Abstract
Human plasma von Willebrand Factor (VWF) plays essential roles in primary hemostasis in cooperation with other coagulations factors. There is ample indication that glycosylation affects many biological phases during the protein life cycle. However, comprehensive characterization of all probable N-glycosites simultaneous with O-glycosites is still not fully revealed. Thus, the intention of this exploration was to estimate the occupancy of all canonical N-glycosites besides simultaneous characterization of N- and O-glycoforms. An RP–LC–MS/MS system functionalized with CID and HCD tandem mass was utilized to analyze VWF. N-Glycosite occupancy varied along the protein backbone chain. Out of 257 HCD spectra, 181 characterized glycoforms were specified as either N- or O-glycosites. Sequential cleavage of glycosidic bonds along with Human Database mass matching have confirmed the glycoform structures. A total of 173 glycoforms represented most commonly biantennary and infrequently tri- and tetra-antennary N-glycans beside high mannose, hybrid, ABH antigen-terminated, and sulfated N-glycans. Many glycoforms were common across all N-sites. Noteworthy, previously unreported N-glycosites within domain D′(TIL′-E′) showed glycosylation. Moreover, sialylated core 1 and core 2 O-glycans were detected on 2298T. Given subtle characterization of site-specific glycoforms, we can attain a profound understanding of the biological roles of VWF as well as facilitate the production of VWF-based therapeutics.
Keywords: plasma von Willebrand Factor, VWF, occupancy, glycosylation, N-glycan, O-glycan, microheterogeneity, CID, HCD, mass spectrometry
Graphical Abstract
INTRODUCTION
Among the wide range of post-translational modifications (PTMs), O- and most frequently N-linked glycosylations have been attracting the most attention.1 Protein glycosylation is common in all living eukaryotic cells and plays essential roles covering a spectrum from those imperceptible to those critical for an organism to survive.2 For instance, von Willebrand factor (VWF), a large plasma glycoprotein, plays two essential roles in hemostasis,3 besides other coagulating factors, such as FV, FVII, FVIII, FIX, and FXI. That is, VWF mediates platelet adhesion at an injured vascular area4 and maintains the plasma FVIII level by preventing premature proteolytic degradation and clearance by active protein C.5 Insufficiency in one or more coagulating factors is correlated with many bleeding illnesses.6 The bleeding disorder, von Willebrand Disease (VWD), commonly treated by plasma VWF concentrate, is caused by a deficiency or defect in VWF.7 Recombinant VWF (rVWF) has also been synthesized and developed for treating VWD.8,9 However, industrial production of this therapeutic protein is restrained by its large size and intrinsic complexity derived from extensive glycosylation and disulfide bond oligomerization.10 One of the critical challenges of clotting factor expression includes addressing PTMs, e.g., glycosylation, and the choice of host cell line.10 For maximal fidelity, the quality of the glycosylation pattern of rVWF should be evaluated to manifest glycosylation of the counterpart plasma-derived VWF. Thus, comprehensive glycoproteomic analysis of plasma concentrate VWF is indispensable.
For in vivo biosynthetic production, VWF is strictly expressed in megakaryocytes and endothelial cells (ECs). Plasma VWF, however, is entirely derived from ECs.11 It is then either directly secreted into the plasma or stored in organelles, so-called Weibel-Palade bodies (WPBs).11 Within ECs, VWF undergoes complex PTMs, including glycosylation with N- and O-linked oligosaccharides.12 Briefly, VWF is initially synthesized in the endoplasmic reticulum (ER) as a prepropolypeptide containing 2813 amino acids (AAs) organized into four homologous domains (D1-2-D′-D3-A1-3-D4-B1-3-C1-C2-CK).12 Recently, all D domains have been reannotated to VWD, cysteine 8 (C8), and trypsin-inhibitor-like (TIL) domains followed by an extra segment (E), except for D4 uniquely preceded by a bridging region (D4N). The C-termini is further characterized to six homologous repeats (C1-C6) along with the dimerization domain CTCK.13
Once the signaling peptide (22 AAs) is removed and N-glycosites are occupied with high mannose oligosaccharides, the monomeric VWF units dimerize by disulfide bond formation within CK domains. In the Golgi apparatus, the dimer undergoes further modification by a series of glycosidases and glycosyltransferases to produce complex N-glycans in addition to ten O-glycosites.11 Eventually, another round of PTMs, including disulfide bond formation and proteolytic cleavage to remove pro-peptide (741 AAs), occur in the trans-Golgi to produce mature VWF multimers.14 Carbohydrate moieties represent approximately 20% of the total molecular weight of the mature monomer VWF (2050 AAs).15
The influence of defect glycosylation in the VWF life cycle has attracted prolonged attention from researchers.16 Alterations in the glycosylation pattern or absence of a glycosite might affect protein multimerization,17,18 platelet adhesion,19,20 ADAMTS13 preoteolysis,21,22 as well as biosynthesis and secretion of VWF.23 Hence, well-known glycosylation positions, as well as glycoform structures, are critical for the plausible production of such therapeutic proteins. The initial studies performed on the VWF glycome were conducted over 20 years ago.24–26 The main N-glycan structure was complex. Interestingly, some of characterized N-glycans terminated with H (Fuc(α1–2) Gal(β1–4) GlcNAc β1-→), A (GalNAc-(α1–3) [Fuc(α1–2)] Gal(β1–4) GlcNAc β1→), and B (Gal(α1–3) [Fuc(α1–2)] Gal(β1–4) GlcNAc β1→) antigens.26 These antigens may have significant effects on the level of plasma VWF and associated with different rates of the protein clearance in ABO individuals.27 The primary study on the mature VWF sequence highlighted 11 consensus N-glycosites (N-X-S/T, where X is any AA except P) and one infrequent sequon (N1147SC). The glycosites N857, N1147, N1231, N1515, N1574, N2223, N2290, N2357, N2400, N2546, N2585, and N2790 were reported to be occupied with the exception of N2635.15 However, a recent study has proven that even N2635 is heavily glycosylated.28 The trypsin-digested VWF glycopeptides carrying consensus N-glycosites were off-line separated and mapped by LC–MALDI-TOF/TOF. The tryptic digest was affinity-enriched and treated with a series of glycosidases prior to MS analysis. Bi-, tri- and tetra-antennary complex glycans were observed besides high mannose glycans. Additional modifications, such as antennae fucose, lactosaminic extension, sulfation, and ABH-antigen termination, were detected.28
On the other hand, the initial characterization of the VWF O-glycome showed that disialylated core 1 (T antigen; NeuAc-(α2–3) Gal(α1–3)-[NeuAc(α2–6)] GalNAc) was the predominant form.29,30 Recently, the heterogeneity of the VWF O-glycome was confirmed with di-, tri-, and tetrasialylated core 1 and core 2 in addition to minor ABH antigen-carrying O-glycans.31 Furthermore, a recent O-glycoproteomic study of plasma VWF has shown that all O-glycosites (T1248, T1255, T1256, S1263, T1468, T1477, S1486, T1487, T1679, and T2298) were occupied, and the predominant O-glycoform is T antigen.32 The proteolytically digested O-glycopeptides were affinity-enriched and subjected to successive glycosidase-based digestions and offline separation before MALDI-TOF/TOF analysis. Although the glycosylation pattern of VWF has been extensively investigated for potential effects on the protein life cycle,17,23 full characterization of glycan microheterogeneity at all possible VWF N-glycosites has yet to be accomplished. Using modern analytical instruments reveals noteworthy structural information about the protein.28,32 Thus, further investigation of VWF microheterogeneity is crucial to lighten the glycosites that are shaded behind the instrument limitations. Moreover, the protein macroheterogeneity (N-glycosite occupancy) is still not known either. Thus, full characterization of VWF micro- and macroheterogeneity is important for understanding the protein biosynthesis and functions, such as the effective interaction with the accompanied coagulating factor FVIII,33 beside evaluating the quality of the biopharmaceutical products.10
For successful site-specific characterization, the digested glycopeptides are usually enriched before analysis because of their low abundance compared to that of nonglycosylated peptides.34 Various types of lectin-based approaches are widely accepted for glycoprotein and glycopeptide enrichment.35 However, these approaches are selective and often accompanied by successive enrichment steps as well as glycosidase digestions.32,36 Unlike lectin affinity that targets specific glycan structures, HILIC is a promising enrichment approach that has attracted more attention in the past few years. In recent studies, HILIC-based enrichments have been employed for site-specific analysis of intact glycopeptides, such as those derived from erythropoietin (EPO)37 and α-1-antitrypsin (A1AT).38 HILIC enrichment allows glycoprotein/glycopeptide isolation independent of the glycan structure.35 Thus, we constructed a simple homemade HILIC SPE microtip column to effectively enrich and purify trypsin-digested VWF peptides carrying either a neutral or negatively charged glycan.
Alongside effective enrichment approaches, sensitive structural characterization and glycan profiling have become attainable because of the major developments in mass spectrometry (MS) in the past decade.39 Although glycoproteomic analysis can be performed on MALDI-TOF, prior glycosidase-based treatments and off-line separation are required to obtain reliable results.28,32 In contrast, LC–MS, escorted with complementary or single MS/MS tandem mass, is widely acceptable because of the simplicity and high accuracy for characterizing carbohydrate moieties in conjunction with site specificity.39 For instance, α-1-acid glycoprotein (AGP)-derived N-glycopeptides, separated on various reversed-phase (RP-C18) columns, from controls and human plasma hepatocellular carcinoma (HCC) were characterized by either collision-induced dissociation (CID) or higher-energy collisional dissociation (HCD) or both.40 Single tandem mass, CID, was used to label-free quantify low abundance site-specific N-glycoforms derived from various types of immunoglobulin G (IgG1, IgG 2/3, and IgG4) in healthy plasma controls and patients with cirrhosis (liver).41 Furthermore, the highly accurate Orbitrap analyzer functionalized with HCD, so-called beam-type CID, with high energy and short activation time, allowed concurrent characterization of N- and O-glycopeptides in EPO.37 Electron-transfer dissociation (ETD) is preferable for characterizing small modifications, such as O-glycosylation and phosphorylation.42,43 To date, a combination of state-of-the-art MS with an efficient enrichment approach is of the essence for successful microheterogeneity analysis.
