Abstract
ColE1-like plasmid vectors are widely used for expression of recombinant genes in E. coli. For these vectors, segregation of individual plasmids into daughter cells during cell division appears to be random, making them susceptible to loss over time when no mechanisms ensuring their maintenance are present. Here we use the plasmid pGFPuv in a recA relA strain as a sensitized model to study factors affecting plasmid stability in the context of recombinant gene expression. We find that in this model, plasmid stability can be restored by two types of genetic modifications to the plasmid origin of replication (ori) sequence: point mutations and a novel 269 nt duplication at the 5’ end of the plasmid ori, which we named DAS (Duplicated Anti-Sense) ori. Combinations of these modifications produce a range of copy numbers and of levels of recombinant expression. In direct contradiction with the classic random distribution model, we find no correlation between increased plasmid copy number and increased plasmid stability. Increased stability cannot be explained by reduced levels of recombinant gene expression either. Our observations would be more compatible with a hybrid clustered and free distribution model, which has been recently proposed based on detection of individual plasmids in vivo using super-resolution fluorescence microscopy. This work suggests a role for the plasmid ori in the control of segregation of ColE1 plasmids that is distinct from replication initiation, opening the door for the genetic regulation of plasmid stability as a strategy aimed at enhancing large-scale recombinant gene expression or bioremediation.
Keywords: Plasmid stability, recombinant expression, high copy number plasmid, plasmid segregation, origin of replication, antisense RNA, ColE1 plasmid, bioremediation, biotechnology
Introduction
Plasmids are self-replicating, extrachromosomal genetic elements found across a variety of microbial taxa. They facilitate horizontal gene transfer and microbial adaptation to specific niches by providing genes contributing to virulence, resistance to antibiotics, resistance to heavy metals, etc. (Frost et al., 2005; Smalla et al., 2015; Thomas and Nielsen, 2005).
The accurate segregation of plasmids into two separate daughter cells following cell division depends largely on autonomous (chromosome segregation-independent) mechanisms, but segregation strategies differ depending on the copy number of the plasmids involved: low-copy number plasmids largely rely on active partitioning mechanisms (Gerdes et al., 2010; Salje, 2010), whereas the segregation of high copy number (hcn) plasmids appears to be random (Cooper et al., 1987; De Gelder et al., 2007; Ponciano et al., 2007).
Several mathematical models have been put forth to describe the dynamics of plasmid loss for hcn plasmids. Plasmid copy number, replication efficiency, post-segregational killing, and an effective decrease in partition units due to concatemerization or clustering are all variables that have been considered (Bergstrom et al., 2000; Cooper et al., 1987; De Gelder et al., 2007; Lau et al., 2013; Lili et al., 2007; San Millan et al., 2014; Werbowy et al., 2017). Given that naturally-occurring plasmids typically experience fluctuating selective pressures and would be susceptible to loss over long periods of time, these plasmids typically have mechanisms modulating one or more of these variables to enhance their stability (Li et al., 2015; Lobato-Marquez et al., 2016; Werbowy and Kaczorowski, 2016). These stabilizing mechanisms include plasmid addiction systems that kill plasmidless daughter cells (Kroll et al., 2010; Li et al., 2015; Mruk and Kobayashi, 2014; Werbowy and Kaczorowski, 2016), multimer resolution systems to suppress the formation of concatemers by homologous recombination (Summers et al., 1993; Werbowy et al., 2015), and the presence of multiple redundant mechanisms for replication initiation (Cesareni et al., 1991).
Derivatives of hcn plasmids are used as vectors for recombinant gene expression because plasmid copy number is largely proportional to the level of recombinant gene expression. These vectors frequently carry a minimized ori sequence, without natural addiction or multimer resolution systems, and sometimes even lacking primosome assembly sites, making these plasmids especially unstable. Plasmid loss can be exacerbated by an increase of empty tRNAS associated with expression of recombinant genes because of differences in codon usage between the recombinant sequence that of the host’s. The presence of empty tRNAs can interfere with antisense RNA regulation, a key regulatory element that helps plasmids maintain a stable plasmid copy number because anticodons can hybridize with the stem-loops in early pre-primer ori RNA, blocking antisense RNA binding (Wang et al., 2002; Wang et al., 2006; Yavachev and Ivanov, 1988). As a result, plasmid instability is one of the main factors limiting the efficiency of large-scale recombinant gene expression or bioremediation, applications where the use of selectable markers is impractical (Grabherr and Bayer, 2002; Kroll et al., 2011).
Here we use pUC19, which is a derivative of the ColE1 plasmid ori pMB1, as a model to study the stability of plasmids in the context of recombinant gene expression. pUC replication starts with transcription of origin of replication (ori) sequence, generating an RNA pre-primer known as RNAII. The 3’ end of RNAII forms a stable hybrid with ori DNA (R-loop), guided by complementary GC-rich stretches. RNaseH processing of R-loop RNA generates a free 3’–OH terminus that is extended by DNA polymerase I (Pol I), initiating leading-strand synthesis (Itoh and Tomizawa, 1980). The nascent leading strand remains hybridized to its template, forming a D-loop. Unwinding of ori duplex DNA also uncovers a hairloop initiation signal on the lagging-strand, known as primosome assembly site or pas. Both D-loops and pas recruit PriA, leading to recruitment of the host replisome machinery and to the coordinated replication of both strands (reviewed in (Lilly and Camps, 2015)).
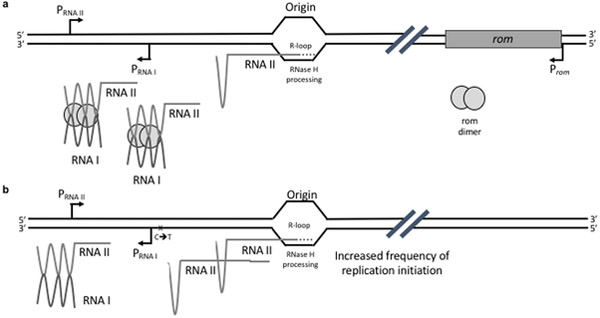
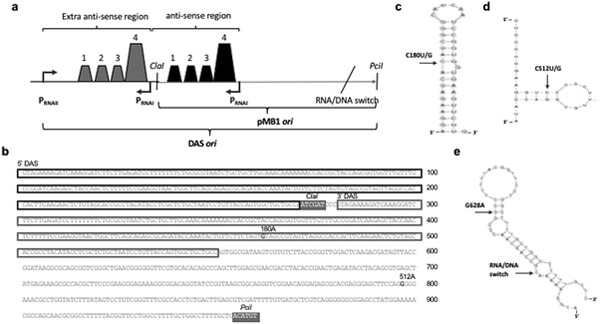
Plasmid copy number is largely regulated at the level of replication initiation through an antisense RNA mechanism, which maintains copy number at a constant level for a given physiological condition (reviewed in (Brantl, 2014; Camps, 2010; Polisky, 1988)) (Fig. 1). The 108nt antisense RNA (RNAI) is transcribed from a strong promoter located in the opposite orientation from the sense (RNAII) promoter. RNAI is short-lived and forms a stable hybrid with the nascent RNAII, blocking downstream R-loop formation by altering the folding of the RNAII (Cesareni et al., 1991; Polisky, 1988). Suppression of RNAII’s replication initiation by RNAI’s “action at a distance” depends on hybridization of three complementary stem-loop structures (SL1–3) that are present both within antisense RNAI and within pre-primer RNAII, leading to the formation of a fourth stem-loop in RNAII (SL4), which prevents R-loop formation. RNAII transcription past the RNAI promoter results in the formation of an additional stem-loop, making the RNAII refractory to RNAI hybridization. Two predicted hairpins toward the 3’ end of ori sequence mediate R-loop formation and recruitment of RNaseH (reviewed in (Camps, 2010)). Mutations within all these functional elements of the ori sequence have been shown to modulate plasmid copy number and compatibility properties, presumably through alterations in antisense RNA regulation (Camps, 2010; Million-Weaver et al., 2012; Polisky et al., 1990).
