Abstract
Background & Aims:
Previous studies reported an association of the bacteria Helicobacter pylori, the primary cause of gastric cancer, and risk of colorectal cancer (CRC). However, these findings have been inconsistent, appear to vary with population characteristics, and may be specific for virulence factor VacA. To more thoroughly evaluate the potential association of H pylori antibodies with CRC risk, we assembled a large consortium of cohorts representing diverse populations in the United States.
Methods:
We used H pylori multiplex serologic assays to analyze serum samples from 4063 incident cases of CRC, collected before diagnosis, and 4063 matched individuals without CRC (controls) from 10 prospective cohorts for antibody responses to 13 H pylori proteins, including virulence factors VacA and CagA. The association of sero-positivity to H pylori proteins, as well as protein-specific antibody level, with odds of CRC was determined by conditional logistic regression.
Results:
Overall 40% of controls and 41% of cases were H. pylori sero-positive (odds ratio [OR], 1.09; 95% CI, 0.99–1.20). H pylori VacA-specific sero-positivity was associated with an 11% increased odds of CRC (OR, 1.11; 95% CI, 1.01–1.22), and this association was particularly strong among African Americans (OR, 1.45; 95% CI, 1.08–1.95). Additionally, odds of CRC increased with level of VacA antibody in the overall cohort (P=.008) and specifically among African Americans (P=.007).
Conclusion:
In an analysis of a large consortium of cohorts representing diverse populations, we found serologic responses to H pylori VacA to associate with increased risk of CRC risk—particularly for African Americans. Future studies should seek to understand whether this marker is related to virulent H pylori strains carried in these populations.
Keywords: gastrointestinal cancers, epidemiology, cohort studies

INTRODUCTION
It is currently estimated that at least 15% of all incident cancers are caused by infection, and the bacterium Helicobacter pylori (H pylori) is the leading single carcinogenic infectious agent, responsible for 770,000 cancer cases worldwide each year due to its established association with gastric cancer.1 Over 50% of the world’s population is infected with this bacterium, and the prevalence of H pylori varies widely by geographic location, with the highest prevalence in East Asia, Africa, and parts of South America, and lowest prevalence in the United States, Oceania, and Western Europe.2 However, even within the United States, there remains great variation in both H pylori prevalence and gastric cancer incidence by race/ethnicity.3,4
The mechanism by which infection with H pylori induces gastric cancer is understood to be due at least in part to chronic inflammation of the gastric mucosa. Several studies have investigated whether H pylori might also increase risk for developing colorectal cancer (CRC). The mechanism underlying this possible association has not yet been delineated.5 Nonetheless, the results of two meta-analyses found significant 30% to 50% increased odds for CRC among those individuals with evidence of a current or past H pylori infection.6,7
H pylori strains are genetically very diverse8, yet the majority of published studies examining the association between H pylori and CRC risk did not take into account this heterogeneity. H pylori strains exhibit variation in the presence or absence of virulence factors (such as the cag pathogenicity island, which encodes the oncogenic effector protein CagA), as well as allelic variation in the Vacuolating cytotoxin A (VacA)9. As the majority of the world’s population is infected with H pylori, but only a small proportion of those individuals develop gastric cancer, it is important that both bacterial virulence factors and host responses, such as the individual’s immune response to H pylori infection, are considered. Our group has utilized multiplex serology to assess immune response to 15 different H pylori proteins in a primarily low-income population in the southeast US, and found an exceptionally high prevalence of antibodies to H pylori, with significantly higher prevalences among African Americans as compared to whites for both CagA and VacA.10 We also found that sero-positivity to VacA was associated with a significant 84% increased CRC risk in this population, and there was also a strong dose-response association for CRC by increasing quartile of VacA antibody level.11
In this study, we sought to thoroughly evaluate the novel association of Helicobacter pylori protein-specific antigen response and the odds of CRC through use of a consortium of nested case-control studies. We modeled the associations of serologic response of individual antigens with CRC incidence, with consideration of the heterogeneity of the outcome (by histologic site, stage, and age at onset of cancer), the time from antibody status assessment to cancer diagnosis, and the potential dose-response relationship between level of serological response and risk of disease. Therefore, we created a consortium of ten prospective cohort studies specifically chosen to highlight the diversity of the US population, including over 4,000 prospectively ascertained CRC cases and 1:1 matched controls. To date, this consortium provides the largest nested case-control study with pre-diagnostic serum to examine the association between H pylori and odds of CRC that addresses both H pylori protein expression diversity as well as the racial/ethnic diversity in the US.
MATERIALS AND METHODS
Study population
This consortium was built using a nested case-control study design that includes prospectively ascertained colorectal cancer cases (and 1:1 matched controls) with available pre-diagnostic serum. These studies represent diverse populations within the US, including: primarily low-income African Americans and whites in the southeast recruited from community health clinics (Southern Community Cohort Study – SCCS, excluding CRC cases and controls participating in the hypothesis-generating earlier study11); individuals of Native Hawaiian, Japanese, and European ancestry in Hawaii and individuals of African and Latino ancestry in Los Angeles, California (Multiethnic Cohort Study – MEC12); medical professionals from over 14 states (Health Professionals Follow-up Study – HPFS13, Nurses’ Health Study – NHS14, and Physicians’ Health Study – PHS15); women visiting a breast screening center in New York City (New York University Women’s Health Study – NYU WHS16); postmenopausal women recruited at 40 Clinical Centers nationwide as part of a long-term national health study (Women’s Health Initiative – WHI17); residents of suburban Washington County, Maryland, recruited using mobile trailers (Campaign Against Cancer and Stroke – CLUE II18); healthy individuals from 21 states recruited by the American Cancer Society (Cancer Prevention Study-II CPS II 19); and participants in a large, nation-wide cancer screening trial with recruitment sites in Washington DC, Pittsburgh, Birmingham, St. Louis, Detroit, Marshfield WI, Minneapolis, Denver, Salt Lake City, and Honolulu (Prostate, Lung, Colorectal, and Ovarian Screening Study – PLCO 20). The participants in these cohorts vary substantially by demographic and lifestyle factors (see Supplementary Table S1), and with this knowledge cases and controls in this consortium are matched not only by age, sex, and race, but also within each cohort.
Cases included incident cancers of the colon and rectum as defined based on the International Classification of Diseases for Oncology (ICD-O-3, codes C180-189, C199, and C209). Controls were selected using incidence density sampling, with one control chosen at random for each CRC case from the appropriate risk sets consisting of all cohort members, who were alive and free of cancer (except non-melanoma skin cancer) at the time of diagnosis of the index case, and had provided blood specimens. Matching criteria included cohort, sex, race, date of birth, and date of blood collection.