The central theme of this study is proteomic and glycoproteomic analyses of plasma VWF on a state-of-the-art RP-UPLC system interfaced with an LTQ-Orbitrap Elite mass analyzer. Accompanied with online separation, the eluted O18-labeled N-deglycopeptides and HILIC-enriched glycopeptides were directly subjected to tandem mass to demonstrate VWF macroheterogeneity (N-glycosite occupancy) and heterogeneous population of N- and O-glycosites, respectively. In this work, we developed a systematic strategy for comprehensive and simultaneous characterization of N- and O-glycopeptides based on highly accurate and efficient resolution MS. Along with effective enrichment on a homemade HILIC microcolumn, the sample complexity was minimized by online RP separation on a nanoLC column. In silico deglycosylation of targeted peptides as well as the distinctive appearance of HCD spectra were used to confirm the peptide sequence and characterize the structure of the attached N- or O-glycoforms, respectively. Besides previously reported glycosylated sites, we have discovered and characterized glycoform microheterogeneity of formerly unconsidered N-glycosites with acceptable sequences. In light of this new detection, further research might reveal new insight into VWF biological roles and functions.
METHODS
Chemicals and Materials
Human plasma VWF (HCVWF-0191 VWF-VIII “free”) was acquired from Haematologic Technologies Inc. (HTI, Essex, VT, USA). Fetuin from fetal bovine serum (Sigma-Aldrich) was used as a model glycoprotein. Sequencing-grade modified porcine trypsin was purchased from Promega (Madison, WI, USA). Peptide-N-glycosidase F (PNGase F) was acquired from New England Biolabs (Ipswich, MA, USA). HILIC material (Click Mal, 5 μm, 100 Å) was obtained from ACCHROM (Beijing, China). Reduction and alkylation reagents, dithio-threitol (DTT), and iodoacetamide (IAM), respectively, were purchased from Acros Organics (Morris Plains, NJ, USA). Formic acid (FA) was obtained from Sigma-Aldrich (St. Louis, MO, USA). Tris-HCl buffer was purchased from US Biological (Swampscott, MA, USA). HPLC-grade acetonitrile (ACN) was purchased from J. T. Baker Chemicals (Avantor Performance Materials, Inc., Center Valley, PA, USA). Water-18O, 97 atom % 18O, was purchased from Sigma-Aldrich (St. Louis, MO, USA), and deionized water was produced using a Milli-Q A 10 system from Millipore (Bedford, MA, USA). Microcon-30 kDa Centrifugal Filter (YM-30, 0.5 mL) with an Ultracel low-binding regenerated cellulose membrane was purchased from Millipore. The 3M Empore C8 disk was purchased from 3M Bioanalytical Technologies (St. Paul, MN, USA). Other materials, including urea, sodium dodecyl sulfate (SDS), and ammonium bicarbonate (ABC; NH4HCO3), were purchased from Sigma-Aldrich.
Sample Preparation and Trypsin Digestion
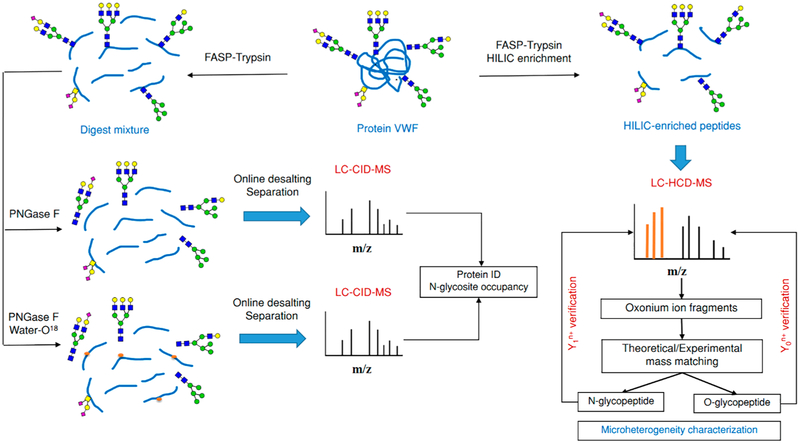
Approximately 100 μg of VWF concentrate, or 200 μg of the model glycoprotein, was subjected to a filter-aided sample preparation (FASP) procedure as previously reported with minor modifications.44 Briefly, the raw therapeutic protein was formerly subjected to ultrafiltration to remove accompanied contents including Na citrate, NaCl, and glycine. Then, the purified protein was thermally denatured and reduced by 50 μL of lysis buffer (20 mM Tris-HCl, 4% (v/v) SDS, 10 mM DTT, pH 7.6) for 5 min at 95 °C. On the room-temperature chilled sample mixture, approximately 200 μL of UA buffer (8 M urea in 0.1 M Tris/HCl, pH 8.5) was added, and the mixture was loaded into a 30 kDa Microcon filtration device. The sample was concentrated by a microcentrifuge at 13,000g at 24 °C until the dead volume was ~10 μL. After washing twice with 200 μL of UA buffer, the glycoprotein was alkylated by 100 μL of 50 mM IAM for 30 min in the dark at room temperature. After centrifugation for 20 min, the concentrate was rewashed with 200 μL of UA buffer twice. Afterward, the buffer was exchanged by 100 μL of ABC buffer (40 mM, pH 8) before adding sequencing-grade trypsin at an enzyme-to-protein ratio of 1:50 (w/w). The digestion was performed at 37 °C overnight. Subsequently, the proteolytically digested peptides were collected by centrifugation at 13,000g for 20 min and washed six times with 50 μL of 50 mM NH4HCO3. The collected peptide and glycopeptide digest were then subjected to protein identification (ID), N-glycosite occupancy, and intact glycopeptide analysis.
Treatment with PNGase F for Protein ID
For protein sequencing, a portion of the proteolytic digest (~10 ug) was subjected to in-solution N-deglycosylation by Peptide-N-glycosidase F (PNGase F) after protease digestion.45 Because the protease had been removed from the sample by ultrafiltration prior to adding PNGase F enzyme, the heating step was excluded. Briefly, the digest was dried and reconstituted in 100 μL of ABC (pH ~8.0) to ensure the complete solubility of the peptide mixture. Then, 5 uL of PNGase F (2 U μL−1) was added to keep the ratio of enzyme to sample at 1:1, and the sample was incubated overnight at 37 °C. The sample was then dried and frozen at −20 °C for future use. Upon MS analysis, the peptide mixture was redissolved in 25 μL of 2% ACN containing 0.1% FA and subjected to online purification and separation.
N-Glycosylation Site Analysis
Approximately 5 ug of the tryptic digest was subjected to glycosylation site-specific stable isotope tagging with H218O according to a previously published protocol.46 In short, the digest was fully dried in a Vacufuge Concentrator and resuspended in 10 uL of 50 mM ammonium bicarbonate dissolved in H218O (97 atom % 18O, Sigma). Deglycosylation of intact N-glycans was performed with PNGase F at a ratio of 1:1 (enzyme:protein, w/w), and the mixture was incubated at 37 °C for 12–14 h. Afterward, the deglycosylated peptide mixture was dried and frozen at −20 °C, awaiting online purification and MS analysis. Upon analysis, the mixture of peptides and 18O-labeled N-deglycosylated peptides was redissolved in 5 μL of 2% ACN containing 0.1% FA before being automatically injected into a nano-LC system.
Enrichment and Desalting of Intact Glycopeptides
The rest portion of the digest was desalted and enriched by HILIC media according to Ma’s procedure with minor modifications.47 Concisely, a homemade SPE column was constituted by inserting a small piece of the C8 disk into a 200 μL tip. Approximately 5 mg of HILIC media previously suspended in 100 μL of ACN was transferred into the microcolumn and subsequently washed twice with 50 μL of the washing buffer (WB; 10% ACN containing 0.1% FA). The microcolumn was next washed twice and equilibrated with the binding buffer (BB; 80% ACN containing 1.0% FA) for 10 min. The proteolytically digested peptide mixture was dried and resuspended in 30 μL of BB before being loaded into the microcolumn. With a suitable syringe, the enriched glycopeptides were desalted and purified with 50 μL of BB five times. Later, the bound glycopeptides were eluted with 100 μL of the elution buffer (EB; water contacting 1.0% FA) twice. The HILIC-enriched glycopeptides were then dried and stored at −20 °C pending MS analysis.
LC–MS/MS Analysis of N-Deglycosylated/18O-Labeled and Intact Glycopeptides
The RP separation of the PNGase F deglycosylated, the isotopically labeled peptides, or the intact glycopeptides was performed on a Dionex Ultimate 3000 RSLCnano System (Thermo Fisher, Waltham, MA, USA). The system was interfaced with an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher) equipped with an EASY-spray source (1.6 kV). Both deglycosylated peptides and intact glycopeptides were resuspended in 2% ACN containing 0.1% FA prepared for online separation and MS analysis. By a temperature-controlled autosampler, 5 μL of the deglycosylated peptide, 5 μL of the intact glycopeptides, or 3 μL of the 18O-labeled N-deglycosylated peptides was loaded into a Nano Trap column packed with Acclaim PepMap100 C18 (2 cm × 75 μm I.D., 3 μm). The flow rate was adjusted to 5 μL/min for 10 min. Then, the sample was introduced onto the Easy-spray PepMap C18 Column (15 cm × 75 μm I.D., 3 μm, 100 Å) for separation. Briefly, the 10 min trapping/washing step was performed using Mobile Phase A (2% ACN, 0.1% FA) followed by a 30 min linear gradient from 3 to 40% Mobile Phase B (80% ACN, 0.1% FA) at a flow rate of 300 nL/min. Subsequently, the column was washed with 99% B for 10 min followed by 5 min of reconditioning to 1.0% B for the next run.
The operation of the LTQ Orbitrap mass spectrometer was tuned in data-dependent mode with an automatic switch between MS and MS2 acquisition. The survey of the full MS scan was performed in the Orbitrap mass analyzer with the following parameters: mass range, m/z 400–1600; automatic gain control target (AGC), 106 ions; resolution at m/z 400, 6 × 104; maximum ion accumulation time, 50 ms. The ten most intense ions in MS1 were subjected to CID in the ion trap analyzer for deglycosylated peptide analysis or HCD in the Orbitrap analyzer for intact glycopeptide analysis. The MS/MS scan model was set as the centroid. For CID-MS, the default charge state was 3, the isolation width was m/z 3.0, normalized collision energy (NCE) was 35%, Activation Q was 0.25, and activation time was 5.0 ms. For HCD, the resolution of the Orbitrap analyzer was 15,000 at m/z 400; AGC was 10,000 ions, and maximum ion accumulation time was 200 ms. All MS parameters were the same as CID, except the isolation width was m/z 2.0 and the activation time was 0.5 ms. The precursor ions transferred to the HCD cell were exposed to fragmentation with 27% NCE.