Fig. 1. ColE1-like plasmid copy number regulation by antisense RNA.
a. Original pMB1 plasmid. b. pUC19 origin of replication present in pGFPuv. ColE1 plasmid replication is regulated by sequence elements known as the plasmid origin of replication (ori). Replication starts with the transcription of an RNA pre-primer. This RNA pre-primer (RNAII) forms a stable hybrid at its 3’ end with ori DNA (R-loop). Processing of this R-loop RNA by RNaseH generates a free 3’–OH end that is extended by DNA polymerase I (Pol I), initiating leading-strand synthesis (Itoh and Tomizawa, 1980). Replication initiation is negatively regulated by an antisense RNA mechanism that maintains copy number constant for a given physiological condition (Brantl, 2014; Camps, 2010). The 108 bp antisense RNA (RNAI) is transcribed from a strong promoter located downstream and in the opposite orientation from the promoter for RNAII. RNAI is short-lived and forms a stable hybrid with the pre-primer RNA, blocking downstream R-loop formation by irreversibly altering the folding of the pre-primer (Cesareni et al., 1991; Polisky, 1988). In the original pMB1 origin of replication, the hybridization between RNAI and RNAII is facilitated and stabilized by a small dimeric protein, Rom (for RNA one modulator) (Lacatena et al., 1984; Tomizawa and Som, 1984). pUC19 is a pMB1-derivative lacking rom and harboring a C→T mutation in the ori, immediately downstream of the RNAI promoter, leading to a G→A substitution in the RNAII. This mutation appears to alter the secondary structure of pUC19’s RNAII, promoting normal folding of the pre-primer. As a result, pUC19 exhibits a several-fold increase in plasmid copy number relative to its parental pMB1 (Lin-Chao et al., 1992).
In the original pMB1 ori, the hybridization between RNAI and nascent RNAII is facilitated and stabilized by a small dimeric protein, Rom (for RNA one modulator) (Lacatena et al., 1984; Tomizawa and Som, 1984) (Fig. 1a). The pMB1 derivative that we use in the present work (pUC19) lacks rom (Vieira and Messing, 1982) and has a C→T mutation immediately downstream of the RNAI promoter (Lin-Chao et al., 1992). The resulting G→A mutation in RNAII appears to alter its secondary structure, promoting normal folding of this pre-primer RNA (Fig. 1b). As a consequence, pUC19 exhibited a several-fold increase in plasmid copy number relative to its parental pMB1 (Lin-Chao et al., 1992).
Here we describe a hcn plasmid model system that is unusually unstable likely due to inefficient plasmid segregation. We find that in this system, plasmid stability is under the control of plasmid origin of replication (ori) sequence. Specifically, two point-mutations, and a novel 269 nt duplication at the 5’ end of the plasmid ori (which we called Duplicated Anti-Sense or DAS ori) synergize to restore plasmid stability. In direct contradiction with a purely random distribution model, we found that these stabilizing effects do not consistently correlate with plasmid copy number. Our observations are more compatible with a hybrid clustered- and free-distribution model, which has been recently proposed based on detection of individual plasmids at the single-cell level using super-resolution fluorescence microscopy (Wang, 2017; Wang et al., 2016). This work suggests a role for the plasmid ori in the control of segregation of ColE1 plasmids that is distinct from replication initiation and thus opens the door for the genetic regulation of plasmid stability as a strategy aimed at enhancing large-scale recombinant gene expression or bioremediation.
Experimental Procedures
Strains and plasmids.
Hosts were Top10 [F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG ] cells, a commercial (Invitrogen) strain frequently used for cloning that is endA1 (deficient in a non-specific endonuclease) and recA (deficient in recombinase). All engineered variants are derivatives of pGFPuv (Clontech Laboratories), which contains pUC (a ColE1-like origin of replication), TEM β-lactamase as a selectable marker, and “cycle 3 GFP” (a GFP mutant selected for improved fluorescence in E. coli) (Crameri et al., 1996; Standley et al., 2017) under lac promoter control as a reporter for expression. The sequence for this plasmid can be found in (Standley et al., 2017).
Plasmid transformation.
Bacteria were transformed by the heat-shock method previously described in detail in (Standley et al., 2017). Briefly cells were made competent by resuspension in CaCl2, mixed with DNA at 4°C and heat-shocked in a water bath at 42°C for two minutes, allowed to recover, and plated in the presence of 100 μg/ml of carbenicillin.
β-lactamase marker retention.
Cells from a fresh transformation were washed directly from plates containing between 100 and 5000 colonies with 2 ml LB media, normalized to OD=1 by OD600 absorbance, and diluted 1:105 into LB containing carbenicillin. Note: for unknown reasons, the cell density of the first transformation is critical: semi-lawns produce the strongest plasmid loss in WT pGFPuv. Cells were expanded to saturation in the presence of carbenicillin (passage 0), and 1:105 dilutions were serially passaged for 22h in 4 ml of LB at 37˚C in the absence of carbenicillin selection (passages 1–13). To determine the fraction of plasmids lost at different passages, cells were withdrawn, serially diluted and plated in the presence or absence of carbenicillin. Colonies arising after 16 hours of incubation at 37°C were counted. Results are expressed as the percentage of viable cells retaining carbenicillin resistance.
Plasmid copy number quantification.
Relative plasmid copy numbers were determined based upon fluorescence intensity of agarose gel bands corresponding to linearized plasmid DNA harvested from identical numbers of cells. Three colonies were picked from a fresh transformation and grown to saturation in LB broth containing 100 μg/ml of carbenicillin. The cultures were normalized according to OD600 absorbance in order to start with constant dry weight of cells. Plasmids were prepped from these normalized suspensions using a plasmid miniprep kit (Machery-Nagel, cat. #740499.250) and 10 μl of prep were linearized with the DpnI enzyme, which has a single recognition site on the pGFPuv plasmid. Linear fragments were separated in a 1% agarose gel, stained with Ethidium Bromide, and visualized using a UVP BioSpectrum 300 LM26E transilluminator equipped with a EtBr filter (570–640 nm). The shutter speed was set to 126 ms. Camera aperture and image contrast were adjusted so that none of the fluorescent bands saturated the image. Band intensity was measured by densitometry using Image J software, with fixed area size for all the samples. The results were expressed as fold increase relative to background.
Flow cytometry analysis.