Covariates collected from participating cohorts included standard demographic and anthropometric variables, such as education as a measure of socio-economic status, and height and weight. Data were harmonized for smoking variables (current/former/never, as well as pack-years); physical activity (METs/day); hormone therapy (HT); diabetes history; CRC screening history; and family history of CRC. Where available, we collected data on use of antibiotics, personal history of inflammatory bowel disease and colorectal polyps, and dietary intake of fruit, vegetables, and red meat, and average daily total energy intake. Clinical information ascertained for cases included tumor site, stage, histology, and date of diagnosis, as available.
Of the 8,420 samples that were assayed by multiplex serology (4,210 cases and 4,210 controls), 100 samples (and their 100 matched case or control counterparts) were excluded due to technical issues of insufficient volume (n=27), pipetting errors (n=52), and invalid measurements due to insufficient bead counts (n=21). We also excluded from analyses 47 pairs that were mis-matched by race and/or sex, resulting in a final study population of 4,063 cases and 4,063 controls. The median follow-up time for cases with valid serology results was 7 years (range <1 to 40·2 years), the majority of cases were found in the colon (84%), and the median age at diagnosis was 73 years (range 40 to 97 years).
Multiplex serology
Serum samples were sent on dry ice to the German Cancer Research Center (DKFZ, Heidelberg, Germany) and analyzed in a 1:1000 final serum dilution. Multiplex serology was performed as described previously.21 Briefly, multiplex serology is a fluorescent bead-based suspension array. H pylori antigens were expressed as Glutathione-S-transferase (GST)-tagged fusion proteins and affinity-purified on glutathione-casein coupled polystyrene beads (Luminex Corp, Austin, TX, USA) with distinct internal fluorescence. The antigen-loaded bead sets were mixed and incubated with serum. A Luminex flow cytometer distinguished between the bead set, and consequently the loaded antigen, as well as quantified the amount of bound serum antibody by a secondary antibody detecting human IgG, IgA, and IgM and a fluorescent reporter conjugate (Streptavidin-R-phycoerythrin). The output was the median fluorescence intensity (MFI) measured on at least 100 beads per set per sample. Net MFI were calculated by subtracting background values resulting from a serum-free reaction as well as from a bead-set loaded with GST-tag only.
All H pylori antigens, except for GroEL (strain G27), were produced as recombinant proteins derived from genes in strain 26695. This strain contains a type s1m1 vacA gene. Three proteins, HyuA, CagA and VacA were expressed in two parts (N- and C-terminus) due to their large size; MFI values measured for each protein part were added to each other for analyses of the responses to full size proteins (Supplementary table S2). Previous analyses with this assay applied in total 15 H pylori antigens; however, we excluded two previously non-informative antigens (CagD and CagM) from the present study, resulting in a total of 13 H pylori proteins assessed.11
Antigen-specific cut-offs were applied as defined in the previous study and quality assured by visual inspection of percentile plots (Supplementary table S2).11 Overall H pylori sero-positivity (H pylori+) was defined as being positive to any four or more of the included 13 H pylori antigens.21 Individual H pylori antigens were only considered as sero-positive when the sample was simultaneously considered overall H pylori positive (H pylori+ antigen+). This prerequisite was applied to ensure that antigen-specific sero-positivity did not result from cross-reactive antibody responses from infection with other pathogens expressing homologous proteins.
Of 82 duplicates within the WHI study set incorporated as blinded quality control samples, correlations for antibody responses (MFI) to H pylori antigens ranged from 0.92 to 1.0 indicating a good reproducibility of the measured values.
Statistical analysis
Conditional logistic regression was applied to estimate the odds ratios (OR) and 95% confidence intervals (CI) of CRC in relation to overall H pylori sero-positivity as well as to individual H pylori antigens. Potential confounders (apart from the matching variables age, sex and race within each cohort) were defined a priori and included education, smoking, body mass index (BMI), family history of CRC, previous colonoscopy/sigmoidoscopy, HT, daily intake of fruit, vegetable or red meat, aspirin use, and diabetes. While we were limited by values not missing at random for a number of these variables, overall, adjusting for any of them did not alter the main effect estimates by more than 10% and therefore main results are presented without further adjustment. The specific effects on the main results ORs from adjustment for the primary potential confounders of education, smoking, and BMI are shown in Supplementary Table S3.
The matching variable race/ethnicity was strongly associated with H pylori sero-positivity, and as we a priori believed that individuals of different race/ethnicity could be infected with different strains of H pylori, we therefore show results for the total cohort but also stratified by race/ethnicity. Additionally, stratified analyses by participating cohorts were performed to rule out that the difference in the association seen by race/ethnicity was study-specific.
We further explored the dose-response relationship of antibody responses to VacA, which was a priori hypothesized to be specifically associated with CRC risk based on our previous study.11 Antibody responses among H pylori VacA sero-positive individuals were analyzed in race/ethnicity-specific quartiles defined based on the MFI distribution among controls. ORs and 95% CIs were calculated using conditional logistic regression models with individuals not H pylori+ VacA+ as the reference group, in the total cohort as well as separately by race/ethnicity. A Cochrane-Armitage test was applied to test for a linear trend in increasing strength of the association.
We also examined the association of H pylori+ VacA+ the odds of CRC in separate models by certain case characteristics, including tumor site, stage, and time between blood draw and diagnosis. We did not identify substantial differences by case characteristics (data not shown).
RESULTS
Baseline characteristics between CRC cases and controls in the consortium differed with respect to many known CRC risk factors. Specifically, CRC cases were more likely to be obese, diabetic, former smokers, to have a positive CRC family history, and to consume more red meat per day. History of endoscopic screening, regular use of aspirin or HT, as well as higher physical activity per day were less frequent among CRC cases than among their age, race, and sex-matched controls (Table 1).