MS/MS Data Interpretation
For LC–CID–MS data analysis of deglycosylated peptides, pFind software 2.8 (http://pfind.ict.ac.cn)48 was used. FASTA sequence of human VWF (P04275-1) and bovine Fetuin (P12763) were downloaded from UniProt (http://www.uniprot.org) and used for protein identification and sequencing with FDR 1%. The precursor peptide ion tolerance was 20 ppm, and the fragment ion tolerance was 0.5 Da. For human VWF, two FASTA databases were customized for sequencing. The first one included 2813 AAs, representing the full prepro VWF protein (309 kDa), whereas the second represented the mature protein (~226 kDa) containing only 2050 residues (764–2813 AAs). The fixed modification was carbamidomethylation of cysteine (+57.021 Da), and the variable modifications were oxidation of methionine (+15.995 Da) and deamidation for N and Q (+0.984 Da) (spontaneous deamidation in ordinary water). Two maximum missed cleavage sites were selected for trypsin (KR-C).
For CID–MS/MS data analysis of 18O-labeled deglycosylated peptides, the mass of the precursor ion was set between 350 and 6000 Da with S/N threshold peak filter at 3. Trypsin full and two max missed cleavages were selected. The tolerance for the precursor ion was 20 ppm, whereas the fragment tolerance was 0.8 Da. Carbamidomethylated Cys (+57.021 Da) was chosen as a static modification, and oxidized Met (+15.995 Da), deamidated Asn and Gln (+0.984 Da), and O18-deamidated Asn (+2.988 Da) were specified as variable modifications with FDR < 0.1. Peak intensities were obtained by extracted ion chromatogram (XIC). The intensities of equivalent peptides, nonglycosylated and deglycosylated, were manually extracted for occupancy estimation after matching the exact mass and the corresponding retention time.
For HCD–MS/MS data analysis of intact glycopeptides, the MS2 spectra exhibiting diagnostic fragment ions (oxonium ions) at m/z 366 (HexHexNAc1, 1+), m/z 292 (Neu5Ac, 1+), m/z 204 (HexNAc, 1+), m/z 162 (Hex, 1+), and subfragment ions at m/z 186 (HexNAc-H2O, 1+), m/z 168 (HexNAc-2H2O, 1+), m/z 138 (HexNAc-2H2O-CH3OH, 1+), m/z 126 (HexNAc-2H2O-CH3COH, 1+), and m/z 274 (Neu5Ac–H2O, 1+) were selected for further investigation of candidate peptides and corresponding glycan structures. PEPTIDEMASS generated a list of theoretical masses of tryptically digested VWF peptides with maximum two trypsin missed cleavages. The online platform is freely accessible via the ExPASy World Wide server (http://web.expasy.org/peptide_mass).49 Moreover, an online accessible GlycoMod tool (http://web.expasy.org/glycomod) was also employed for intact N- and O-glycopeptide MS data interpretation. The last tool generated a list of potential peptides containing asparagine residues with the consensus sequence (N-X-S/T/C, where X is any AA except proline) besides possible glycan compositions. Several glycan-peptide complements were proposed for even one input of the experimentally generated precursor ion [M + H]+. Unfortunately, O-glycosites lack the consensus sequence such that all serine and threonine residues are theoretically possible for O-glycosylation by GlycoMod. Further investigation and manual efforts herein were required on the picked HCD spectrum to confirm the glycopeptide identity.
Interestingly, HCD–MS spectra of intact N- and O-glycopeptides were characterized with intense Y1n+ ([Pep + GlcNAc]n+) and Y0n+ ([Pep]n+) peaks, respectively, as well as a series of peaks representing the sequential fragmentation of glycosidic bonds. That is, Y1n+ and Y0n+ were employed for cross-referencing the selected spectra with peptide ID for unambiguous identification. Confirmation of O-glycan-containing peptides was straightforward compared to that of N-glycan-containing peptides. The HCD–MS2 spectrum of an O-glycopeptide was uncomplicated as the attached glycan was small such that more energy was available for peptide backbone fragmentation. Hence, more b- and y-fragments were observed in O-glycopeptide HCD–MS2 spectra. In contrast to O-glycopeptides, the MS2 spectra of N-glycan-containing peptides predominated with glycosidic fragments. Once the peptide ID was determined, the glycan mass was calculated from the difference between the precursor ion and the assigned peptide. The calculated mass was then searched against CarbBank50 using GlycoWorkbench software.51 The search parameters were underivatized glycans with free reducing ends. The proposed structures were then specified by subsequent fragmentation of the oligosaccharide illustrated on the HCD spectrum.
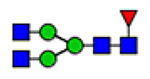
For semiquantitation of the intact glycopeptide, the peak area of each glycopeptide was extracted by XIC regarding the accurate mass and corresponding retention time (RT) of the precursor ion. The extracted area was used for calculating site-specific relative abundance (SRA) of N- and O-glycoforms detected on each site and total relative abundance (TRA) of all N-glycoforms derived from plasma VWF. For SRA, all glycan signal areas assigned (S:N > 3) at each site were summed to generate a total signal area. Then, the peak area of each glycoform was normalized to the total signal at each specific site. A total of 173 N-glycoforms and 8 O-glycoforms were distributed among glycosites and quantified regarding the local glycan signals (for details, see Tables S1–S12). For the representation of the microheterogeneity at each site to be simplified, the glycoforms were categorized into closely related compositions in which the N-glycan core and sialylated O-glycan core 1 (NeuAc(α2–3) Gal(β1–3) GalNAc) are preserved. The microheterogeneity illustration was based on the SRA sum of closely related structures (see N- and O-glycosite Microheterogeneity section, Figure 5).
Figure 5.
Microheterogeneity and relative abundances of N- and O-glycoforms attached to specific glycosites of VWF. (blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), (purple diamond) N-acetylneuraminic acid (Neu5Ac), and (red triangle) fucose.
For total quantification of the VWF N-glycome, the categorized N-glycoforms, detected on all sites, were merged together to obtain a total of 61 N-glycan compostions (Table 1). The TRA of each N-glycan composition was calculated by normalizing the summed peak areas of that particular composition observed on all sites to the total area of all compositions detected on VWF (see Characterization of VWF N-Glycome, Figure 7 (TRA, bars; left side)). Scheme 1 shows the overall workflow for proteomic and glycoproteomic analysis of plasma VWF by RP–LC–ESI–MS/MS.
Table 1.
Total Putative N- and O-Glycoforms Derived from Plasma VWFa
| No. | Sites | Glycan | No. | Site | Glycan | No. | Sites | Glycan | No. | Sites | Glycan |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2290 |  |
16 | 820, 2290 | 31 | 1574 |  |
46 | 2290 |  |
|
| 2 | 847, 1515, 1574, 2290, 2585, 2635 |
 |
17 | 1574, 2290 | 32 | 1574, 2223 |  |
47 | 1515, 1574, 2585 | ||
| 3 | 2223 |  |
18 | 2290 | 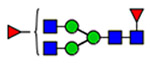 |
33 | 2290 | 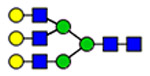 |
48 | 820 | 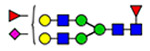 |
| 4 | 820, 1515 | 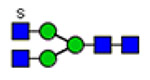 |
19 | 820, 857, 1515, 1574, 2223,2290, 2546, 2585, 2635 |
34 | 2290 | 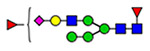 |
49 | 820 | ||
| 5 | 2290 | 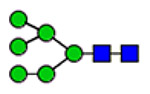 |
20 | 820 | 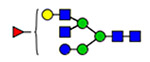 |
35 | 820 | 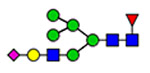 |
50 | 2290 | 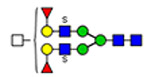 |
| 6 | 2585 | 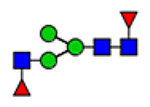 |
21 | 820, 1574 | 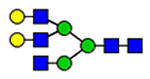 |
36 | 1515 | 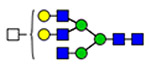 |
51 | 2290 | 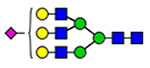 |
| 7 | 820, 1574 | 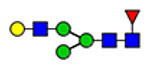 |
22 | 2290 | 37 | 2357 | 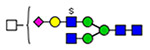 |
52 | 1515, 1574, 2290 | ||
| 8 | 1574, 2290, 2585 |
 |
23 | 820, 1574 | 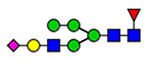 |
38 | 820, 847, 857, 1574, 2223, 2290, 2357, 2585, 2635 |
53 | 2223, 2290 | 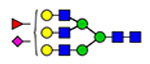 |
|
| 9 | 2585 | 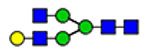 |
24 | 2585 | 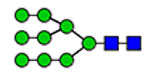 |
39 | 820, 2635 | 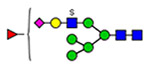 |
54 | 1574 | |
| 10 | 1574, 2290 | 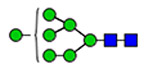 |
25 | 820, 1574, 2290 | 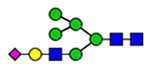 |
40 | 820 | 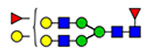 |
55 | 2290 | 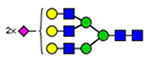 |
| 11 | 820 | 26 | 2290 | 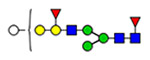 |
41 | 820 | 56 | 2357 | 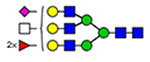 |
||
| 12 | 2290, 2585 | 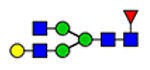 |
27 | 2290 | 42 | 820 | 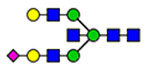 |
57 | 2357 | 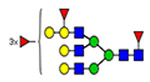 |
|
| 13 | 857, 1515, 1574 | 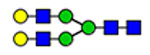 |
28 | 820, 1574, 2290, 2585 |
43 | 2223 | 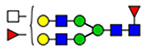 |
58 | 2635 | 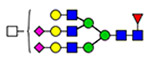 |
|
| 14 | 2290,2585 | 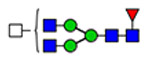 |
29 | 2223, 1635 | 44 | 2223 | 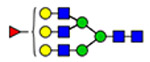 |
59 | 2290 | 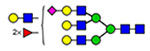 |
|
| 15 | 820,2635 | 30 | 1574 | 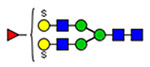 |
45 | 2290 | 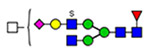 |
60 | 2290 | 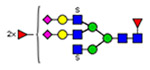 |
|
| 1 | 2298 |  |
2 | 2298 | 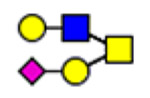 |
3 | 2298 | 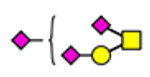 |
61 | 1574 |
(blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), (purple diamond) N-acetylneuraminic acid (Neu5Ac), and (red triangle) fucose.