Cultures were washed into Ca/Mg-free sheath buffer (Phosphate Buffered Saline and 1mM EDTA) and analyzed with a Cytopeia Influx cytometer for GFP fluorescence using 200 mW Coherent 488 laser, 531/40 PMT (constant for all samples). Fluorescence emission and scatter for 250,00 single cells was analyzed by flow cytometry. Single, viable cells were gated based upon forward and side scatter parameters. The subpopulation of plasmidless cells was determined based on the fluorescence emission profile of an untransformed control and conformed by sorting and plating 1000 and 100 cells on LB only (viability control) and on LB with carbenicillin (as an indicator of plasmid retention). Cells that carried no plasmid corresponded to a relative fluorescence intensity of <1.05. The distribution of GFP fluorescence (centered in the geometric mean to allow comparison) was then used to estimate the distribution of plasmid copy number in the population. GFP fluorescence has been shown to be a good indicator of copy number for ColE1 plasmids bearing “cycle 3 GFP” (Million-Weaver et al., 2012). GFP expression was maintained without IPTG induction due the leaky nature of the lac promoter.
Determination of GFP expression by fluorescence.
The impact of several pGFPuv ori modifications on GFP expression was also determined by measuring fluorescence (Million-Weaver et al., 2012). Plasmids with either Q183R OR L18_Stop GFP were also included as non-fluorescent negative controls to establish the background (not shown). After transformation of Top10 cells, ten individual colonies were selected from each transformation, and grown overnight in LB broth media containing carbenicillin at 37 °C with shaking (MaxQ 4000 shaker/incubator, Thermo Scientific). Following incubation, 200 μl of each culture was transferred to a black-walled 96-well microtiter plate (Greiner, cat #655087), and fluorescence (ex: 395 nm, em: 509 nm, autocutoff: 495 nm) and OD600 were measured in tandem on a Spectramax M2e plate reader (Molecular Devices). Settings for the instrument included endpoint read, with 10 seconds of mixing prior to the read (6 reads per well). Fluorescence readings were normalized by OD600 measurements prior to being reported.
Western blot quantification.
GFP expression from several of our engineered plasmid ori variants was also measured directly by Western blot, in duplicate. Top10 cells transformed with these plasmids were plated on LB agar containing carbenicillin to have a final colony density of 1000–3000 colonies/plate. Following incubation at 37 °C for 16 hours, colonies were harvested from the plates into 2 mL of LB broth containing carbenicillin. Cells were then lysed by employing five 15 second bursts with a probe sonicator (allowing cells to cool on ice for one minute between bursts). Cell lysates were then spun at 20,000xg for five minutes, and the soluble fraction (supernatant) was collected. Total protein was quantified with the Pierce BCA Protein Assay Kit (Thermo Scientific, cat# 23225). A total of 15 ug of protein was loaded onto a 12% SDS-Page gel, and run at 60V for 30 minutes, followed by 120V for 1.5 hours. Protein from the gel was transferred to a PVDF membrane at 35V overnight in a cold room (wet transfer method). The following day, the membrane was blocked with 5% nonfat dry milk in PBST, and hybridized with mouse monoclonal anti-GFP (R&D Systems, cat# 454505) in 3% nonfat dry milk in PBST. Following 3×10min washes with PBST, the membrane was hybridized with HRP-conjugated anti-mouse IgG (Promega, cat# W402B). Following 3×10min washes with PBST, the blot was developed with Pierce ECL western blotting substrate (Themo Scientific, cat# 32209). Blots were visualized using a BioRad ChemiDoc XRS+ System and individual band densities were quantified by densitometry using BioRad ChemiDoc XRS+’s Image Lab image acquisition and analysis software.
Results
A model system to study determinants of hcn plasmid stability
We made the observation that when Top10 cells are transformed with the plasmid pGFPuv, this plasmid is very unstable. pGFPuv bears a pUC19 (ColE1-like) origin of replication, TEM β-lactamase, and a cycle3 GFP reporter gene (Crameri et al., 1996) under control of the Plac promoter. Top 10 is a commercial strain engineered to facilitate recombinant expression (see Methods). Specifically, this strain has two genetic alterations which help maintain a high plasmid copy number under conditions of prolonged culture: ΔrelA (preventing induction of the stringent response) and ΔrecA (preventing recombination).
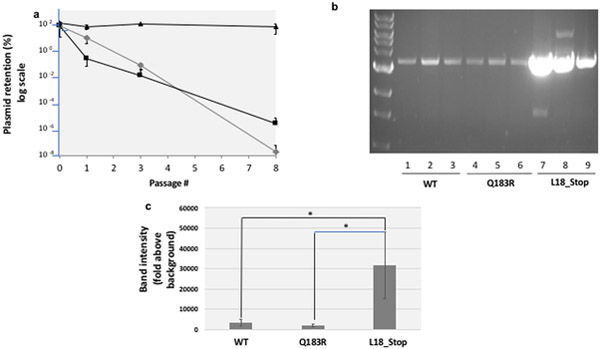
Cells transformed with our reporter plasmid were washed from plates, grown to stationary phase, diluted into fresh medium containing carbenicillin, and grown until they reached saturation. Following one, two and eight passages (with each passage representing roughly 30 generations) in the absence of antibiotic, we quantified percent plasmid retention by determining the number of colonies that remained sensitive to carbenicillin. Fig. 2 shows that the pGFPuv plasmid was rapidly lost in the absence of antibiotic selection (Fig. 2, grey diamonds). Following three passages, fewer than 0.01% of cells within the population retained the plasmid; after 8 passages, that number had dropped to less than 10−6% of cells. We looked to see whether this instability was due to toxicity associated with fluorescence by expressing a GFP mutation (Q183R), which abolishes GFP fluorescence, and found that absence of GFP fluorescence did not make a significant difference (Fig. 2, black squares). Finally, we looked to see whether instability could be attributed to recombinant gene expression and we found that indeed it was: placing a stop codon at position 18 of GFP completely stabilized pGFPuv in Top10 cells (Fig. 2, black triangles).
Fig. 2. The pGFPuv retention model of plasmid stability.
a. Plasmid stability. Cells transformed with plasmids harboring wild-type pGFPuv (grey diamonds), a Q183R mutant of GFP (which abolishes fluorescence, black squares), and a L18* truncated mutant (black triangles) were washed from plates with carbenicillin, normalized for cell number and expanded in liquid culture in the presence of carbenicillin. After the first passage (passage 0), these cells were passaged 8 more times through a 1:105 dilution and growth for 22h in LB at 37˚C in the absence of carbenicillin. To monitor plasmid retention, at specific time-points cells were withdrawn, serially diluted and plated in the presence or absence of carbenicillin. CFUs arising after 16 hours incubation at 37˚C were quantified. b Plasmid copy number. Plasmids from a normalized number of cells bearing the WT pGFPuv plasmid, the Q183R (non-fluorescent) mutant and the L18_Stop (truncated) mutant were recovered, linearized, and run on an agarose gel as described in the Methods. c Gel band density quantification. Band intensities in the gel shown in section b were measured by densitometry using Image J software, with fixed area size for all the samples, and averaged. Error bars represent the standard deviation for three replicates. The difference between L18_Stop and WT and between L18_Stop and Q183R was confirmed by a 2-tailed unpaired Student t test (p<0.05) (asterisks).