Table 1:
Baseline characteristics of the cohorts participating in this study.
| Variable | Total (n=8126) | Controls (n=4063) | Cases (n=4063) |
|---|---|---|---|
| Study, n (%) | |||
| CLUE | 982 (12) | 491 (12) | 491 (12) |
| CPSII | 722 (9) | 361 (9) | 361 (9) |
| HPFS | 302 (4) | 151 (4) | 151 (4) |
| MEC | 1510 (19) | 755 (19) | 755 (19) |
| NHS | 576 (7) | 288 (7) | 288 (7) |
| NYUWHS | 572 (7) | 286 (7) | 286 (7) |
| PHS | 360 (4) | 180 (4) | 180 (4) |
| PLCO | 1240 (15) | 620 (15) | 620 (15) |
| SCCS | 252 (3) | 126 (3) | 126 (3) |
| WHI | 1610 (20) | 805 (20) | 805 (20) |
| Age at blood draw | |||
| Median (range) | 64 (18-89) | 64 (18-88) | 64 (18-89) |
| Sex, n (%) | |||
| Female | 5112 (63) | 2556 (63) | 2556 (63) |
| Male | 3014 (37) | 1507 (37) | 1507 (37) |
| Race/Ethnicity, n (%) | |||
| White | 6134 (75) | 3067 (75) | 3067 (75) |
| African American | 798 (10) | 399 (10) | 399 (10) |
| Asian American | 614 (8) | 307 (8) | 307 (8) |
| Latino | 422 (5) | 211 (5) | 211 (5) |
| Other/unknown/multiracial | 158 (2) | 79 (2) | 79 (2) |
| Education, n (%) | |||
| Less than HS | 972 (12) | 468 (12) | 504 (13) |
| Completed HS or GED | 1668 (21) | 823 (20) | 845 (21) |
| Post HS training other than college | 362 (5) | 183 (5) | 179 (4) |
| Some college | 1672 (21) | 845 (21) | 827 (21) |
| College graduate | 1483 (19) | 756 (19) | 727 (18) |
| Graduate school | 1861 (23) | 946 (24) | 915 (23) |
| Missing | 108 | 42 | 66 |
| BMI1, n (%) | |||
| <20 | 194 (3) | 99 (3) | 95 (3) |
| 20-24.9 | 2314 (33) | 1226 (35) | 1088 (31) |
| 25-29.9 | 2852 (40) | 1456 (41) | 1396 (40) |
| >=30 | 1691 (24) | 748 (21) | 943 (27) |
| Missing | 1075 | 534 | 541 |
| Smoking, n (%) | |||
| Never | 3628 (45) | 1853 (46) | 1775 (44) |
| Former | 3358 (42) | 1622 (40) | 1736 (43) |
| Current | 1051 (13) | 546 (14) | 505 (13) |
| Missing | 89 | 42 | 47 |
| Family history of CRC2, n (%) | |||
| No | 5167 (85) | 2638 (87) | 2529 (84) |
| Yes | 907 (15) | 408 (13) | 499 (16) |
| Missing | 2052 | 1017 | 1035 |
| CRC Screening, n (%) | |||
| No | 3243 (53) | 1554 (51) | 1689 (55) |
| Yes | 2853 (47) | 1494 (49) | 1359 (45) |
| Missing | 2030 | 1015 | 1015 |
| Polyps, n (%) | |||
| No | 4216 (86) | 2095 (86) | 2121 (87) |
| Yes | 667 (14) | 349 (14) | 318 (13) |
| Missing | 3243 | 1619 | 1624 |
| METS/day3, n (%) | |||
| Q1 | 1312 (24) | 612 (23) | 700 (26) |
| Q2 | 1358 (25) | 667 (25) | 691 (26) |
| Q3 | 1352 (25) | 692 (26) | 660 (24) |
| Q4 | 1374 (25) | 723 (27) | 651 (24) |
| Missing | 2730 | 1369 | 1361 |
| Fruit intake/day4, n (%) | |||
| Q1 | 1307 (25) | 627 (24) | 680 (26) |
| Q2 | 1321 (25) | 668 (25) | 653 (25) |
| Q3 | 1328 (25) | 657 (25) | 671 (25) |
| Q4 | 1350 (25) | 707 (27) | 643 (24) |
| Missing | 2820 | 1404 | 1416 |
| Vegetable intake/day4, n (%) | |||
| Q1 | 1316 (25) | 652 (25) | 664 (25) |
| Q2 | 1312 (25) | 629 (24) | 683 (26) |
| Q3 | 1345 (25) | 699 (26) | 646 (24) |
| Q4 | 1333 (25) | 679 (26) | 654 (25) |
| Missing | 2820 | 1404 | 1416 |
| Red meat intake/day1, n (%) | |||
| Q1 | 1679 (24) | 880 (25) | 799 (23) |
| Q2 | 1733 (25) | 897 (26) | 836 (24) |
| Q3 | 1764 (26) | 835 (24) | 929 (27) |
| Q4 | 1735 (25) | 849 (25) | 886 (27) |
| Missing | 1215 | 602 | 613 |
| Ever regular Aspirin use5, n (%) | |||
| No | 1302 (44) | 614 (41) | 688 (46) |
| Yes | 1690 (56) | 884 (59) | 806 (54) |
| Missing | 5134 | 2565 | 2569 |
| Ever HT use6, n (%) | |||
| No | 2273 (51) | 1083 (48) | 1190 (54) |
| Yes | 2164 (49) | 1150 (52) | 1014 (46) |
| Missing | 3689 | 1830 | 1859 |
| Diabetes1, n (%) | |||
| No | 6475 (91) | 3279 (92) | 3196 (91) |
| Yes | 602 (9) | 271 (8) | 331 (9) |
| Missing | 1049 | 513 | 536 |
all studies except CLUE (NA)
all studies except NYUWHS, CLUE (NA), SCCS (<75% of data available)
all studies except CLUE, PLCO, PHS (NA)
all studies except WHI, CLUE (NA)
all studies except WHI, CLUE, CPSII, NYUWHS (NA)
all studies except HPFS, PHS (NA), CLUE (<75% of data available); women only;
HS = High school; GED = General Educational Development Test; METS = Metabolic Equivalent of Task (measure of physical activity); HT = hormone therapy
In the consortium overall, 40% of controls and 41% of CRC cases were assessed as H pylori sero-positive, resulting in an odds ratio of 1.09 (95% CI: 0.99-1.20). The prevalence of general H pylori sero-positivity varied substantially by race/ethnicity with H pylori sero-prevalence being lowest in whites (33% of controls and 35% of cases) and Asian Americans (39% of controls and 46% of cases) and higher in African Americans (65% of controls and 71% of cases) and Latinos (77% of controls and 74% of cases) (chi-square p-value for difference in prevalence by race <0.0001). The magnitude of the association of H pylori sero-positivity with CRC risk also differed across race/ethnicity. We found no association of H pylori sero-positivity with odds of CRC among whites (OR: 1.06, 95% CI: 0.95-1.18) or Latinos (OR: 0.84, 95% CI: 0.55-1.30), whereas suggestions of increased odds were seen with Asian Americans (OR: 1.30; 95% CI: 0.94-1.81) and African Americans (OR: 1.30, 95% CI: 0.97-1.76) (Table 2), although none reached statistical significance.
