation and attenuates parathyroid hormone bone anabolic effects. J. Bone Miner. Res . 25, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y-C et al. (2016) Integrative Analysis of Genomics and Transcriptome Data to Identify Potential Functional Genes of BMDs in Females. J. Bone Miner. Res . 31, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 68.Leiserson MDM et al. (2013) Network analysis of GWAS data. Curr. Opin. Genet. Dev . 23, 602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pearl J (1985) Bayesian Networks: A Model of Self-activated Memory for Evidential Reasoning,
- 70.Zhang B et al. (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mäkinen V-P et al. (2014) Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 10, e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Su C et al. (2013) Using Bayesian networks to discover relations between genes, environment, and disease. BioData Min. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B et al. (2016) Characterization of Genetic Networks Associated with Alzheimer’s Disease. Methods Mol. Biol . 1303, 459–477 [DOI] [PubMed] [Google Scholar]
- 74.Kanis JA et al. (2005) Assessment of fracture risk. Osteoporos. Int . 16, 581–589 [DOI] [PubMed] [Google Scholar]
- 75.Stone KL et al. (2003) BMD at multiple sites and risk of fracture of multiple types: long-term results from the Study of Osteoporotic Fractures. J. Bone Miner. Res . 18, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 76.Flint J and Eskin E (2012) Genome-wide association studies in mice. Nat. Rev. Genet . 13, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farber CR et al. (2011) Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 7, e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rat Genome Sequencing and Mapping Consortium et al. (2013) Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat. Genet 45, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jepsen KJ et al. (2009) Phenotypic integration of skeletal traits during growth buffers genetic variants affecting the slenderness of femora in inbred mouse strains. Mamm. Genome 20, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jepsen KJ (2009) Systems analysis of bone. Wiley Interdiscip. Rev. Syst. Biol. Med . 1, 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boyle EA et al. (2017) An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fisher SRA (1918) The Correlation Between Relatives on the Supposition of Mendelian Inheritance, Royal Society of Edinburgh. [Google Scholar]
- 83.Wray NR et al. (2018) Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 173, 1573–1580 [DOI] [PubMed] [Google Scholar]
- 84.Devikanniga D and Joshua Samuel Raj R (2018) Classification of osteoporosis by artificial neural network based on monarch butterfly optimisation algorithm. Healthc Technol Lett 5, 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kavitha MS et al. (2013) The combination of a histogram-based clustering algorithm and support vector machine for the diagnosis of osteoporosis. Imaging Sci. Dent . 43, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shioji M et al. (2017) Artificial neural networks to predict future bone mineral density and bone loss rate in Japanese postmenopausal women. BMC Res. Notes 10, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shaikhina T and Khovanova NA (2017) Handling limited datasets with neural networks in medical applications: A small-data approach. Artif. Intell. Med . 75, 51–63 [DOI] [PubMed] [Google Scholar]
- 88.Libbrecht MW and Noble WS (2015) Machine learning applications in genetics and genomics. Nat. Rev. Genet . 16, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Camacho DM et al. (2018) Next-Generation Machine Learning for Biological Networks. Cell 173, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 90.Basile AO and Ritchie MD (2018) Informatics and machine learning to define the phenotype. Expert Rev. Mol. Diagn . 18, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Min X et al. (2017) Predicting enhancers with deep convolutional neural networks. BMC Bioinformatics 18, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young JD et al. (2017) Unsupervised deep learning reveals prognostically relevant subtypes of glioblastoma. BMC Bioinformatics 18, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swan AL et al. (2015) A machine learning heuristic to identify biologically relevant and minimal biomarker panels from omics data. BMC Genomics 16 Suppl 1, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu T et al. (2018) An evolutionary learning and network approach to identifying key metabolites for osteoarthritis. PLoS Comput. Biol . 14, e1005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma J et al. (2018) Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods 15, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]