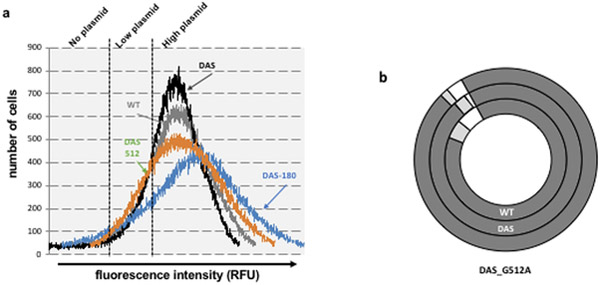
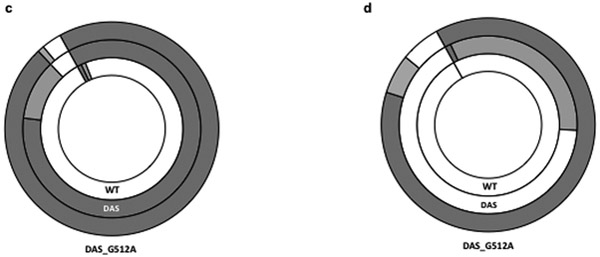
We confirmed these results using GFP fluorescence in individual cells as a readout for plasmid retention. Baseline GFP fluorescence in the absence of inducer arises due to leaky expression from the Plac promoter, which should occur at a relatively consistent level across a population. We again transformed cells with the pGFPuv plasmid, washed plates into fresh medium, then proceeded to monitor fluorescence following different passages. In the presence of carbenicillin, fluorescence emission followed a normal Gaussian distribution, centered on the mean (Fig. 3a, grey line). We next measured the fluorescence emission and side-scatter properties of untransformed cells and used parameters from the control population to define three fractions: designated as “no plasmid”, “low plasmid”, and “high plasmid”. We have previously shown a good correlation between plasmid copy number and GFP expression (Million-Weaver et al., 2012). We used fluorescence measurements of FACS-sorted cells as an indicator of plasmid copy number in individual cells, with the understanding that this is likely a simplification because of the compounded effects of heterogeneous GFP transcription and translation in individual cells (Gustafsson et al., 2012; Munsky et al., 2012; Welch et al., 2009). The results of passaging our cultures in the absence of antibiotic indeed paralleled the results of our marker retention experiments. While most cells classify into the “high plasmid” category in the presence of carbenicillin (Fig. 3b, inner circle), after a single passage in the absence of carbenicillin, 98% of cells sorted into the “no plasmid” category (Fig. 3c, inner circle). After four passages without selection, we were unable to detect cells still displaying GFP fluorescence, indicating that the overwhelming majority of the population had lost the plasmid by then (Fig. 3d, inner circle).
Fig. 3. Plasmid copy number distribution and plasmid retention by flow cytometry.
a. Fluorescence emission distribution in the presence of antibiotic, as an approximate indication of plasmid copy number distribution in the population. Cells transformed with WT pGFPuv (WT, grey line), DAS-pGFPuv (DAS, black line), pGFPuv DAS-G180A (DAS180A, blue line), or pGFPuv DAS-G512A (DAS512, red line), were grown in the presence carbenicillin and analyzed by flow cytometry. Fluorescence emission and scatter for 250,000 single cells was analyzed by flow cytometry as described in Methods. All distributions were centered in the geometric mean to allow comparison. Three subpopulations were defined based on the fluorescence emission profile as described in Methods. The results show one representative experiment. b. Plasmid retention. Cells carrying the WT pGFPuv plasmid (WT), the pGFPuv-DAS plasmid (DAS) or the pGFPuv-DAS plasmid with the G512A substitution (DAS_G512A) were grown in the presence of carbenicillin, analyzed by flow cytometry, and placed into one of the three categories following the parameters defined for the control: no plasmid (white), low plasmid (light grey), or high plasmid (dark grey). c. Same analysis performed after a single passage in the absence of carbenicillin as described in the legend for Fig. 2 and in Methods. d. Same analysis performed after four passages in the absence of carbenicillin as described in the legend for Fig. 2 and in Methods.
Plasmid instability is likely due to a defect in segregation
We next investigated the mechanism of plasmid instability by looking at plasmid copy number and also by looking at retention in the presence of drug.
The rationale for looking at plasmid copy number is that, according to the random distribution model, low plasmid copy numbers increase the chances of segregation of plasmid-less daughter cells. Fig. 2b shows the plasmid content for cells transformed with pGFPuv (lanes 1,2,3), pGFPuv-Q183R (lanes 4,5,6), and pGFP-uv L18_Stop (lanes 7,8,9), with the corresponding band intensity quantification in Fig. 2c. These gels indicate that GFP expression (regardless of whether the protein is functional or not) is associated with a substantial decrease in pGFPuv plasmid copy number.
To see whether the observed decrease in plasmid number leads to an increased production of daughterless cells during segregation, we looked to see the fraction of plasmidless cells that we could detect in the presence of carbenicillin. Under these conditions, plasmidless cells would not have had an opportunity to expand, even if the loss of plasmid provides them with a selective advantage so they have to represent plasmidless daughter cells. We did find that a fraction of cells (~3%) grown in the presence of carbenicillin sorted into the “no plasmid” category (Fig 3b, inner circle). To confirm that these were indeed plasmidless, viable cells, we sorted 50,000 GFP-negative cells and then plated 100 and 1000 of these cells on LB (as a control for viability) or on LB + carbenicillin (to monitor plasmid retention). 50–60% of the plated cells were viable, and only 5 to 10% of these were resistant to carbenicillin, confirming both our fluorescence-based classification and the presence of a significant fraction of plasmidless cells in cultures grown in the presence of carbenicillin. Thus, using FACS sorting we were able to identify a population of cells that are plasmidless in the presence of a positive selection for plasmid maintenance. We also noted that when we plate cells bearing the WT plasmid grown in the presence of carbenicillin, we observe fewer colonies on carb plates than on LB plates (a 35% decrease on average, see complete statistical analysis further below).
Overall, then, it seems that GFP expression leads to a substantial decrease in plasmid copy number and likely also to poor segregation, both of which could be major contributors to plasmid instability.
Modulation of plasmid copy number does not correlate with plasmid stability
To check whether decreased plasmid copy number was indeed contributing to plasmid instability, we introduced two mutations that modulate plasmid copy number. The first mutation is a G→A transition at position 180 of the plasmid ori according to ColE1 numbering (Ohmori and Tomizawa, 1979), resulting in a C→U mutation in RNAII (Million-Weaver et al., 2012). The second point mutation is a G→A mutation at position 512, resulting in a C→U mutation in RNAII (Sam Million-Weaver 2012). C180U maps to the fourth predicted stem loop structure (SL4), which forms upon hybridization between sense and antisense RNAs, preventing R-loop formation, whereas C512U is located in a hairpin loop that facilitates R-loop formation in the wild-type pUC19 origin (Camps, 2010). Henceforth, we’ll refer to these two mutations using the template (DNA) ori numbering (G180A and G512A) for consistency with another ori modification described below that is outside the area transcribed into RNAII.
We also introduced a duplication in the 5’ end to modulate plasmid copy number by altering the ratio of RNA I to RNA II. A detailed map of this 269nt duplication is shown in Fig. 6a; note that this area contains most of the sequence elements involved in antisense regulation of replication initiation, including SL1–4, and promoters for the pre-primer (PRNAII) and antisense (PRNAI) RNAs. We therefore named this construct DAS_GFPuv, for GFPuv plasmid with a Duplicated Antisense Sequence. The full nucleotide sequence of the DAS ori is shown in Fig. 6b, highlighting the duplicated sections. Finally, we combined the plasmid copy number-modulating mutations G180A and G512A with the DAS ori by cloning the mutations using the ClaI PciI sites flanking the WT origin, leaving the duplicated section intact. The resulting variety of genetic ori modifications that we generated produced a range of plasmid copy numbers that allowed us to address whether plasmid copy number is a key determinant of plasmid stability in our system (Fig. 4a,b and 5a,b).