Table 2:
Sero-positivity to H pylori proteins and CRC risk, among all and by race/ethnicity.
| All | White | African American | Asian American | Latino | Other/unknown/multiracial | |
|---|---|---|---|---|---|---|
| N, cases/controls | 4063 / 4063 | 3067 / 3067 | 399 / 399 | 307 / 307 | 211 / 211 | 79 / 79 |
| N (%) H pylori+1 | 1682 (41) / 1606 (40) | 1067 (35) / 1027 (33) | 282 (71) / 259 (65) | 140 (46) / 121 (39) | 156 (74) / 163 (77) | 37 (47) / 36 (46) |
| OR (95% CI)2 | 1.09 (0.99-1.20) | 1.06 (0.95-1.18) | 1.30 (0.97-1.76) | 1.30 (0.94-1.81) | 0.84 (0.55-1.30) | 1.05 (0.57-1.94) |
| N (%) H pylori+1 VacA+ | 1434 (35) / 1350 (33) | 876 (29) / 830 (27) | 267 (67) / 233 (58) | 123 (40) / 111 (36) | 134 (64) / 145 (69) | 34 (43) / 31 (39) |
| OR (95% CI)2 | 1.11 (1.01-1.22) | 1.08 (0.96-1.21) | 1.45 (1.08-1.95) | 1.20 (0.85-1.69) | 0.80 (0.53-1.19) | 1.18 (0.62-2.25) |
| N (%) H pylori+1 GroEL+ | 1557 (38) / 1467 (36) | 996 (32) / 957 (31) | 268 (67) / 239 (60) | 114 (37) / 96 (31) | 146 (69) / 143 (68) | 33 (42) / 32 (41) |
| OR (95% CI)2 | 1.11 (1.01-1.22) | 1.06 (0.95-1.19) | 1.39 (1.03-1.87) | 1.32 (0.93-1.87) | 1.07 (0.70-1.64) | 1.06 (0.55-2.01) |
| N (%) H pylori+1 Omp+ | 1400 (34) / 1316 (32) | 870 (28) / 812 (26) | 255 (64) / 240 (60) | 116 (38) / 100 (33) | 127 (60) / 137 (65) | 32 (41) / 27 (34) |
| OR (95% CI)2 | 1.11 (1.01-1.22) | 1.10 (0.98-1.24) | 1.18 (0.88-1.57) | 1.26 (0.90-1.76) | 0.83 (0.56-1.21) | 1.31 (0.69-2.52) |
| N (%) H pylori+1 HcpC+ | 1407 (35) / 1325 (33) | 880 (29) / 847 (28) | 250 (63) / 223 (56) | 114 (37) / 98 (32) | 127 (60) / 128 (61) | 36 (46) / 29 (37) |
| OR (95% CI)2 | 1.10 (1.00-1.22) | 1.06 (0.94-1.19) | 1.33 (1.00-1.77) | 1.27 (0.90-1.79) | 0.98 (0.68-1.43) | 1.39 (0.76-2.55) |
| N (%) H pylori+1 CagA+ | 1139 (28) / 1074 (26) | 645 (21) / 609 (20) | 244 (61) / 221 (55) | 115 (37) / 95 (31) | 104 (49) / 120 (57) | 31 (39) / 29 (37) |
| OR (95% CI)2 | 1.10 (0.99-1.22) | 1.08 (0.95-1.23) | 1.28 (0.96-1.70) | 1.39 (0.97-1.98) | 0.72 (0.49-1.08) | 1.12 (0.58-2.15) |
| N (%) H pylori+1 HP0231+ | 782 (19) / 729 (18) | 552 (18) / 509 (17) | 126 (32) / 105 (26) | 41 (13) / 36 (12) | 51 (24) / 62 (29) | 12 (15) / 17 (22) |
| OR (95% CI)2 | 1.09 (0.98-1.22) | 1.10 (0.97-1.26) | 1.27 (0.94-1.70) | 1.17 (0.71-1.92) | 0.78 (0.51-1.19) | 0.67 (0.30-1.48) |
| N (%) H pylori+1 HP0305+ | 886 (22) / 870 (21) | 565 (18) / 541 (18) | 163 (41) / 161 (40) | 64 (21) / 58 (19) | 73 (35) / 91 (43) | 21 (27) / 19 (24) |
| OR (95% CI)2 | 1.03 (0.92-1.15) | 1.06 (0.93-1.21) | 1.02 (0.77-1.36) | 1.14 (0.76-1.72) | 0.71 (0.48-1.04) | 1.17 (0.54-2.52) |
| N (%) H pylori+1 UreA+ | 883 (22) / 869 (21) | 574 (19) / 577 (19) | 130 (33) / 133 (33) | 74 (24) / 61 (20) | 90 (43) / 78 (37) | 15 (19) / 20 (25) |
| OR (95% CI)2 | 1.02 (0.92-1.14) | 0.99 (0.87-1.13) | 0.97 (0.72-1.29) | 1.28 (0.87-1.89) | 1.27 (0.86-1.89) | 0.74 (0.37-1.47) |
| N (%) H pylori+1 HyuA+ | 802 (20) / 789 (19) | 530 (17) / 533 (17) | 112 (28) / 108 (27) | 57 (19) / 58 (19) | 85 (40) / 74 (35) | 18 (23) / 16 (20) |
| OR (95% CI)2 | 1.02 (0.91-1.14) | 0.99 (0.87-1.14) | 1.06 (0.77-1.46) | 0.98 (0.66-1.46) | 1.26 (0.84-1.88) | 1.15 (0.55-2.43) |
| N (%) H pylori+1 Cad+ | 593 (15) / 582 (14) | 386 (13) / 380 (12) | 119 (30) / 110 (28) | 23 (7) / 29 (9) | 55 (26) / 55 (26) | 10 (13) / 8 (10) |
| OR (95% CI)2 | 1.02 (0.90-1.16) | 1.02 (0.88-1.19) | 1.13 (0.82-1.55) | 0.77 (0.43-1.38) | 1.00 (0.64-1.57) | 1.25 (0.49-3.17) |
| N (%) H pylori+1 Catalase+ | 905 (22) / 910 (22) | 604 (20) / 603 (20) | 145 (36) / 142 (36) | 52 (17) / 59 (19) | 86 (41) / 88 (42) | 18 (23) / 18 (23) |
| OR (95% CI)2 | 0.99 (0.89-1.10) | 1.00 (0.88-1.14) | 1.03 (0.77-1.39) | 0.85 (0.56-1.30) | 0.96 (0.65-1.42) | 1.00 (0.48-2.10) |
| N (%) H pylori+1 HpaA+ | 628 (15) / 638 (16) | 374 (12) / 383 (12) | 140 (35) / 136 (34) | 47 (15) / 45 (15) | 55 (26) / 61 (29) | 12 (15) / 13 (16) |
| OR (95% CI)2 | 0.98 (0.87-1.11) | 0.97 (0.83-1.13) | 1.05 (0.78-1.40) | 1.05 (0.68-1.63) | 0.87 (0.57-1.33) | 0.91 (0.69-2.14) |
| N (%) H pylori+1 NapA+ | 785 (19) / 810 (20) | 501 (16) / 552 (18) | 122 (31) / 115 (30) | 55 (18) / 41 (13) | 89 (42) / 87 (41) | 18 (23) / 15 (19) |
| OR (95% CI)2 | 0.96 (0.86-1.07) | 0.89 (0.78-1.02) | 1.09 (0.80-1.47) | 1.39 (0.91-2.13) | 1.04 (0.70-1.56) | 1.25 (0.59-2.67) |
overall H pylori positive (≥4 antigens positive)
Conditional logistic regression model, controls are matched to cases by age, sex and race/ethnicity; significant associations at p<0.05 are marked in bold font
Sero-positivity to four individual H pylori proteins included in the multiplex serology – VacA, GroEL, Omp, and HcpC – were significantly associated with a 10 to 11% increased odds of CRC in the total consortium. Again, the strength of the association differed between races/ethnicities, with the strongest association of H pylori+ VacA+ with odds of CRC found among African Americans (OR: 1.45, 95% CI: 1.08-1.95). No association of the dichotomous measure of H pylori+ VacA+ and odds of CRC was found among Asian Americans (OR: 1.20, 95% CI: 0.85-1.69), whites (OR: 1.08, 95% CI: 0.96-1.21), or Latinos (OR: 0.80, 95% CI: 0.53-1.19) (Table 2). This race/ethnicity-specific association was generally consistent across different cohorts (Figure 1), although it only reached statistical significance for African Americans in the MEC (OR: 1.91, 95% CI: 1.19-3.20).