Figure 7.
General characterization of plasma VWF N-glycome (61 compositions). (A) Total relative abundance (TRA) (bars; left side) and site-specific relative abundance (SRA (heat map; right side) of N-glycan compositions). (B) Distribution of N-glycan complexity (the outer circle represents the percentage of sulfated glycans of each type). (blue square) N-Acetylglucosamine (GlcNAc, N), (yellow square) N-acetylgalactosamine (GalNAc, N), (green circle) mannose (Man, H), (yellow circle) galactose (Gal, H), (purple diamond) N-acetylneuraminic acid (Neu5Ac, S), (red triangle) fucose (Fuc, F), and sulfate groups (s).
Scheme 1. Schematic Depiction of the Overall Workflow for Proteomic/Glycoproteomic Analysis of Plasma VWF by RP–LC–ESI–MS/MSa.
aY1n+ ([peptide + GlcNAc]n+), Yon+([peptide]n+); (blue square) N-acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), and (purple diamond) N-acetylneuraminic acid (Neu5Ac).
RESULTS AND DISCUSSION
Peptide Fragment Fingerprinting (PFF) and Protein Identification
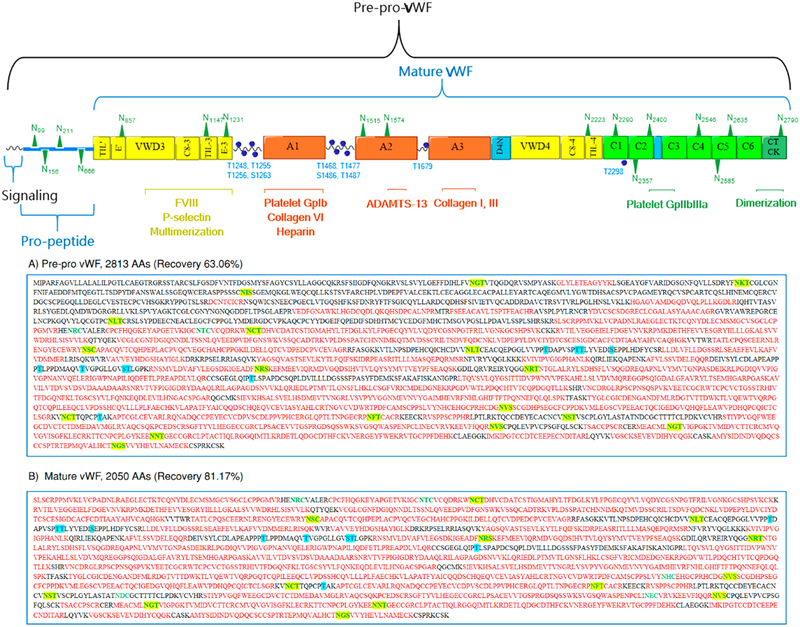
Plasma VWF glycoprotein was purified from excipients and subsequently tryptic digested by FASP. Then, a portion of the proteolytic digest was subjected to PNGase F treatment before bottom-up analysis. The deglycosylated peptides were online separated on an RP column to determine the peptide mass and the peptide fragment fingerprinting (PFF) by the Orbitrap and Ion Trap analyzers, respectively. All peptides were fully eluted within 40 min under RP conditions, and most of the abundant peptides showed mass to charge ratios lower than m/z 1200 within a full MS scan from m/z 400 to 1600. Proteomic analysis of VWF tryptic digest confirmed the identity of plasma VWF. As shown in Figure 1A, the sequence coverage of the full glycoprotein (prepro VWF, 2813 AAs, 309 kDa) is 63.06% with minimal protein impurities. A total of 17 N-glycosites and 10 O-glycosites are involved in the full-length VWF. This form of the glycoprotein includes the signaling sequence (1–22 AAs), the propolypeptide sequence (23–763 AAs), and the basic subunit with 2050 AAs. However, the detected peptides in the range 1–763 AAs, representing the signaling peptide as well as the propolypeptide, generated only one or two matched MS spectra with low scores. This result indicates that the protein is in its fully mature form as a result of prosequence removal of 763 AAs.11
Figure 1.
Proteomic analysis of therapeutic VWF by RPLC–ESI–CID–MS. The upper depiction represents the full length VWF including distinct domains, binding sites, and N- and O-glycosites. The lower part represents the coverage of (A) full length VWF (2813 AAs) and (B) mature VWF (2050 AAs).
Herein, we customized a FASTA sequence to include only the mature form of VWF. The input proteomic parameters were similar for both FASTA databases. As represented in Figure 1B, proteomic analysis of the trypsin-digested VWF peptides shows a coverage of 81.17%, and most of the identified peptides have more than two matched MS/MS spectra with acceptable scores. Hence, the plasma VWF concentrate is most likely in its mature form (764–2813 AAs) in which 13 N-glycosites and 10 O-glycosites are distributed over the functional domains. These sites were later screened for N-glycosite occupancy as well as N- and O-glycoform microheterogeneity. Although the proteolytically digested peptide, treated with PNGase F in advance, can be used for monitoring incomplete glycosylation at specific N-glycosites; the increment of +1 Da, as a result of Asn → Asp conversion, is indistinguishable. Indeed, spontaneous deamidation of unoccupied Asn in aqueous solution causes an increment of +0.984 Da. Therefore, we conducted a separate experiment for accurate determination of N-glycosite occupancy. Moreover, the peptide fragments including 8 O-glycosites, flanking both sides of Domain A1, modulate the interaction with the platelet receptors,52 and two discrete O-glycosites on Domain A3 and D4 were inaccessible for in silico sequencing. Indeed, sequence matching of O-glycan-containing peptides by proteomic software is still an algorithmic challenge. With various O-glycan structures, in silico sequencing of O-glycan-containing peptides is beyond the ability of most proteome software. Besides the absence of definable glycosite motifs, these O-glycopeptides also lack a suitable deglycosylation approach that maintains the peptide integrity.53
Occupancy of N-Glycosites
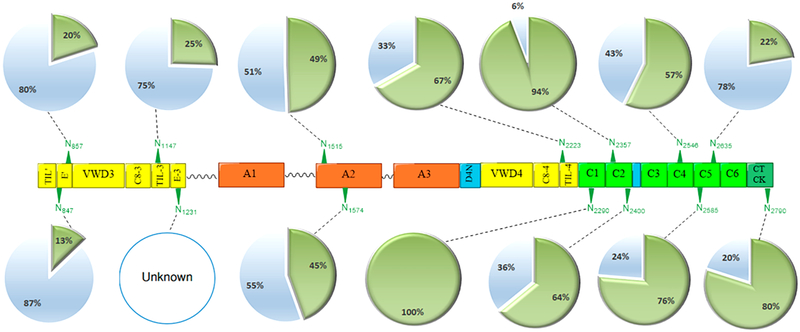
To determine glycosylation of all VWF asparagine residues involved in consensus sequence motifs, the tryptic digest derived from plasma VWF was isotopically labeled with 18O-water. The canonical N-site motif is N-X-S/T/C, where X is any amino acid except proline. Generally speaking, PNGase F cleaves the amide linkage (-N-CO-) between the first N-acetylglucosamine (GlcNAc) of the N-glycan core, resulting in Asn residue conversion into aspartic acid (Asp). The amide group (-CO-NH2) is replaced with a carboxyl group (-COOH). This replacement results in approximately +1 Da increment. However, in aqueous solutions, unoccupied Asn is spontaneously deamidated, which results in a mass change of 0.984 Da. Considering the deamidated Asn residues, the PNGase F deglycosylated sites are indistinguishable from those naturally deamidated. Thus, estimation of the glycosylation occupancy is imprecise depending only on regular conversion of Asn → Asp. Thus, we performed PNGase F digestion in 18O-water, which leads to a mass increment of 2.988 Da at the deglycosylated site. Herein, N-glycosites can be readily and confidently differentiated from free deamidated Asn residues in ordinary aqueous solutions. To avoid 18O-integration into the C-terminus of the peptide, the proteolytic digestion was performed before N-glycan release in 18O-water. Furthermore, the protease was excluded from the mixture by FASP in advance.
The intensity of 18O-labeled peptide (+2.988 Da mass shift) is compared with the counterpart that lacks glycosylation at the canonical N-sites. For the N-deglycosylated peptide identity to be confirmed, b- and y-ions generated by CID enable glycosite recognition. The CID spectra confirm all expected glycosylation sites: N857CT, N1147SC, N1515RS, N1574RT, N2223VS, N2290CT, N2357FT, N2400ST, N2546VS, N2585GT, N2635NT, and N2970GS. However, one glycosite, N1231LT, is inaccessible for glycosite monitoring. This site is positioned on a long tryptic-digested peptide (R1204–R1274, 70 AAs), which carries multiple O-glycosites. As aforementioned, the O-glycan-containing peptides are still problematic for most available proteomics software.53 Furthermore, trypsin-based digestion is unable to isolate the N1231 glycosite from the region containing the O-glycan clusters because of the absence of R or K residues.
Noteworthily, we have detected an Asn residue with a canonical sequon that has as yet been unreported as a glycosylated site. The 18O-labeled peptide (IGCN→dTCVC-QDR) containing an acceptable N-motif, N847TC on the D′(E′) domain, shows approximately 13% glycosylation possibility, as illustrated in Figure 2. Furthermore, relative quantification of 18O-labeled peptides and corresponding nonglycosylated peptides has indicated that N2290CT and N2357FT are highly occupied (>90%). Because the counterpart peptide of KVN→dCTTQPCPTAK is absent, and the de-N-glycopeptide shows an increment of 2.988 Da, we have assumed that N2290CT is completely glycosylated. Furthermore, the results indicate that more than half of the monitored N-glycosites in our study, seven sites, are >50% pre-N-glycan occupied. That is, the occupancy of N2223VS, N2290CT, N2357FT, N2400ST, N2546VS, N2585GT, and N2970GS are 67, 100, 94, 64, 57, 76, and 80%, respectively. However, N857CT, N1147SC, N1515RS, N1574RT, and N2635NT glycosites have less N-glycan inhabitation with 20, 25, 49, 45, and 22%, respectively. Moreover, N-glycosylation occupancies are noticeably light toward the N-terminus compared to the middle or C-terminus of mature VWF. The exception at the C-terminal side is N2635NT, which has only 22% occupancy. Regardless of the light occupancy, our glycoproteomic analysis has manifested slightly high microheterogeneity at this site with various glycoform structures. Our results have confirmed the previously claimed glyco-heterogeneity at this site.28
Figure 2.