Fig. 6. Map of modifications increasing pMB1 stability.
a Duplicated antisense sequence Schematic showing the duplication of antisense sequence (DAS). Shown are promoters PRNAI and PRNAII; the stem-loop structures that are important for antisense regulation; the two restriction sites used for cloning of the plasmid ori mutants; and the position of the RNA/DNA switch. b. DAS ori sequence. Duplicated sequences are boxed; mutations tested for increased stability are highlighted in bold, denoting nucleotide positions (relative to the start of the ori following standard ColE1 numbering (Ohmori and Tomizawa, 1979)) and base pair substitutions. The two restriction sites used for cloning of the plasmid ori mutants are denoted and highlighted in grey boxes. c Location of the G180A/C mutation. This mutation is mapped on a secondary structure for stem-loop number four of RNAII, which comprises nucleotides 172 through 207. The secondary structure shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−12.20 kcal/mol. d Location of the G512A/C mutation. This mutation is mapped on a secondary structure for hairpin 2 of RNAII, which comprises nucleotides 508 through 525. The model shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−7.3 kcal/mol. e Location of the G628A mutation. This mutation is mapped on a secondary structure for single-stranded DNA in area surrounding the RNA/DNA switch. The model shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−11.27 kcal/mol.
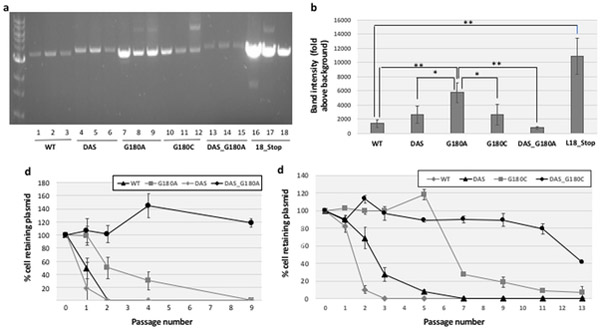
Fig. 4. Effects of G180 ori point mutations (with and without DAS).
a. Effects on plasmid copy number. The copy number of pGFPuv plasmids containing G180 ori point mutations, both in the presence and absence of DAS, was determined by miniprep extraction, restriction enzyme linearization and agarose gel electrophoresis as described in Methods. b. Quantitative Measurement. Band intensities in the gel shown in section a were measured by densitometry using Image J software, with fixed area size for all the samples, and averaged. Error bars represent the standard deviation for three replicates. Statistically significant differences were confirmed by a 2-tailed, unpaired Student t test, with one asterisk indicative of p<0.05 and two asterisks indicative of p<0.01. c. Plasmid stability of G180A-containing variants and d. Plasmid stability of G180C-containing variants. The stability of the pGFPuv plasmids included in sections a and b was determined by serial passage in liquid culture in the absence of carbenicillin, and determining the % cells retaining carbenicillin resistance as described in Methods. Error bars represent the standard deviation of three biological replicates. The legend is shown on the upper right corner of the figure.
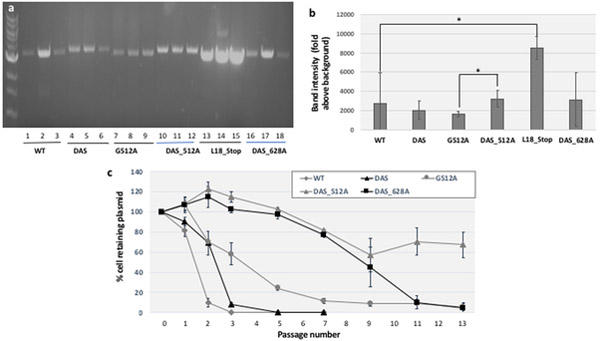
Fig. 5. Effects of G512 ori point mutations with and without DAS on plasmid copy number and on plasmid stability.
a. Effects on plasmid copy number. The plasmid copy number of pGFPuv containing point-mutations at the 512 position, both in the presence and absence of DAS, and of DAS_G628A ori was determined by miniprep extraction, restriction enzyme linearization and agarose gel electrophoresis as described in Methods. b. Quantitative measurement. Band intensities in the gel shown in section a were measured by densitometry using Image J software, with fixed area size for all the samples, and averaged. Error bars represent the standard deviation for three biological replicates. Statistically significant differences were confirmed by a 2-tailed, unpaired Student t test, with one asterisk indicative of p<0.05. c. Plasmid stability. The stability of the pGFPuv plasmids included in sections a and b was determined by serial passage in liquid culture in the absence of carbenicillin, and determining the % cells retaining carbenicillin resistance as described in Methods. Error bars represent the standard deviation of three biological replicates. The legend is shown on the upper right corner of the figure.
We find that the G180A mutation induces a substantial increase in plasmid copy number (Fig. 4a,b), whereas G512A has no significant impact (Fig. 5a,b). DAS has no significant effect on plasmid copy number either (Fig. 4a,b and Fig. 5a,b). However, in combination with the G180A, DAS substantially decreases plasmid copy number (Fig. 4b), while in combination with G512A, it has the opposite effect (Fig. 5b).
The impact of these ori modifications on plasmid stability are shown in Fig. 4c (G180A-containing variants), Fig. 4d (G180C-containing variants) and Fig. 5c (G512A-containing variants). Both the G180A and G512A mutations substantially increased plasmid stability. While DAS had a much more modest effect on stability on its own, it was strongly synergistic with the two point mutations and DAS (Fig. 4c, and Fig. 5c).
We confirmed these observations by flow cytometry. After a single passage in the absence of carbenicillin selection, only 4% of cells bearing the pDAS construct sorted into the “no plasmid” category, compared to 98% of cells transformed with the parental construct at this time-point (Fig. 2c). Even after four passages in the absence of selection, 37% of cells still retained GFP fluorescence, while fluorescent cells originally transformed with the parental plasmid were not detectable by FACS (Fig. 2d). We also saw a dramatic increase in plasmid stability for the DAS plus G512A combination: after four passages, 88% of G512A plasmids still expressed a high level of GFP fluorescence, compared to only 1% for DAS-pGFPuv (Fig. 2d). Thus, flow cytometry confirmed both a stabilizing effect of DAS, and a much more profound stabilization by the DAS_G512 combination.
Comparing our plasmid copy number and our plasmid stability results, we found no correlation, as both variants with a clear increase in copy number relative to WT such as 18_Stop and G180A (Fig. 4b) and the variant with the lowest copy number (DAS_G180A) (Fig. 4b) produced comparable increases in plasmid stability (Fig. 2A and Fig 4c).
Specificity of point mutation effects
Both G180A and G512A involve C→U mutations at stem portions of stem-loops of the pre-primer RNA (Fig. 6c,d). We wondered: 1) whether the specific substitution was important or just breaking base-pairing of the stem-loop was; 2) whether the substitutions had to be located in the pre-primer RNA or could also be located beyond the RNA/DNA switch. To address the first question, we introduced A→C substitution at positions 180 (G180C) and 512 (G512C), resulting in C→G (instead of C→U) mutations in RNAII. Unlike G180A, the G180C mutation didn’t significantly increase plasmid copy number relative to WT pUC19 (Fig. 4a lanes 10,11, 12, Fig. 4b), yet G180C produced a similar effect on plasmid stability (Fig. 4d, gray squares), once more decoupling plasmid copy number from plasmid stability. Again, DAS strongly enhanced G180C’s plasmid stability phenotype (Fig. 4d, black circles). In the case of G512, the C mutation also led to a strong increase in plasmid stability, at least in the DAS background (not shown). Taken together, these results strongly suggest that disruption of the stem-loop rather than the specific replacement of a G by an A is the change needed at these two positions to increase plasmid stability.