Figure 1: Forest plot of sero-positivity to H pylori VacA and CRC risk by race/ethnicitiy and study.
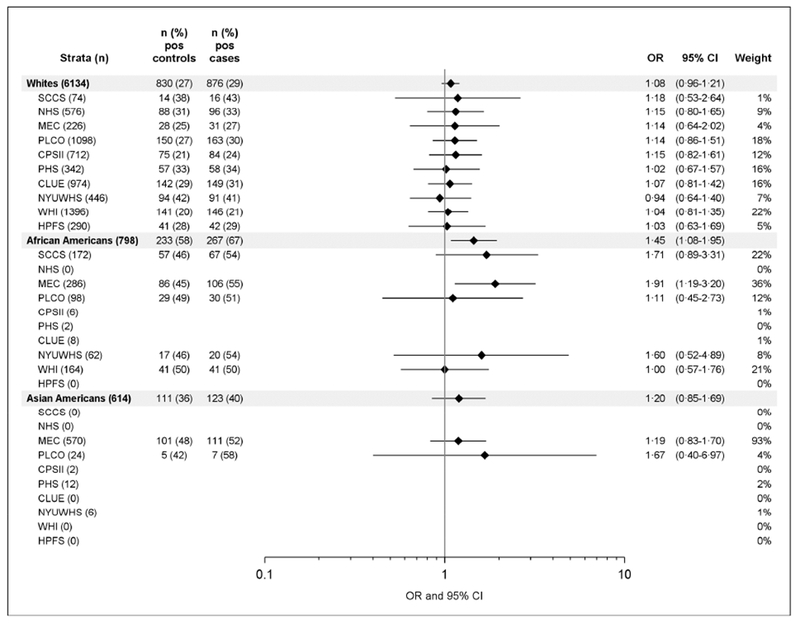
Conditional logistic regression models were applied to determine OR (diamonds) and 95% CI (horizontal lines), controls are matched to cases by study, age, sex and race/ethnicity. No values are given when total case numbers are below 20. Weight shows the contribution of each study to the number of participants per race/ethnicity in %. The vertical line at an OR of 1 serves as a reference for null association. Pos = antibody positive.
In analyses of a dose-response relationship of VacA antibodies in quartiles with CRC risk, we found a significant trend of rising odds of CRC with increasing antibody response to VacA among all races/ethnicities combined (p=0.008) and among African Americans alone (p=0.007). Among all individuals, compared to individuals not H pylori+ VacA+, those in the fourth quartile of antibody response to VacA were at a 25% greater odds of developing CRC (OR: 1.25, 95% CI: 1.07-1.47). This association was particularly strong among Asian Americans (OR: 1.86, 95% CI: 1.06-3.25) and African Americans (OR: 1.70, 95% CI: 1.12-2.58) (Table 3).
Table 3:
CRC risk by strength of antibody response to H pylori VacA, among all and by race
| All | White | African American | Asian American | |||||
|---|---|---|---|---|---|---|---|---|
| N (%) cases / controls | OR (95% CI)4 | N (%) cases / controls | OR (95% CI)4 | N (%) cases / controls | OR (95% CI)4 | N (%) cases / controls | OR (95% CI)4 | |
| H pylori1 or VacA − | 2629 (65) / 2713 ( 67) | 1.00 (ref) | 2191 (71) / 2237 (73) | 1.00 (ref) | 132 (33) / 166 (42) | 1.00 (ref) | 184 (60) / 196 (64) | 1.00 (ref) |
| H pylori+1 VacA+ MFI Q12 | 330 (8) / 338 (8) | 1.02 (0.87-1.20) | 196 (6) / 208 (7) | 0.96 (0.78-1.18) | 58 (15) / 59 (15) | 1.23 (0.81-1.88) | 29 (10) / 28 (9) | 1.12 (0.63-2.00) |
| H pylori+1 VacA+ MFI Q22 | 342 (8) / 337 (8) | 1.06 (0.90-1.25) | 214 (7) / 207 (7) | 1.06 (0.87-1.30) | 65 (16) / 58 (15) | 1.44 (0.94-2.21) | 21 (7) / 28 (9) | 0.80 (0.43-1.49) |
| H pylori+1 VacA+ MFI Q32 | 359 (9) / 338 (8) | 1.11 (0.95-1.30) | 226 (7) / 208 (7) | 1.11 (0.91-1.35) | 67 (17) / 58 (15) | 1.48 (0.97-2.26) | 29 (10) / 28 (9) | 1.12 (0.64-1.96) |
| H pylori+1 VacA+ MFI Q42 | 403 (10) / 337 (8) | 1.25 (1.07-1.47) | 240 (8) / 207 (7) | 1.18 (0.97-1.44) | 77 (19) / 58 (15) | 1.70 (1.12-2.58) | 44 (14) / 27 (9) | 1.86 (1.06-3.25) |
| P(trend)3 | 0.008 | 0.061 | 0.007 | 0.107 | ||||
overall H pylori positive (≥4 antigens positive);
Conditional logistic regression model, controls are matched to cases by age, sex and race/ethnicity;
Cochrane-Armitage trend test ;
race-specific quartile borders based on MFI distribution in controls (All: 25th percentile: 162 MFI, 50th percentile: 404 MFI, 75th percentile: 1153 MFI; white: 25th percentile: 149 MFI, 50th percentile: 354 MFI, 75th percentile: 1009 MFI; African American: 25th percentile: 199 MFI, 50th percentile: 533 MFI, 75th percentile: 1313 MFI; Asian American: 25th percentile: 165 MFI, 50th percentile: 442 MFI, 75th percentile: 1027 MFI); significant associations at p<0.05 are marked in bold font
DISCUSSION
In this large prospective US cohort consortium, we found no overall association of H pylori sero-positivity and odds of colorectal cancer. In line with our pilot data, we did find a significant 11% increased odds of developing CRC for individuals with antibody responses to the H pylori virulence factor VacA. When assessing this association separately by race/ethnicity, we found that the association was particularly strong for African Americans, among whom odds of CRC was significantly elevated by 45% and there was a strong dose-response-relationship between VacA antibody level and the odds of developing CRC. We also found a significant VacA-associated increased odds among Asian Americans, notably those in the highest quartile of VacA antibody level. These findings suggest that serological immune responses to H pylori proteins could serve as a marker for individuals at increased risk of developing CRC, possibly due to differences in strains carried by different ethnic groups as discussed further below.