N-Glycosite occupancy of VWF analyzed by RP LC–ESI–CID–MS. The glycoprotein was tryptic digested and then treated with PNGase F in the presence of 18O-water. The asparagine residues are converted into aspartic acid with a mass increment of +2.988 Da, signaling pre-N-glycosylation. (green triangle) N-glycosites, (open blue circle) nonscreened sites, (green circle) occupied, and (blue circle) unoccupied.
Unlike N- and C-termini, the midmost domain A2, carrying N1515RS and N1574RT, have exhibited moderate glycan occupancy with 49 and 45%, respectively. Glycosylation at these two N-glycosites plays an essential and protective role in the protein life cycle.22 Glycosylation on domain A2 modulates the interaction of plasma VWF with ADAMTS13 protease and thus preserves the level of VWF in the blood.22,54 According to McKinnon et al., mutations at N1574 increased the susceptibility of VWF to ADAMTS13 proteolysis because of the conformational changes in addition to steric hindrance by the glycan moieties.22 All 18O-labeled deglycosylated peptides, sequenced in the ion trap analyzer with 35.0 NCE, demonstrate predominant b- and y-ions in CID–MS/MS spectra of VWF. The peptide carrying an acceptable sequon, IGCN847TCVC-QDR, generates two distinct fragments, (y8+1) at m/z 1055.49 and (b4+1) at m/z 448.18 in the collision cell (Figure S1). Both fragments represent an increment of 2.988 Da as a result of 18O incorporation at the preglycosylated site compared to that of the unlabeled peptide. To our knowledge, this has not yet been reported as a glycosylated site. The glycoproteomic analysis of the intact VWF glycopeptides, meanwhile, has confirmed the glycosylation at this site with various glycoforms (Table S2). CID sequencing of all de-N-glycosylated peptides subsequently 18O-labeled, indicating prior glycosylation at canonical sites, is represented in Figures S1 and S2.
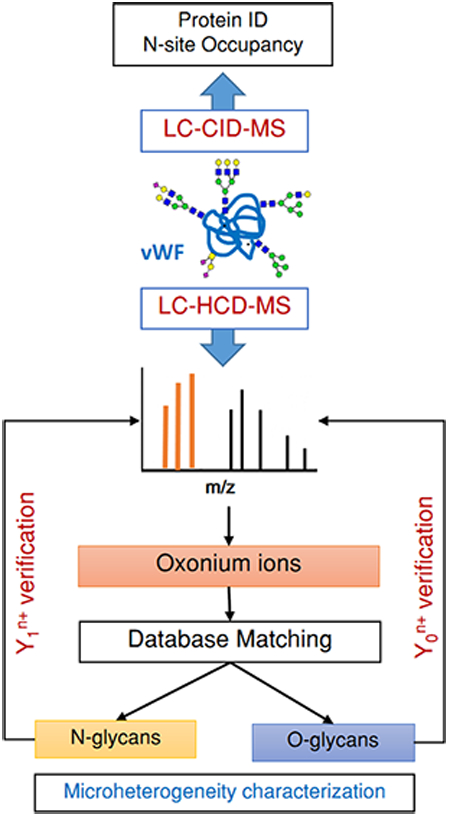
Strategy for VWF Microheterogeneity Characterization
Glycosylation is a highly complex modification that influences protein functions and hence is critical for adequate activities of the key hemostatic proteins.16 Plasma VWF is a heavily glycosylated protein, and our SDS-PAGE experiment (data not shown) has implied that the glycosylation portion might represent approximately 20% of the whole protein molecular weight. The therapeutic VWF concentrate was purified from excipients and subjected to tryptic FASP digestion. Subsequently, the protein digest was desalted and enriched on a homemade HILIC SPE column before online RP separation. All eluted glycopeptides were directed into a high-precision Orbitrap Elite instrument in which the 10 most intense glycopeptide ions were oriented to the collision cell at the far side of the C-trap for HCD fragmentation. The fragments were then transferred back to the C-trap and analyzed by Orbitrap analyzer with high resolution and accuracy. The HCD technique surmounts the problems of the low mass cutoff of ion trap fragmentation and dramatically improves the quality of MS/MS spectra because of the high accuracy at both the precursor mass and fragment levels.55
All HCD spectra with signature peaks were manually picked and cross-referenced with available online tools and databases to specify the peptide and glycan portions. Ultimately, Y0n+ (peptide) and Y1n+ (peptideY0 + GlcNAc) were manually verified by sequential glycoside-bond cleavages to ensure accurate assignment of the glycopeptide ID. Furthermore, [Y0 — NH3]n+ and [Y0 – H2O]n+ were also noticeable in O-glycopeptide HCD spectra. A total of 515 HCD spectra were diagnosed with oxonium ion fragments. However, only 257 spectra showed clear Y1n+ and Y0n+ ions with subsequent cleavages representing the glycosidic bonds within the carbohydrate portion. Out of 257 HCD spectra, approximately 70% (181 spectra) were specified to either N- or O-glycosites, whereas ~30% (76 spectra) with acceptable glycosylation compositions were inaccessible for coordination to a certain VWF glycosite. The specified HCD spectra were categorized into 173 N-glycopeptides and eight O-glycopeptides. Although some of the unspecified HCD spectra showed acceptable N- or O-glycan compositions, those unsynchronized Y1n+ and Y0n+ ions to a specific canonical VWF glycosite were excluded from our results.
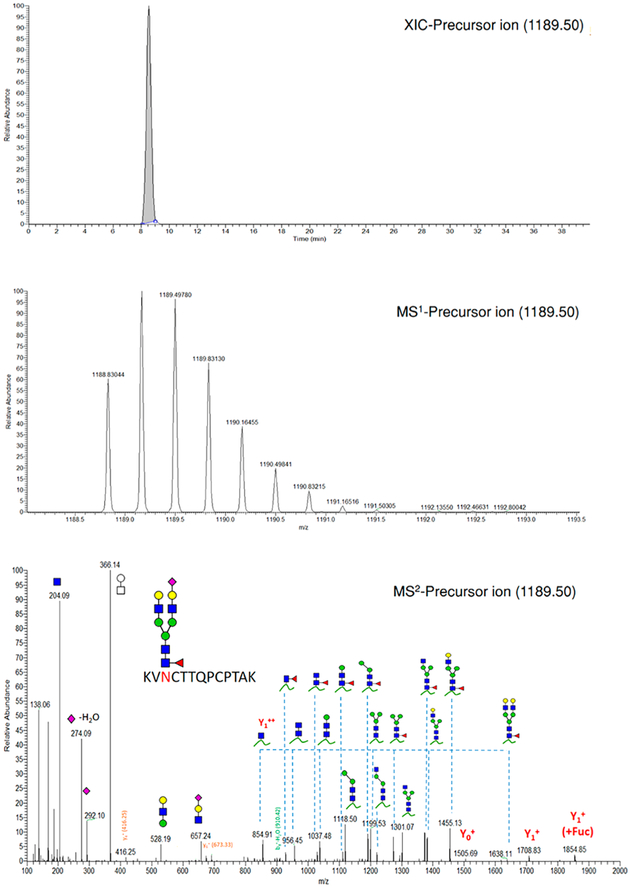
Interestingly, the Y1n+ + Fuc (peptide Y0 + GlcNAc + Fuc) ion was also observed with high intensity compared to Y1n in some core-fucosylated N-glycopeptides. HCD fragmentation of these core-fucosylated glycopeptides most likely generates two synchronous peaks corresponding to the core-fucosylated and the corresponding nonfucosylated glycopeptide. Figure 3 shows the phenomenon above for the (KVN2290CTTQPCPTAK) peptide carrying a core-fucosylated biantennary glycan. As shown in HCD-MS2, the cluster of peaks at downstream m/z represent the diagnostic fragments: m/z 204 (HexNAc, 1+), m/z 162 (Hex, 1+), m/z 186 (HexNAc-H2O, 1+), m/z 168 (HexNAc-2H2O, 1+), m/z 138 (HexNAc-2H2O-CH3OH, 1+), m/z 366 (HexHexNAc1, 1+), m/z 292 (Neu5Ac, 1+), and m/z 274 (Neu5Ac-H2O, 1+). The daughter ions at m/z 854.91 and 1708.83 represent Y12+ and Y11+, respectively. Although the intact glycopeptide (m/z 1189.50, MS1) is triply charged, the predominant fragments are either doubly or singly charged. The identity of the precursor ion is confirmed from its isotopic pattern together with Y1n+ and Y0n+ and available peptide-backbone fragments. The mass difference between the precursor ion and the corresponding peptide determines the glycan composition subsequently proposed by GlycoWork-bench. For stipulating the glycan structure, sequential fragmentation of the glycosidic bonds, as shown in Figure 3, affirms the glycan integrity.
Figure 3.
Extracted ion chromatogram (XIC), MS1, and HCD-MS2 spectra of (KVN2290CTTQPCPTAK) glycopeptide derived from VWF. Y1n+ ([peptide + GlcNAc]n+), Yon+([peptide]n+). (blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), (purple diamond) N-acetylneuraminic acid (Neu5Ac), and (red triangle) fucose.