To address whether plasmid-stabilizing mutations can also be located outside RNAII, i.e. beyond the RNA/DNA switch, we introduced a G→A mutation at position G628, which is in the stem portion of a stem-loop structure that forms when the DNA duplex opens up during replication initiation (Fig. 6e). To facilitate detection of an effect on plasmid stability, we introduced this mutation in the DAS background, which synergized with mutations at the other two positions we tried (G180 and G512). DAS_G628A showed a highly stable phenotype (Fig. 5c, black squares). Again, this observation clearly decoupled plasmid stability from plasmid copy number because DAS_G628A no detectable effect on copy number relative to DAS or to WT (Fig. 5a, lanes 16,17,18, Fig. 5b).
Effects of stabilizing point mutations on the viability of cells grown on carbenicillin
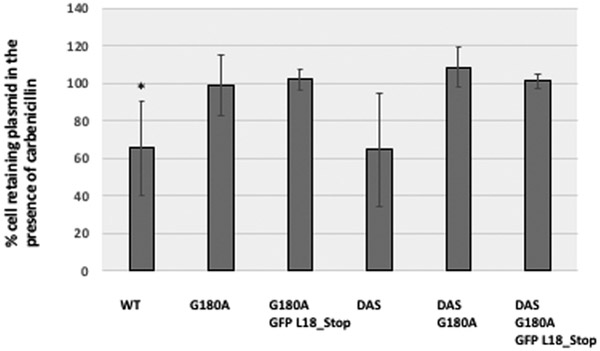
We mentioned that cells bearing the WT plasmid showed decreased viability when plated on carbenicillin plates. We thought that it would be informative to know whether stabilizing mutations eliminate this phenotype. Stabilizing mutations also serve as an important specificity control for the decreased viability we observed. We compared plasmids bearing WT and DAS oris (which are unstable and moderately stabilized, respectively) with plasmids bearing the G180A and GFP L18_stop mutations (which are highly stable). Fig. 7 shows the average ratios of growth on carbenicillin vs. LB. We calculated the ratio of carb/LB grown cells for each of our samples, and found the WT’s to be statistically significant by a single-tailed, paired Student t-test (p<0.012), and DAS’ to be trending significant (p<0.074). Importantly, the average ratio for these two samples was considerably lower than that of the rest of the samples (average 65.4% and 64.5%, respectively), whereas the average for the other four samples was within a narrow interval close to 100% (99–109%). These results suggest that DAS has little effect on the viability of cultures grown in carbenicillin, but that the G180A mutation, alone or in combination with DAS, markedly improves it.
Fig. 7. Effects of stabilizing mutations on viability in carbenicillin.
Here we compare WT and DAS oris (which are unstable and moderately stable, respectively) with plasmid ori variants of these two oris containing G180A and G180A with GFP_L18Stop (both of which are highly stable). For each sample, six cultures were grown on carbenicillin and plated on carbenicillin or LB plates (for DAS we only had 5 samples, and for G180A and for DAS_G180A GFP_L18_stop only 3). The ratio of colonies grown in the presence of carbenicillin vs. LB was averaged out. To account for differences in n between samples, the error bars represent standard error of the mean, not standard deviation. Significant decreases in ratio were established through a one-tailed, paired Student’s t-test. For WT, the p-value was <0.012 (asterisk), and for DAS, p<0.074.
Effect of recombinant gene expression
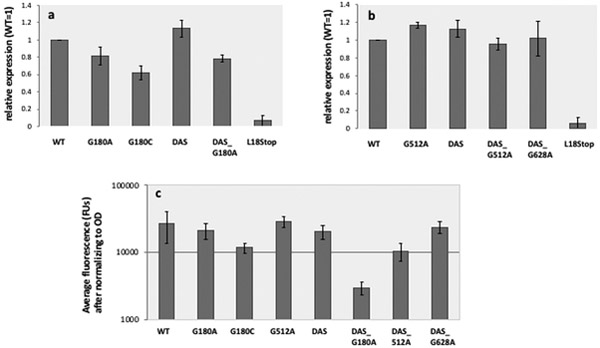
Given that early truncation of GFP expression resulted in a dramatic increase in plasmid stability, we considered whether increased plasmid stability could be attributed to decreased recombinant GFP expression and determined the impact of our ori modifications on the level of GFP expression. Fig. 8 shows GFP expression as measured by western blot, for G180-containing plasmids (Fig. 8a), and for G512-containing plasmids and for DAS_G628A (Fig. 8b) as measured by fluorescence (Fig. 8c). We find inconsistent effects in our engineered ori variants, with some of them (DAS and G512A) resulting in a slight increase in GFP expression, while others (G180C and DAS_G180A) clearly showing decreased expression (Fig. 8a,c). Other engineered variants, particularly DAS_G628A did not appear to affect expression (Fig. 8b,c). Given that all these engineered plasmids, regardless of their effect on GFP expression, increase plasmid stability, we conclude that modulation of GFP expression cannot explain the observed effects on plasmid stability.
Fig. 8. Quantification of GFP expression.
a. Western blot values. GFP expression for cells expressing the engineered variants listed in the X axis is shown, as band intensity on a Western blot, determined by densitometry, normalized to that of WT. Error bars represent standard deviation of two biological replicas. b. Fluorescence values. The fluorescence of cultures expressing the engineered variants listed on the X axis was measured as described in Methods and plotted (in log scale) on the Y-axis. Error bars represent standard deviation of 10 biological replicas.
Discussion
The stable vertical transmission of ColE1-like plasmids is an important determinant of yield for large-scale recombinant gene expression (Grabherr and Bayer, 2002; Kroll et al., 2011) and is essential for bioremediation approaches. Here we use the plasmid pGFPuv in a recA relA host strain as a model to study factors affecting the stability of these vectors. The recA mutation suppresses homologous recombination, minimizing multimer formation (Summers, 1998). The relA knockout eliminates the stringent response that shifts cell metabolism toward conservation, in order to preserve a high plasmid copy number even when cultures run low on nutrients and/or cells run low on loaded-tRNAs (Jain et al., 2006; Kanjee et al., 2012; Nazir and Harinarayanan, 2016).
We find that in this model system, the plasmid pGFPuv is extremely unstable (Fig. 2a). This allowed us to investigate factors modulating plasmid stability. We found that rapid plasmid loss in the absence of positive selection was connected with the translation of cycle3 GFP, because truncation of the ORF restored stability (Fig. 2a, black triangles).
Interestingly, truncating GFP also resulted in a dramatic increase in plasmid copy number. It would be tempting to speculate that the increase in plasmid copy number and/or suppression of recombinant gene expression (with its concomitant alteration in levels of loaded tRNAs) (Wang et al., 2002; Wang et al., 2006; Yavachev and Ivanov, 1988) explain the stability of plasmids carrying GFP with a premature stop codon. However, given that in the engineered ori variants that we studied here we found no correlation between plasmid copy number or level of pGFPuv expression and plasmid stability, the mechanism of stabilization for GFP-truncated mutants remains to be established. We noted an increase in transformation efficiency for the L18_Stop mutants relative to WT and Q183R (p<0.03 in both cases), suggesting the mechanism of stabilization may involve increased replication initiation or improved replication efficiency. These mechanisms would also be consistent with the dramatic increase in plasmid copy number observed (Fig. 2b, c, 4b and 5b).