Previously published findings on the potential association between H pylori and CRC have varied widely, with results seemingly dependent on the racial/ethnic make-up of the study population. For example, four separate US studies reported different results according to the race/ethnicity examined. As mentioned above, our pilot study, in an independent nested case-control population comprised of 77% African Americans within the SCCS, found no association overall between H pylori sero-positivity and CRC, but did find significant associations with VacA (OR: 1.84, 95% CI 1.17-2.89), HP 0231 (OR: 1.74, 95% CI: 1.14-2.66), NapA (OR: 1.67, 95% CI: 1.10-2.53), HcpC (OR: 1.66, 95% CI: 1.13-2.43), and HP 0305 (OR, 1.63, 95% CI: 1.13-2.35).11 In a retrospective clinic-based study of African American patients undergoing bidrectional endoscopy on the same day, individuals with H pylori identified using immunohistochemistry staining on gastric biopsies were at greater odds of also having a colorectal adenoma (OR: 1.5, 95% CI, 1.1-2.0) or colorectal polyp (OR: 1.5, 95% CI, 1.1-2.0).22 In a nested case-control study of colorectal cancer risk among primarily elderly white Americans, no overall association was found with H pylori sero-positivity, although one antigen, GroEL, did reach significance dichotomously (OR: 1.32, 95% CI: 1.03-1.70) but not in a dose-response manner by quartile.23 And, in another retrospective clinic-based study in the US, this time among Hispanic patients undergoing bidirectional endoscopy, no association was found between H pylori found on biopsies and presence of colorectal adenoma.24 In addition, Asian studies have tended to report associations, whereas those in Western European populations have not.6, 7, 25 Our findings reinforce the notion that the association of H pylori with odds of CRC was most pronounced among African Americans, and that the strongest associations were specifically for antibody responses to the H pylori protein VacA, as we did not find strong dose-response associations for the other three antigens found to be associated with odds of CRC dichotomously.
H pylori VacA is a secreted toxin that causes, amongst other effects, vacuolation in host cells.26 While every H pylori strain harbors a vacA gene, the encoded proteins vary in amino acid sequences. Subtypes of VacA proteins differ in avidity of host cell binding, ability to form membrane channels, and ability to induce cell vacuolation, and are associated with different levels of gastric cancer risk.27 Heterogeneity in these vacA variants by race/ethnicity in the US has been explored in one study, which found that African Americans, Latinos, and Asian Americans tend to harbor the most active forms of VacA. This could explain why the association of odds of CRC with antibody responses to VacA detected in African Americans and Asian Americans was stronger as compared to that in whites; however, it would not explain the lack of association in Latinos. Expression of other virulence factors, like CagA, could be an explanation for this observation. Previously, Parsonnet et al. found that African Americans more often harbored CagA-positive strains than whites or Latinos.28 Indeed, in our study sero-prevalence to CagA among CRC cases was 61% among African Americans compared to 49% in Latinos, although overall H pylori sero-prevalence was similar (71% and 74%, respectively). Consequently, the observed race/ethnicity-dependent difference in association of antibody response to H pylori with odds of developing CRC might depend on the infecting strain. Typing of the infecting strain from gastroscopy samples could help to verify this hypothesis, though such samples/information are not available for the participating studies in this consortium. Additionally, genetics of the host, including host markers of inflammation such as polymorphisms in IL-1β, TNF-α, and IL-10, could be considered in this complex interplay of the bacterium and host’s immune response and would add another level of information on the observed race/ethnicity-dependent differences in the association between antibodies to H pylori and odds of CRC.
It remains unclear how H pylori, or as suggested above potentially more virulent strains of the species, could increase risk of developing CRC. It is hypothesized that, if H pylori is causally involved in colorectal carcinogenesis, it could be mediated via direct effects. This may be particularly true for VacA, a known gastric cancer virulence factor, which may also have the potential to exert its effects beyond the stomach, such as through cellular vacuolation, interference with cellular pathways, effects on epithelial permeability, as well as immunomodulatory and pro-inflammatory effects.26 Precedent for the ability of selected H pylori virulence factors to exert their effects beyond the stomach have already been shown in murine models of colitis.29 In contrast to other major H pylori virulence factors such as CagA which requires direct contact between the bacteria and host cells, VacA may disseminate more freely beyond the gastric niche. Antigens GroEl, OMP, and HcpC, which were significantly associated with odds of CRC as well, are reported to be released into the extracellular space, similar to VacA.30,31 Their potential biological function in carcinogenesis, and the reasons why antibody responses to this group of antigens specifically were associated with CRC risk, remain to be elucidated.