Noteworthily, N-acetylneuraminic acid residues (Neu5Ac) are imperceptible in the m/z range of sequential glycosidic bond cleavages, especially in HCD spectra of N-glycopeptides. The glycosidic bond of Neu5Ac is extremely vulnerable and susceptible to full precleavage in the dissociation cell before detection by the Orbitrap analyzer. The existence of Neu5Ac is, however, endorsed by oxonium ion peaks at m/z 292 (Neu5Ac, 1+) and m/z 274 (Neu5Ac-H2O, 1+), as represented in Figure 3. Moreover, for further confirmation of the corresponding peptide, the peptide sequence is confirmed by pLabel (http://pfind.ict.ac.cn),48 standalone software, once peptide backbone fragments are available. Peptide fragments, b- and y-ions, are assigned with the corresponding peaks, as shown in HCD spectrum of (KVN2290CTTQPCPTAK). Unlike O-glycopeptides, the glycosidic bond cleavages are predominate in the N-glycopeptide HCD spectrum so that a few peaks are correlated to the peptide backbone fragments. Once the peptide portion and the carbohydrate moiety are characterized, the intact glycopeptide is semiquantified via the normalized peak area obtained by XIC.
N- and O-Glycosite Microheterogeneity
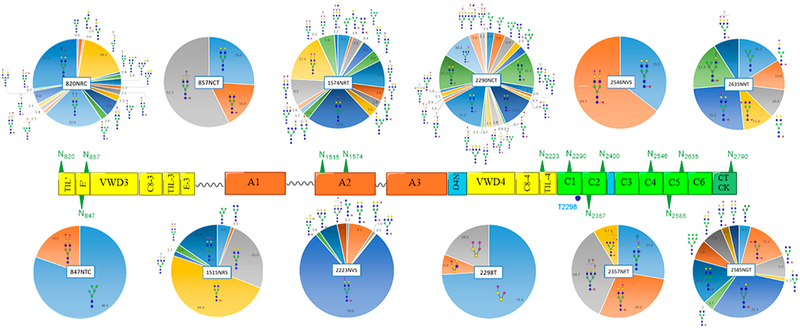
To attain site-specific characterization of plasma-derived VWF N- and O-glycosites, we performed trypsin proteolysis, offline HILIC enrichment, and RP–LC–MS/MS via HCD fragmentation. Manual characterization was simultaneously carried out on N- and O-glycopeptide HCD spectra exhibiting signature fragments of oxonium ions. All observed N- and O-glycoform compositions are summarized in Table 1. To simplify the data display, we categorized 181 detected glycoforms according to the glycan compositions in which N- and O-glycan cores are preserved. A total of 61 N-glycan compositions, representing 173 N-glycan structures, are assigned to 11 specific N-glycosites. That is, 32, 4, 3, 11, 32, 15, 44, 5, 2, 17, and 8 N-glycan structures have been assigned to 820NRC, 847NTC, 857NCT, 1515NRS, 1574NRT, 2223NVS, 2290NCT, 2357NFT, 2546NVS, 2585NGT, and 2635NNT, respectively. Besides N-glycoforms, a total of 8 O-glycoforms are associated with 2298T, all of which are classified into three O-glycan compositions.
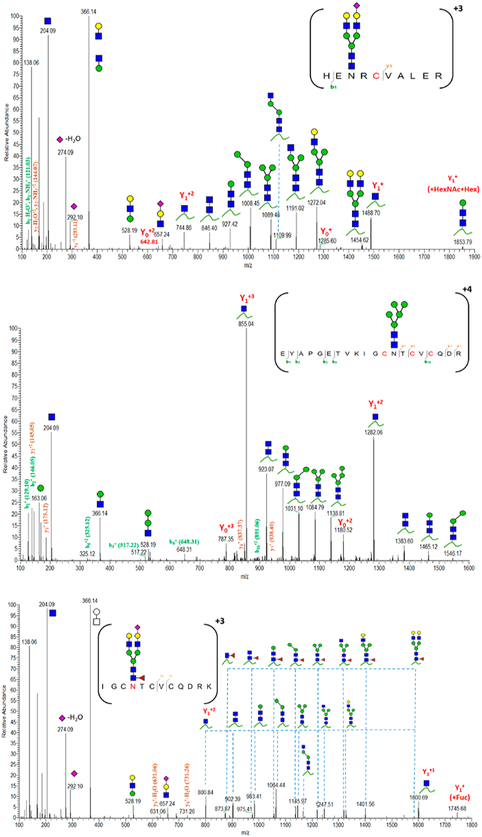
We have considered all glycopeptides carrying consensus sequences (N-X-S/T/C) for glycosylation investigation of HCD spectra. Once the identity of the glycopeptide ions (Y0n+ and Y1n+) are defined, the glycan composition is specified by glycosidic bond cleavages, and the precursor ion is confirmed by mass matching and peptide backbone fragments, the N-site is considered as glycosylated. Notably, 820NRC carried on HEN820RCVALER and 847NTC correlated to IGCN847TCVCQDRK or EYAPGETVKIGCN847TCVCQDR disclose various degrees of glycosylation, as shown in Table 1 and Figure 4. Our results indicate that 820NRC is even heavily glycosylated with 32 glycoforms, whereas 847NTC shows less microheterogeneous peptides eluted at different RT. Similarly, Canis et al. have reported that 2635NNT was heavily glycosylated, although this site had been reported as unoccupied.28 The employment of advanced MS increases the ability to uncover newly glycosylated sites even for well-known glycoproteins. Hence, regarding the N-site occupancy, 13% of 847NTC-carrying peptides are occupied with four different N-glycoforms compared to those of the counterpart peptides. In contrast, the highly abundant nonglycosylated peptides might have suppressed the ionization efficiency of 820N-containing peptides. The glycoproteomic analysis, however, verifies high glycosylation at this site. Figure 4 shows in-depth sequencing of the detected glycopeptides containing either 820NRC or 847NTC. Although glycosidic cleavages most likely predominate HCD spectra, Y1n+ and Yon+ ions, as well as peptide backbone fragments, assist the confirmation of the glycopeptide identity. Similarly, other N- and O-glycopeptides are underlined by the presence of correlated fragment ions.
Figure 4.
Characterized HCD spectra of intact VWF glycopeptides carrying 820NRC or 847NTC N-glycosites. Y1n+ ([peptide + GlcNAc]n+), Yon+([peptide]n); y- and b-ions are peptide backbone fragments. (blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), (purple diamond) N-acetylneuraminic acid (Neu5Ac), and (red triangle) fucose.
The most likely detected N-glycoforms are biantennary glycans. Tri- and tetra-antennary, as well as those glycoforms including lactosaminic extension, ABH antigens, and sulfation, are also observed. Our results are harmonious with those previously published about Asn-attached carbohydrate moieties of VWF.26,28 However, HILIC-based enrichment is unbiased to specific glycan structures such that we have also detected hybrid glycans across multiple N-glycosites. Figure 5 shows the relative abundance of these detected VWF N- and O-glycoforms. Detailed structures and site-specific relative abundance of detected glycoforms are summarized in Tables S1–S12. Briefly, 820NRC, 1574NRT, 2290NCT, and 2585NGT represent the highest microheterogeneity in the level of glycan composition or structure. For instance, a total of 32 glycan structures specified to 820NRC are categorized into 19 glycan compositions. These glycoforms mostly represent complex glycans with or without ABH antigens. Hybrid and sulfated glycans are also tangible. The highly abundant glycoforms are H5N5S1F1, H5N4S1F1, and H4N3S1F1 with 27, 22, and 18%, respectively. The lowest abundant glycan (<1%) was the sulfated glycan (H3N4S1). The hybrid glycans represent <3% of the total identified glycans at this site.
Moreover, the terminal 847NTC site displays two distinct compositions, high mannose and complex glycans, carried on two different peptides shown in Figure 4. Two glycoforms represent high mannose (H5N2) covalently linked to (EYA-PGETVKIGCNTCVCQDR), whereas the other two complex glycans (H5N4S1F1) are covalently attached to (IGCNTCV-CQDRK). The high mannose carrying glycopeptides, eluted at 11.71 and 11.77 min, represent approximately 80% of the detected glycoforms on this site. The shorter glycopeptides, eluted at 9.77 (11%) and 10.42 (9%) min, carry either a core fucosylated biantennary glycans or the counterpart fucose-terminated glycoform, respectively. The lowest number of detected glycoforms are observed on 857NCT (three glyco-forms) and 2546NVS (two glycoforms). Both sites are modified with biantennary complex glycoforms, as illustrated in Figure 5. Glycosylation of the 857NCT site is in fact essential for appropriate folding of the newborn protein and hence affects VWF expression.23 Less than ten glycoforms are detected on 2357NFT and 2635NNT, most of which are biantennary complex glycoforms. Highly complex glycans, triantennary, are also noticeable as well as blood group antigens (A and H).
The midmost glycosites, 1515NRS (11 glycoforms) and 2223NVS (15 glycoforms), as well as 2585NGT (17 glycoforms) on the VWF C-terminus, reveal <20 glycoforms, most of which are biantennary glycans. On 1515NRS, the asialobiantennary H5N4 and core fucosylated H5N4F1 represent approximately 25 and 49% compared to the counterpart disialylated glycoforms of 2.5 and 15.4%, respectively. This site flanks the N-terminus side of the ADAMTS13 cleavage site (A2 domain). The sialylation pattern of the attached glycoforms might mediate VWF proteolysis by ADAMTS13 as well as clearance from the bloodstream.21 Noticeable, the biantennary core-fucosylated glycan (H5N4S1F1) occupied ~78% of 2223NVS, and the same glycoform represented <3% of the 2585NGT occupancy. The latter site is mostly occupied with H4N4F1 (31.3%).
Furthermore, two canonical N-glycosites, 1574NRT and 2290NCT, exhibit heavy glycosylation with 32 and 44 N-glycoforms, respectively. As shown in Figure 5, the highly abundant glycoforms on 1574NRT are H5N4S1 (22.6%) and H5N4S2 (17%). The fucosylated forms, H5N4S1F1 and H5N4S2F1, represent only 9.5 and <1%, respectively. Furthermore, the highly complex glycan, H8N6S3F2 (<2%), with galactosaminic extension is also observed. Several studies have claimed that sialylated glycans attached to 1574NRT might play an essential role in VWF proteolysis by ADAMTS13.21,22 That is, glycosylation at 1574NRT in conjunction with the Ca2+ binding site and the disulfide bond within domain A2 are necessary components for stable interaction with ADAMTS13.56
Unsurprisingly, the second heavily glycosylated site, 2290NCT, previously demonstrated as a fully occupied site, reveals highly diverse microheterogeneity with 44 glycoforms characterized into 28 glycan structures. Most of the detected glycoforms are attached to (KVNCTTQPCPTAK, 0 or 1 missed cleavages), and only one glycoform (H5N5S2F3s2) is associated with a longer peptide (K2288–R2311, three missed cleavages). Briefly, biantennary core fucosylated glycoforms H5N4S1F1(15.2%), H5N4S2F1 (12.2%), H4N4F1 (12.2%), and H5N4F1 (11.2%) represent the abundant glycoforms. Regarding triantennary glycans, only one core fucosylated glycan (H6N5S1F1) shows slightly high abundance (10.2%), whereas other triantennary glycans with or without H antigen are low abundance (<3.6%). Moreover, H and A antigens are observed on this site besides high mannose (4, 5, 6, and 7 <4%) and hybrid forms (<1%).