Our main finding is that, in addition to a connection with the translation of a plasmid-borne recombinant gene, plasmid stability is also under the control of plasmid origin of replication (ori) sequence and that this control can be independent of the frequency of replication initiation. For example, our DAS_G180A mutant, which displays a very low copy number (Fig. 4b), and our DAS_G628A variant, which has a plasmid copy number comparable to that of the WT pUC19 ori (Fig. 5b) are amongst our most stable ones (Fig. 4c, black circles and Fig. 5c, black squares).
A possible indirect effect suppressing recombinant gene expression does not explain the observed increase in plasmid stability, as the variant yielding the highest level of GFP expression (G512A) (Fig. 8b) is also stable (Fig. 5c, grey circles) and a variant that has no significant effect on GFP expression, DAS_G628A, is one of the most stable ones we have (Fig. 5c, back squares).
The poor correspondence between plasmid stability and plasmid copy number that we observed stands in direct contradiction with a purely random distribution model of plasmid segregation for vertical transmission of hcn plasmids in bacteria. According to this model, the probability of random plasmid loss upon each round of cell division is given by the following formula: ρo=2(1-n), where n equals plasmid copy number (Summers, 1998). Thus, the random generation of plasmidless cells, which is a limiting step for plasmid loss in the absence of a phenotypic selection for plasmid maintenance, should be proportional to plasmid copy number. Further, the detection of a sizeable plasmidless cell population in the presence of carbenicillin selection, which we interpreted as an indication of failed plasmid segregation, is also in direct contradiction with a purely random distribution model. According to the random distribution model, this should be an exceedingly rare event, with a probability several orders of magnitude lower than our observations.
Evidence against the random distribution model for hcn plasmids has been accumulating through intracellular plasmid localization studies, which show significant clustering (Pogliano et al., 2001; Reyes-Lamothe et al., 2014; Yao et al., 2007). The formation of concatemers has also been documented (Summers et al., 1993). Both types of observations imply a substantial decrease in the number of partitioning units relative to the original assumption going into the random distribution model. Our results for the first time provide genetic evidence against the random distribution model and thus complement those earlier localization and concatemerization studies. By allowing modulation of plasmid stability, the ori variants that we described open the door to more detailed mechanistic studies. Our observations are more consistent with a recently proposed hybrid model based on superresolution fluorescence microscopy, where some plasmids are freely diffusing while others are clustered (Wang, 2017; Wang et al., 2016).
We describe three types of plasmid ori stability control elements:
-
1)
Two point-mutations that likely destabilize secondary structures in the pre-primer RNA during plasmid replication initiation (C180U/G and C512U/G) (Fig. 6c,d).
-
2)
A 269 nt duplication at the 5’ end of the plasmid ori, which we called Duplicated Anti-Sense or DAS, that likely alters the ratio of RNAI vs RNAII by providing additional sense and antisense promoters (Fig. 6a,b).
-
3)
G628A, a point mutation downstream of the DNA/RNA switch that also likely destabilizes a stem-loop in the template DNA when it is in single-stranded form during replication initiation (Fig. 6e).
The diversity of elements contributing to plasmid stability, including mutations downstream of the DNA/RNA switch, suggest a rather non-specific mechanism.
The nature of this mechanism remains unclear, though. The increased viability in the presence of carbenicillin shown in Fig. 7 suggests that G180A and DAS_G180A may result in a more accurate plasmid segregation. The flow cytometry data in Fig. 3a, showing a broader plasmid copy number distribution for DAS_180A compared to WT (blue line), and also for DAS_512A (red line) is consistent with an increased number of partition units, which would support more accurate plasmid segregation. However, loss of viability in the presence of carbenicillin is a very indirect measure of plasmid segregation, and there are a number of other possible explanations. Also, there is a gap between the fraction of plasmidless cells detected by sorting (2–3%) and the fraction of cells that die in the presence of carbenicillin (~35% decrease on average, but it can be up to 80%). The long half-life of GFP (Kitsera et al., 2007) may partially explain this gap, but it is unlikely to be the whole explanation. Also, the fraction of plasmidless cells detected by flow cytometry in DAS_G512 cultures (~1.5%) is not that different from the fraction seen for the WT plasmid (~3%), suggesting that improved segregation accuracy is unlikely to be the main mechanism explaining plasmid stabilization. Overall, the mechanism leading to an increased plasmid stabilization by our genetic modifications to the plasmid ori remains unclear but the ability to restore viability in the presence of carbenicillin provides an important clue.
The strong synergy between DAS and our G180 and G512 point mutations for enhancement of plasmid stability (Fig. 4c, d and Fig. 5c) as well as the interplay between DAS and G180A and G512A point mutations and plasmid copy number (Fig. 4b, Fig. 5b) are intriguing and also provide clues to help elucidate the mechanism involved.
Finally, this work also has translational implications. For large-scale recombinant gene expression, plasmid loss is one of the limiting factors for yield (Grabherr and Bayer, 2002; Kroll et al., 2011). Plasmid stability in the absence of selection is also critical for the success of bioremediation approaches. Here we describe plasmid origin of replication modifications that, unlike addiction systems, in principle don’t require specific genetic modifications in the host. This should help with the design of improved vectors for recombinant gene expression and/or bioremediation.
Acknowledgements
This work was supported with start-up funds, with partial support from K08 Award CA116429–04 (NCI), and from the UCSC’s Office of Research funds to MC. The sponsor had no role in the design, collection, analysis or interpretation of the results.