Several indirect mechanisms could be considered as well, including H pylori infection and resulting changes in the gastric microbiome potentially leading to an altered intestinal microbiome that is associated with increased CRC risk.32 Furthermore, chronic gastritis as induced by H pylori leads to increased gastrin production, a peptide hormone that functions as a mitogen.33, 34 Several human studies suggest an association between gastrin and CRC risk, although the findings remain inconclusive.35, 36 Another potential mechanism is that H pylori-induced gastric atrophy leads to increased levels of cyclooxygenase-2 and consequently urinary prostaglandin E2, inflammation-related biomarkers that have been identified as associated with colorectal adenoma and CRC risk.37–40 The finding of a dose-response-relationship of antibody response to VacA and odds of CRC supports a connection between severity of infection, inflammation, and resulting molecular consequences with increased CRC risk. It is noteworthy that the highest MFI values were found among African Americans. Furthermore, we have found that antibody responses to H pylori were higher in active infections among a subset of African Americans in the SCCS, as determined by Urea breath test (unpublished data), and decrease after eradication of the bacterium,41 and thus could be seen as a measure for an active infection. The dose-response relationship could therefore indicate that a more pronounced infection and inflammation induced by H pylori more strongly increases the risk of developing CRC. It is possible that VacA itself may not biologically be conferring risk, but rather, due to its high level of immunogenicity, may be a more sensitive biomarker to indicate a pronounced H pylori infection.
A limitation of this study was that potentially informative variables like recent antibiotic use, and diagnosis of gastritis as well as inflammatory bowel disease were missing for the majority of study subjects. These variables could have added information on active versus past H pylori infections and/or eradication of H pylori infection at the time of blood draw as well as on a potential link between H pylori infection and inflammatory diseases of the bowel. In this context, it is important to note that serological analysis of H pylori infection as a systemic measure for past and/or acute infections does not provide information on the current site-specific infection status of the individual. And, while we were able to adjust for the main potential confounders of socio-economic status in terms of education, BMI, and smoking status, finding that although these factors were associated with H pylori status, their adjustment had little impact on the associations between H pylori VacA and odds of colorectal cancer, there was still a significant amount of missing data on potential confounders such as family history of CRC and CRC screening which we were not able to effectively control for. We chose not to present a multiple testing correction with these analyses based on our a priori VacA-specific findings in the pilot data in the SCCS.11 Thus for the VacA analyses we are comfortable using uncorrected p-values. Nonetheless, we appreciate that a Bonferroni-corrected p-value would be 0.0038 (or 0.05/13), and considering this criteria none of the significant associations found when dichotomizing sero-positivity for the other H pylori-specific antigens would be significant. This cohort consortium, however, also has several strengths: as demonstrated, participants in this study arose from diverse populations, with different underlying risk factors for CRC, varying prevalence of H pylori subtype-specific antibodies, and different baseline risks of disease. The heterogeneity of this population allowed for the potential of greater generalizability of the findings as well as the power to explore sub-analyses among more homogeneous populations and disease types.
In conclusion, we assessed the prospective association of a serological immune response to H pylori proteins with the odds of developing CRC in a large US cohort consortium including over 4,000 CRC cases and their matched controls. We demonstrated that strong antibody responses to H pylori virulence factor VacA were significantly associated with odds of CRC and the association appeared to vary by race/ethnicity, with particularly strong associations observed for African Americans and Asian Americans. Future studies are needed to assess whether this association is causal, and if causally related, how H pylori infection, whose natural habitat is the gastric but not colonic mucosa, mechanistically increases CRC risk. In this context, it will also be important to investigate why this association exists only in certain racial/ethnic populations.
Supplementary Material
ACKNOWLEDGEMENTS
CLUE thank the participants and staff for their contributions, as well as the Maryland Cancer Registry, Center for Cancer Surveillance and Control, Department of Health and Mental Hygiene, 201 W. Preston Street, Room 400, Baltimore, MD 21201, http://phpa.dhmh.maryland.gov/cancer, 410-767-4055.
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II cohort. The authors express sincere appreciation to all Cancer Prevention Study-II participants, and to each member of the study and biospecimen management group. The authors would like to acknowledge the contribution from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries and cancer registries supported by the National Cancer Institute’s Surveillance Epidemiology and End Results Program.
Grant Support: The funding sources played no role in the analysis or interpretation of the data.
The National Cancer Institute funds: this consortium (R01 CA190428, PI: Epplein); the Southern Community Cohort Study (U01 CA202979, PI: Blot); the NYU Women’s Health Study (UM1 CA182934, PI: Zeleniuch-Jaquotte; P30 CA016087, PI: Neel); the NHS/HPFS (U01 CA167552; P01 CA087969; UM1 CA186107; UM1 CA167552); the PHS (R01 CA097193; R01 CA040360; R01 HL034595); and the MEC (U01 CA164973, PI: Le Marchand).
R.M. Peek is supported by R01 DK058587 and R01 CA077955.
T.L. Cover is supported by NIH R01 AI039657, R01 AI118932, P01 CA116087 and the Department of Veterans Affairs BX000627.
M. Song is supported by the American Cancer Society (Grant number MRSG-17-220-01 – NEC to M.S.); the 2017 AACR-AstraZeneca Fellowship in Immuno-oncology Research (Grant Number 17-40-12-SONG to M.S.); the U.S. National Institutes of Health (NIH) (K99 CA215314 to M.S.).
Matthew G. Varga is supported by NIH T32 CA057726.
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts, HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C.
The development of H pylori multiplex serology was funded in part by the Joint Initiative for Innovation and Research of the German Helmholtz Association.
Abbreviations:
- BMI
body mass index
- CagA
Cytotoxin-associated gene A
- CI
confidence interval
- CRC
colorectal cancer
- HT
hormone therapy
- METs
metabolic equivalents
- MFI
median fluorescence intensity
- OR
odds ratio
- VacA
Vacuolating cytotoxin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Study sponsor had no role in the study design or collection, analysis, or interpretation of the data. Authors have nothing to disclose.
Transcript Profiling: NA
Writing Assistance: NA
Author names in bold designate shared co-first authorship.