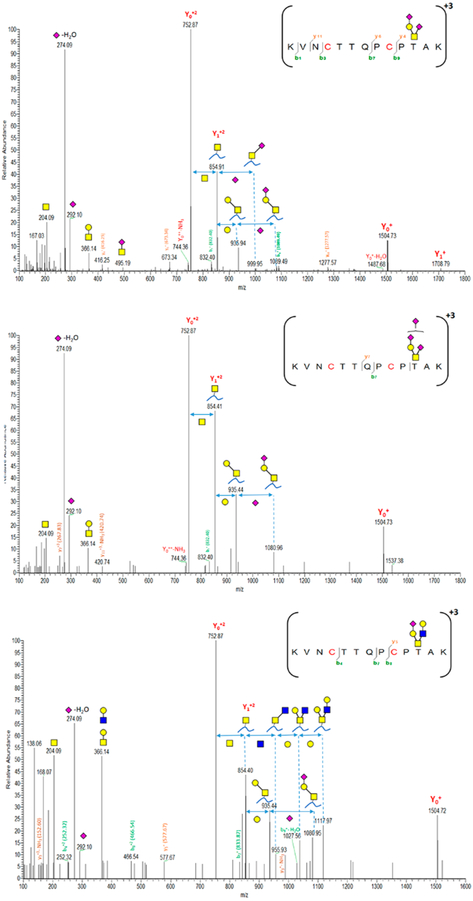
Regarding O-glycan microheterogeneity, a total of 8 HCD spectra are correlated to 2298T-containing peptides. These HCD spectra are characterized to disialylated core 1 (H1N1S2, 74%, five spectra), trisialylated core 1 (H1N1S3, 19%, two spectra), and sialylated core 2 (H2N2S1, 7%, 1 spectrum). The primary structure of disialylated core 1 (T antigen) is (NeuAc(α2–3) Gal(β1–3)-[NeuAc(α2–6)] GalNAc). Di- and trisialylated T antigens were previously reported on VWF 2298T-O-glycoforms.32 Furthermore, monosialylated O-glycan core 2 (H2N2S1, <7%) is also common at this site. Figure 6 shows characterized HCD spectra of di- and trisialylated core 1 as well as monosialylated core 2 attached to 2298T. Of interest, smaller O-glycan structures result in more fragments derived from the peptide backbone being observed in the HCD spectrum. On the other hand, other O-glycosites located on long trypsin-digested peptides are inaccessible for detection. It is reasonable that the effect of multiple hydrophobic amino acid residues might overwhelm the hydrophilic interaction with HILIC. These long O-glycopeptides can barely be enriched by HILIC interaction, especially when such small O-glycans are attached to the peptide. However, this hydrophobic effect could be easily prevented by using multiple enzyme digestions so that an effective HILIC enrichment could be attainable.
Figure 6.
Characterized HCD spectra of di- and trisialylated core 1 and monosialylated core 2 glycoforms attached to the 2298T O-glycosite of VWF. Y1n+ ([peptide + GalNAc]n+), Yon+([peptide]n+); y- and b-ions are peptide backbone fragments. (blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (yellow circle) galactose (Gal), and (purple diamond) N-acetylneuraminic acid (Neu5Ac).
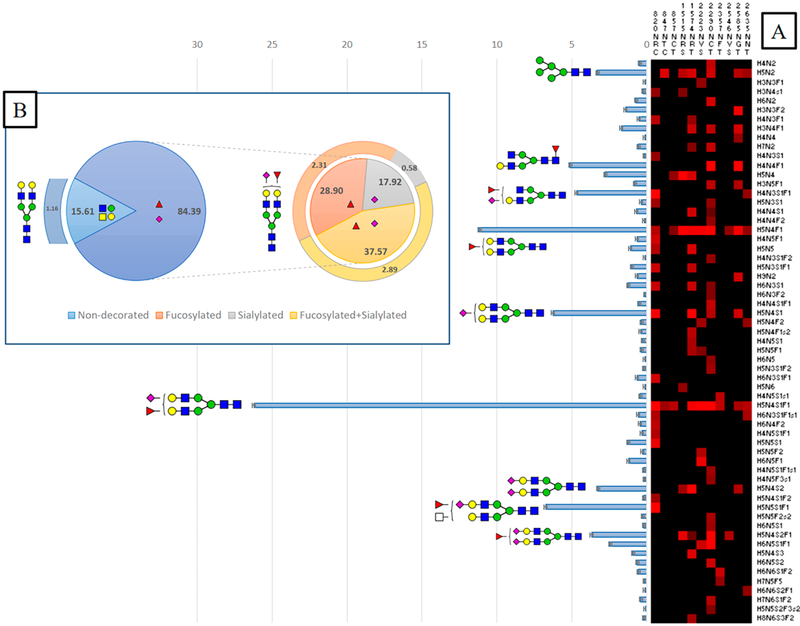
Characterization of VWF N-Glycome
All potential N-glycosites including Asn residues within consensus sequons (N-X-T/S/C) were monitored for glycan microheterogeneity. Eleven N-glycosites are identified with different glycoforms. Regardless, the structural details either at the core or the nonreducing terminus of the glycan, a total of 173 N-glycoforms are categorized into 61 N-glycan compositions in which the N-glycan core is preserved. Figure 7 illustrates the general characterization and distribution of VWF N-glycome based on these 61 N-glycan compositions. Depending on normalized areas, the relative abundance of all N-glycan compositions is determined for the total N-glycome and site-specific. Interestingly, the most abundant glycan is biantennary. As shown in Figure 7A, the most abundant glycoform is H5N4S1F1 (>26.2%). This N-glycan with either core or terminal fucose is defined on almost all monitored N-glycosites, except 1515NRS and 2546NVS. The second abundant glycoform is the asialo-form (H5N4F1, 11.2%). The last glycoform is distributed among all N-glycosites except 847NRS and 2546NVS.
The abundance of biantennary glycoforms is unsurprising. Our result is in fact consistent with those characterized VWF N-glycans profiled decades earlier.26 Unlike biantennary glycoforms, most of the highly complex glycans (tri- and tetra-antennary) are represented in low abundance. The highest abundant triantennary glycan (H6N5S1F1, 2.5%) is covalently attached to Asn of 2223NVS and 2290NCT. Furthermore, the hybrid glycan H6N3S1 is distributed among three glycosites, 820NRC, 1574NRT, and 2290NCT, with a total of 1.23% abundance. The counterpart core fucosylated hybrid glycan is even low abundance (<0.7%, 820NRC). Moreover, sulfated N-glycans represent only <1% of the total detected glycoforms as well as high mannose (4–9 Man, <1%). An exception is H5N2 (3.3%). As shown in Figure 7B, most of the detected N-glycans (~84%) are decorated with fucose (28.9%), N-acetylneuraminic (17.92%), or both (37.57%), whereas the nondecorated forms represent only ~16%. The VWF N-glycome profile is highly fucosylated. Furthermore, most sulfated glycans are accompanied by fucosylation, sialylation, or both. Sulfated glycoforms, nevertheless, are infrequent.
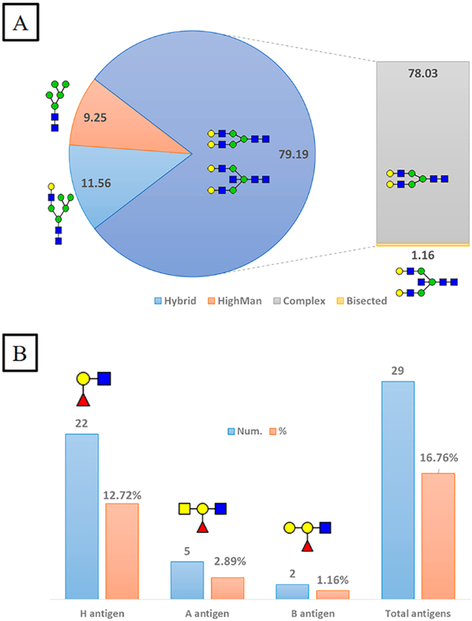
Furthermore, we have classified individual N-glycoforms detected at each site based on the glycan type. A total of 173 glycoforms are categorized into complex, hybrid, or high mannose. As illustrated in Figure 8A, complex glycans represent ~79% of the total glycoforms, only ~1% of which are bisected. Unlike complex glycans, high mannose and hybrid glycans exist in noticeably lower abundance. It is worth mentioning that the homemade HILIC column is efficient and unbiased for enriching all glycan structures, including those carrying ABH antigens. A total of 29 glycoforms are characterized as A, B, or H antigen carrying glycans, as demonstrated in Figure 8B. Approximately 17% of total N-glycoforms carry fucose residues on their nonreducing termini. The H antigen (13%) is highly abundant compared to A (3%) or B antigens (1%). Our result is predictable because it was estimated that 13% of N-glycans were terminated with H antigens.26 However, considering the unmonitored N-glycosites, the abundance of H-antigen-carrying glycoforms might be underestimated. Hence, further analysis is required to determine blood group antigens at the unscreened sites. After all, rational understanding of glycoform microheterogeneity is indeed crucial for meaningful manufacturing and quality control in the biopharmaceutical industry.
Figure 8.
Classification of 173 N-glycoforms specified into VWF glycosites. (A) Relative abundance of VWF N-glycoform types. (B) Number and relative abundance of ABH antigens containing N-glycoforms. (blue square) N-Acetylglucosamine (GlcNAc), (yellow square) N-acetylgalactosamine (GalNAc), (green circle) mannose (Man), (yellow circle) galactose (Gal), (purple diamond) N-acetylneuraminic acid (Neu5Ac), and (red triangle) fucose.