Abbreviations:
- hcn
high copy number
- ori
plasmid origin of replication
- R-loop: DNA
stable RNA hybrid with template DNA, and the associated non-template single-stranded DNA
- D-loop
Displacement loop
- pas
primosome assembly site
- SL
Stem Loop
- Rom
RNA one modulator
- DAS
Duplicated Anti-Sense
- LB
Luria-Bertami media
- FACS
Fluorescence-Activated Cell Sorting
References
- Bergstrom CT, Lipsitch M, and Levin BR (2000). Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S (2014). Plasmid Replication Control by Antisense RNAs. Microbiology spectrum 2, PLAS-0001–2013. [DOI] [PubMed] [Google Scholar]
- Camps M (2010). Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent patents on DNA & gene sequences 4, 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G, Helmer-Citterich M, and Castagnoli L (1991). Control of ColE1 plasmid replication by antisense RNA. Trends Genet 7, 230–235. [DOI] [PubMed] [Google Scholar]
- Cooper NS, Brown ME, and Caulcott CA (1987). A mathematical method for analysing plasmid stability in micro-organisms. J Gen Microbiol 133, 1871–1880. [DOI] [PubMed] [Google Scholar]
- Crameri A, Whitehorn EA, Tate E, and Stemmer WP (1996). Improved green fluorescent protein by molecular evolution using DNA shuffling. Nature biotechnology 14, 315–319. [DOI] [PubMed] [Google Scholar]
- De Gelder L, Ponciano JM, Joyce P, and Top EM (2007). Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153, 452–463. [DOI] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, and Toussaint A (2005). Mobile genetic elements: the agents of open source evolution. Nature reviews Microbiology 3, 722–732. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Howard M, and Szardenings F (2010). Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942. [DOI] [PubMed] [Google Scholar]
- Grabherr R, and Bayer K (2002). Impact of targeted vector design on Co/E1 plasmid replication. Trends in biotechnology 20, 257–260. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Minshull J, Govindarajan S, Ness J, Villalobos A, and Welch M (2012). Engineering genes for predictable protein expression. Protein expression and purification 83, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, and Tomizawa J (1980). Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proceedings of the National Academy of Sciences of the United States of America 77, 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Kumar M, and Chatterji D (2006). ppGpp: stringent response and survival. Journal of microbiology 44, 1–10. [PubMed] [Google Scholar]
- Kanjee U, Ogata K, and Houry WA (2012). Direct binding targets of the stringent response alarmone (p)ppGpp. Molecular microbiology 85, 1029–1043. [DOI] [PubMed] [Google Scholar]
- Kitsera N, Khobta A, and Epe B (2007). Destabilized green fluorescent protein detects rapid removal of transcription blocks after genotoxic exposure. BioTechniques 43, 222–227. [DOI] [PubMed] [Google Scholar]
- Kroll J, Klinter S, Schneider C, Voss I, and Steinbuchel A (2010). Plasmid addiction systems: perspectives and applications in biotechnology. Microbial biotechnology 3, 634–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J, Klinter S, and Steinbuchel A (2011). A novel plasmid addiction system for large-scale production of cyanophycin in Escherichia coli using mineral salts medium. Applied microbiology and biotechnology 89, 593–604. [DOI] [PubMed] [Google Scholar]
- Lacatena RM, Banner DW, Castagnoli L, and Cesareni G (1984). Control of initiation of pMB1 replication: purified Rop protein and RNA I affect primer formation in vitro. Cell 37, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Lau BT, Malkus P, and Paulsson J (2013). New quantitative methods for measuring plasmid loss rates reveal unexpected stability. Plasmid 70, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Spychala CN, Hu F, Sheng JF, and Doi Y (2015). Complete nucleotide sequences of bla(CTX-M)-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob Agents Chemother 59, 3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lili LN, Britton NF, and Feil EJ (2007). The persistence of parasitic plasmids. Genetics 177, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly J, and Camps M (2015). Mechanisms of Theta Plasmid Replication. Microbiology spectrum 3, PLAS-0029–2014. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S, Chen WT, and Wong TT (1992). High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Molecular microbiology 6, 3385–3393. [DOI] [PubMed] [Google Scholar]
- Lobato-Marquez D, Molina-Garcia L, Moreno-Cordoba I, Garcia-Del Portillo F, and Diaz-Orejas R (2016). Stabilization of the Virulence Plasmid pSLT of Salmonella Typhimurium by Three Maintenance Systems and Its Evaluation by Using a New Stability Test. Front Mol Biosci 3, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million-Weaver S, Alexander DL, Allen JM, and Camps M (2012). Quantifying plasmid copy number to investigate plasmid dosage effects associated with directed protein evolution. Methods in molecular biology 834, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I, and Kobayashi I (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic acids research 42, 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B, Neuert G, and van Oudenaarden A (2012). Using gene expression noise to understand gene regulation. Science 336, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir A, and Harinarayanan R (2016). (p)ppGpp and the bacterial cell cycle. J Biosci 41, 277–282. [DOI] [PubMed] [Google Scholar]
- Ohmori H, and Tomizawa J (1979). Nucleotide sequence of the region required for maintenance of colicin E1 plasmid. Molecular & general genetics : MGG 176, 161–170. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Ho TQ, Zhong Z, and Helinski DR (2001). Multicopy plasmids are clustered and localized in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 98, 4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B (1988). ColE1 replication control circuitry: sense from antisense. Cell 55, 929–932. [DOI] [PubMed] [Google Scholar]
- Polisky B, Zhang XY, and Fitzwater T (1990). Mutations affecting primer RNA interaction with the replication repressor RNA I in plasmid CoIE1: potential RNA folding pathway mutants. The EMBO journal 9, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponciano JM, De Gelder L, Top EM, and Joyce P (2007). The population biology of bacterial plasmids: a hidden Markov model approach. Genetics 176, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Tran T, Meas D, Lee L, Li AM, Sherratt DJ, and Tolmasky ME (2014). High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic acids research 42, 1042–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J (2010). Plasmid segregation: how to survive as an extra piece of DNA. Critical reviews in biochemistry and molecular biology 45, 296–317. [DOI] [PubMed] [Google Scholar]
- San Millan A, Pena-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, and MacLean RC (2014). Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun 5, 5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla K, Jechalke S, and Top EM (2015). Plasmid Detection, Characterization, and Ecology. Microbiology spectrum 3, PLAS-0038–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley M, Allen J, Cervantes L, Lilly J, and Camps M (2017). Fluorescence-Based Reporters for Detection of Mutagenesis in E. coli. Methods in enzymology 591, 159–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D (1998). Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Molecular microbiology 29, 1137–1145. [DOI] [PubMed] [Google Scholar]
- Summers DK, Beton CW, and Withers HL (1993). Multicopy plasmid instability: the dimer catastrophe hypothesis. Molecular microbiology 8, 1031–1038. [DOI] [PubMed] [Google Scholar]
- Thomas CM, and Nielsen KM (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature reviews Microbiology 3, 711–721. [DOI] [PubMed] [Google Scholar]
- Tomizawa J, and Som T (1984). Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell 38, 871–878. [DOI] [PubMed] [Google Scholar]
- Vieira J, and Messing J (1982). The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19, 259–268. [DOI] [PubMed] [Google Scholar]
- Wang Y (2017). Spatial distribution of high copy number plasmids in bacteria. Plasmid 91, 2–8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Penkul P, and Milstein JN (2016). Quantitative Localization Microscopy Reveals a Novel Organization of a High-Copy Number Plasmid. Biophys J 111, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Le G, Shi Y, Wegrzyn G, and Wrobel B (2002). A model for regulation of ColE1-like plasmid replication by uncharged tRNAs in amino acid-starved Escherichia coli cells. Plasmid 47, 69–78. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yuan Z, Xiang L, Shao J, and Wegrzyn G (2006). tRNA-dependent cleavage of the ColE1 plasmid-encoded RNA I. Microbiology 152, 3467–3476. [DOI] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, and Gustafsson C (2009). Design parameters to control synthetic gene expression in Escherichia coli. PloS one 4, e7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbowy O, Boratynski R, Dekowska A, and Kaczorowski T (2015). Genetic analysis of maintenance of pEC156, a naturally occurring Escherichia coli plasmid that carries genes of the EcoVIII restriction-modification system. Plasmid 77, 39–50. [DOI] [PubMed] [Google Scholar]
- Werbowy O, and Kaczorowski T (2016). Plasmid pEC156, a Naturally Occurring Escherichia coli Genetic Element That Carries Genes of the EcoVIII Restriction-Modification System, Is Mobilizable among Enterobacteria. PloS one 11, e0148355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbowy O, Werbowy S, and Kaczorowski T (2017). Plasmid stability analysis based on a new theoretical model employing stochastic simulations. PloS one 12, e0183512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Helinski DR, and Toukdarian A (2007). Localization of the naturally occurring plasmid ColE1 at the cell pole. Journal of bacteriology 189, 1946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavachev L, and Ivanov I (1988). What does the homology between E. coli tRNAs and RNAs controlling ColE1 plasmid replication mean? Journal of theoretical biology 131, 235–241. [DOI] [PubMed] [Google Scholar]
- Zuker M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]