REFERENCES
- 1.Plummer M, de Martel C, Vignat J, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 2016;4:e609–16. [DOI] [PubMed] [Google Scholar]
- 2.Hooi JKY, Lai WY, Ng WK, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017;153:420–429. [DOI] [PubMed] [Google Scholar]
- 3.Grad YH, Lipsitch M, Aiello AE. Secular Trends in Helicobacter pylori Seroprevalence in Adults in the United States: Evidence for Sustained Race/Ethnic Disparities. Am J Epidemiol 2012;175:54–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 5.de Korwin JD, Ianiro G, Gibiino G, et al. Helicobacter pylori infection and extragastric diseases in 2017. Helicobacter 2017;22 Suppl 1. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Wang X, Wang Y. Helicobacter pylori infection and colorectal carcinoma risk: A meta-analysis. J Cancer Res Ther 2016;12:15–18. [DOI] [PubMed] [Google Scholar]
- 7.Zumkeller N, Brenner H, Zwahlen M, et al. Helicobacter pylori infection and colorectal cancer risk: a meta-analysis. Helicobacter 2006;11:75–80. [DOI] [PubMed] [Google Scholar]
- 8.Blaser MJ. The biology of cag in the Helicobacter pylori-human interaction. Gastroenterology 2005;128:1512–5. [DOI] [PubMed] [Google Scholar]
- 9.Cover TL. Helicobacter pylori Diversity and Gastric Cancer Risk. MBio 2016;7:e01869–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epplein M, Signorello LB, Zheng W, et al. Race, African ancestry, and Helicobacter pylori infection in a low-income United States population. Cancer Epidemiol Biomarkers Prev 2011;20:826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epplein M, Pawlita M, Michel A, et al. Helicobacter pylori Protein-Specific Antibodies and Risk of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 2013;22:1964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol 2000;151:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei EK, Giovannucci E, Fuchs CS, et al. Low plasma adiponectin levels and risk of colorectal cancer in men: a prospective study. J Natl Cancer Inst 2005;97:1688–94. [DOI] [PubMed] [Google Scholar]
- 14.Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer 2005;5:388–96. [DOI] [PubMed] [Google Scholar]
- 15.Lee JE, Wei EK, Fuchs CS, et al. Plasma folate, methylenetetrahydrofolate reductase (MTHFR), and colorectal cancer risk in three large nested case-control studies. Cancer Causes Control 2012;23:537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toniolo PG, Levitz M, Zeleniuch-Jacquotte A, et al. A prospective study of endogenous estrogens and breast cancer in postmenopausal women. J Natl Cancer Inst 1995;87:190–7. [DOI] [PubMed] [Google Scholar]
- 17.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 18.Huang HY, Alberg AJ, Norkus EP, et al. Prospective study of antioxidant micronutrients in the blood and the risk of developing prostate cancer. Am J Epidemiol 2003;157:335–44. [DOI] [PubMed] [Google Scholar]
- 19.Calle EE, Rodriguez C, Jacobs EJ, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer 2002;94:500–11. [DOI] [PubMed] [Google Scholar]
- 20.Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials 2000;21:349S–355S. [DOI] [PubMed] [Google Scholar]
- 21.Michel A, Waterboer T, Kist M, et al. Helicobacter pylori multiplex serology. Helicobacter 2009;14:525–535. [DOI] [PubMed] [Google Scholar]
- 22.Brim H, Zahaf M, Laiyemo AO, et al. Gastric Helicobacter pylori infection associates with an increased risk of colorectal polyps in African Americans. BMC Cancer 2014;14:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blase JL, Campbell PT, Gapstur SM, et al. Prediagnostic Helicobacter pylori Antibodies and Colorectal Cancer Risk in an Elderly, Caucasian Population. Helicobacter 2016;21:488–492. [DOI] [PubMed] [Google Scholar]
- 24.Patel S, Lipka S, Shen H, et al. The association of H. pylori and colorectal adenoma: does it exist in the US Hispanic population? J Gastrointest Oncol 2014;5:463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H, Park JJ, Park YM, et al. The association between Helicobacter pylori infection and the risk of advanced colorectal neoplasia may differ according to age and cigarette smoking. Helicobacter 2018:e12477. [DOI] [PubMed] [Google Scholar]
- 26.McClain MS, Beckett AC, Cover TL. Helicobacter pylori Vacuolating Toxin and Gastric Cancer. Toxins (Basel) 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira RM, Machado JC, Figueiredo C. Clinical relevance of Helicobacter pylori vacA and cagA genotypes in gastric carcinoma. Best Pract Res Clin Gastroenterol 2014;28:1003–15. [DOI] [PubMed] [Google Scholar]
- 28.Parsonnet J, Replogle M, Yang S, et al. Seroprevalence of CagA-positive strains among Helicobacter pylori-infected, healthy young adults. J Infect Dis 1997;175:1240–2. [DOI] [PubMed] [Google Scholar]
- 29.Luther J, Owyang SY, Takeuchi T, et al. Helicobacter pylori DNA decreases pro-inflammatory cytokine production by dendritic cells and attenuates dextran sodium sulphate-induced colitis. Gut 2011;60:1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn BE, Vakil NB, Schneider BG, et al. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun 1997;65:1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snider CA, Voss BJ, McDonald WH, et al. Growth phase-dependent composition of the Helicobacter pylori exoproteome. J Proteomics 2016;130:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noto JM, Peek RM Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog 2017;13:e1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konturek SJ, Konturek PC, Hartwich A, et al. Helicobacter pylori infection and gastrin and cyclooxygenase expression in gastric and colorectal malignancies. Regul Pept 2000;93:13–9. [DOI] [PubMed] [Google Scholar]
- 34.Sobhani I, Lehy T, Laurent-Puig P, et al. Chronic endogenous hypergastrinemia in humans: evidence for a mitogenic effect on the colonic mucosa. Gastroenterology 1993;105:22–30. [DOI] [PubMed] [Google Scholar]
- 35.Strofilas A, Lagoudianakis EE, Seretis C, et al. Association of helicobacter pylori infection and colon cancer. J Clin Med Res 2012;4:172–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorburn CM, Friedman GD, Dickinson CJ, et al. Gastrin and colorectal cancer: a prospective study. Gastroenterology 1998;115:275–80. [DOI] [PubMed] [Google Scholar]
- 37.Kountouras J, Zavos C, Chatzopoulos D, et al. New aspects of Helicobacter pylori infection involvement in gastric oncogenesis. J Surg Res 2008;146:149–58. [DOI] [PubMed] [Google Scholar]
- 38.Cai Q, Gao YT, Chow WH, et al. Prospective study of urinary prostaglandin E2 metabolite and colorectal cancer risk. J Clin Oncol 2006;24:5010–6. [DOI] [PubMed] [Google Scholar]
- 39.Bezawada N, Song M, Wu K, et al. Urinary PGE-M levels are associated with risk of colorectal adenomas and chemopreventive response to anti-inflammatory drugs. Cancer Prev Res (Phila) 2014;7:758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davenport JR, Cai Q, Ness RM, et al. Evaluation of pro-inflammatory markers plasma C-reactive protein and urinary prostaglandin-E2 metabolite in colorectal adenoma risk. Mol Carcinog 2016;55:1251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, Zhang Y, Su H, et al. Helicobacter pylori antibody responses in association with eradication outcome and recurrence: a population-based intervention trial with 7.3-year follow-up in China. Chin J Cancer Res 2017;29:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