CONCLUSIONS
In summary, implementation of a state-of-the-art mass spectrometry system equipped with a sensitive tandem mass technique is an attractive method for glycoprotein detection and fragmentation. With high resolution and accuracy, the Orbitrap analyzer enabled us to simultaneously and elaborately determine structural diversities of even low abundance VWF N- and O-glycoforms along with site localization. Conformational changes among the detected glycoforms were distinguishable via the sequential cleavage of glycosidic bonds. Furthermore, for delicate assignment of the glycoform to a specific glycosite, Y1n+ and Y0n+ ions were verified on HCD spectra, besides valuable b- and y-fragments derived from the peptide backbone. Together with N-occupancy estimation, the glycopeptide analysis revealed new N-glycosites, one of which showed heavy glycosylation. The homemade SPE column packed with HILIC material endowed an efficient and equitable way to enrich short N- and O-glycopeptides, including those glycoforms terminated with ABH antigens, sulfate groups, as well as lactosaminic extension. Although the one-enzyme digestion system produced some peptides with insufficient hydrophilicity, essential for effective interaction with HILIC, the number of detected glycoforms was significant. A total of 257 HCD spectra were diagnosed with oxonium ions, 181 of which were specified to VWF glycosites. Multiple enzymatic digestions complemented with effective HILIC enrichment is an appropriate method that might reveal even more glycosite microheterogeneity unless very short peptides are produced. Furthermore, our findings on a real concentrate VWF, commonly used to treat bleeding disorders, have lightened the ambiguity shading glycosylation pattern of nonreported canonical N-sites. Such explicit depiction of PTMs, e.g., glycosylation, is of great importance for quality evaluation of rVWF to the maximal fidelity to the counterpart plasma-derived VWF.10 Characterization of N- and O-glycome heterogeneity might provide meaningful data that facilitates the complexity accompanying the industrial production of such a complex clotting factor, VWF. Moreover, given newly discovered N-glycosites, further studies are required to investigate probable effects on VWF properties and functions.
Supplementary Material
ACKNOWLEDGMENTS
We are much obliged to Dr. Xinmiao Liang from the Dalian Institute of Chemical Physics, the Chinese Academy of Sciences, for providing Click Maltose materials. We sincerely thank the Georgia Research Alliance (GRA) and Georgia State University for purchasing the analytical instrument used in this research.
Funding
This project was partially supported by funding from NIH grants HL114152 and HL130871.
ABBREVIATIONS
- AAs
amino acids
- AGC
automatic gain control target
- FASP
filter-aided sample preparation
- HILIC
hydrophilic interaction liquid chromatography
- NCE
normalized collision energy
- PFF
peptide fragment fingerprinting
- PTMs
post-translational modifications
- SPE
solid-phase extraction
- VWF
von Wille-brand Factor
- XIC
extracted ion chromatogram
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteo-me.7b00359.
Figure S1, MS2 fragmentation of 18O-labeled de-N-glycosylated VWF peptides located on Domain D′(E′), D3 (TIL-3), D4 (TIL-4), and C1–C2; Figure S2, MS2 fragmentation of 18O-labeled de-N-glycosylated VWF peptides located on Domain A2, C2–C5, and CTCK; Table S1, detected N-glycoforms associated with the 820NRC site; Table S2, detected N-glycoforms associated with the 847NTC site; Table S3, detected N-glycoforms associated with the 857NCT site; Table S4, detected N-glycoforms associated with the 1515NRS site; Table S5, detected N-glycoforms associated with the 1574NRT site; Table S6, detected N-glycoforms associated with the 2223NVS site; Table S7, detected N-glycoforms associated with the 2290NCT site; Table S8, detected O-glycoforms associated with the 2298T site; Table S9, detected N-glycoforms associated with the 2357NFT site; Table S10, detected N-glycoforms associated with the 2546NVS site; Table S11, detected N-glycoforms associated with the 2585NGT site; and Table S12, detected N-glycoforms associated with the 2635NNT site (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Khoury GA; Baliban RC; Floudas CA Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep 2011, 1, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Varki A; Esko JD; Colley KJ Cellular Organization of Glycosylation In Essentials of Glycobiology, 2nd ed.; Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, Eds.; Cold Spring Harbor: New York, NY, 2009. [PubMed] [Google Scholar]
- (3).Peyvandi F; Garagiola I; Baronciani L Role of von Willebrand factor in the haemostasis. Blood Transfus 2011, 9 (Suppl 2), s3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Baruch D; Bahnak B; Girma JP; Meyer D von Willebrand factor and platelet function. Bailliere’s clinical haematology 1989, 2 (3), 627–72. [DOI] [PubMed] [Google Scholar]
- (5).Wise RJ; Dorner AJ; Krane M; Pittman DD; Kaufman RJ The role of von Willebrand factor multimers and propeptide cleavage in binding and stabilization of factor VIII. J. Biol. Chem 1991, 266 (32), 21948–55. [PubMed] [Google Scholar]
- (6).Versteeg HH; Heemskerk JW; Levi M; Reitsma PH New fundamentals in hemostasis. Physiol. Rev 2013, 93 (1), 327–58. [DOI] [PubMed] [Google Scholar]
- (7).Castaman G; Tosetto A; Rodeghiero F Reduced von Willebrand factor survival in von Willebrand disease: pathophysiologic and clinical relevance. J. Thromb. Haemostasis 2009, 7 (Suppl 1), 71–4. [DOI] [PubMed] [Google Scholar]
- (8).Fischer BE Recombinant von Willebrand factor: potential therapeutic use. J. Thromb. Thrombolysis 1999, 8 (3), 197–205. [DOI] [PubMed] [Google Scholar]
- (9).Turecek PL; Mitterer A; Matthiessen HP; Gritsch H; Varadi K; Siekmann J; Schnecker K; Plaimauer B; Kaliwoda M; Purtscher M; Woehrer W; Mundt W; Muchitsch EM; Suiter T; Ewenstein B; Ehrlich HJ; Schwarz HP Development of a plasma- and albumin-free recombinant von Willebrand factor. Hamostaseologie 2009, 29 (Suppl 1), S32–8. [PubMed] [Google Scholar]
- (10).Kumar SR Industrial production of clotting factors: Challenges of expression, and choice of host cells. Biotechnol. J 2015, 10 (7), 995–1004. [DOI] [PubMed] [Google Scholar]
- (11).Wagner DD; Marder VJ Biosynthesis of von Willebrand protein by human endothelial cells: processing steps and their intracellular localization. J. Cell Biol 1984, 99 (6), 2123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wagner DD Cell biology of von Willebrand factor. Annu. Rev. Cell Biol 1990, 6, 217–46. [DOI] [PubMed] [Google Scholar]
- (13).Zhou Y-F; Eng ET; Zhu J; Lu C; Walz T; Springer TA Sequence and structure relationships within von Willebrand factor. Blood 2012, 120 (2), 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Vischer UM; Wagner DD von Willebrand factor proteolytic processing and multimerization precede the formation of Weibel-Palade bodies. Blood 1994, 83 (12), 3536–44. [PubMed] [Google Scholar]
- (15).Titani K; Kumar S; Takio K; Ericsson LH; Wade RD; Ashida K; Walsh KA; Chopek MW; Sadler JE; Fujikawa K Amino acid sequence of human von Willebrand factor. Biochemistry 1986, 25 (11), 3171–84. [DOI] [PubMed] [Google Scholar]
- (16).Karlaftis V; Perera S; Monagle P; Ignjatovic V Importance of post-translational modifications on the function of key haemostatic proteins. Blood Coagulation Fibrinolysis 2016, 27 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- (17).Badirou I; Kurdi M; Legendre P; Rayes J; Bryckaert M; Casari C; Lenting PJ; Christophe OD; Denis CV In vivo analysis of the role of O-glycosylations of von Willebrand factor. PLoS One 2012, 7 (5), e37508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Haberichter SL; Budde U; Obser T; Schneppenheim S; Wermes C; Schneppenheim R The mutation N528S in the von Willebrand factor (VWF) propeptide causes defective multimerization and storage of VWF. Blood 2010, 115 (22), 4580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cruz MA; Handin RI; Wise RJ The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX. The role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain protein. J. Biol. Chem 1993, 268 (28), 21238–45. [PubMed] [Google Scholar]
- (20).Fallah MA; Huck V; Niemeyer V; Desch A; Angerer JI; McKinnon TA; Wixforth A; Schneider SW; Schneider MF Circulating but not immobilized N-deglycosylated von Willebrand factor increases platelet adhesion under flow conditions. Biomicrofluidics 2013, 7 (4), 44124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).McGrath RT; McKinnon TA; Byrne B; O’Kennedy R; Terraube V; McRae E; Preston RJ; Laffan MA; O’Donnell JS Expression of terminal alpha2–6-linked sialic acid on von Willebrand factor specifically enhances proteolysis by ADAMTS13. Blood 2010, 115 (13), 2666–73. [DOI] [PubMed] [Google Scholar]
- (22).McKinnon TA; Chion AC; Millington AJ; Lane DA; Laffan MA N-linked glycosylation of VWF modulates its interaction with ADAMTS13. Blood 2008, 111 (6), 3042–9. [DOI] [PubMed] [Google Scholar]
- (23).McKinnon TA; Goode EC; Birdsey GM; Nowak AA; Chan AC; Lane DA; Laffan MA Specific N-linked glycosylation sites modulate synthesis and secretion of von Willebrand factor. Blood 2010, 116 (4), 640–8. [DOI] [PubMed] [Google Scholar]
- (24).Debeire P; Montreuil J; Samor B; Mazurier C; Goudemand M; van Halbeek H; Vliegenthart JF Structure determination of the major asparagine-linked sugar chain of human factor VIII–von Willebrand factor. FEBS Lett 1983, 151 (1), 22–6. [DOI] [PubMed] [Google Scholar]
- (25).Samor B; Michalski JC; Debray H; Mazurier C; Goudemand M; Van Halbeek H; Vliegenthart JF; Montreuil J Primary structure of a new tetraantennary glycan of the N-acetyllactosaminic type isolated from human factor VIII/von Willebrand factor. Eur. J. Biochem 1986, 158 (2), 295–8. [DOI] [PubMed] [Google Scholar]
- (26).Matsui T; Titani K; Mizuochi T Structures of the asparagine-linked oligosaccharide chains of human von Willebrand factor. Occurrence of blood group A, B, and H(O) structures. J. Biol. Chem 1992, 267 (13), 8723–31. [PubMed] [Google Scholar]
- (27).Jenkins PV; O’Donnell JS ABO blood group determines plasma von Willebrand factor levels: a biologic function after all? Transfusion 2006, 46 (10), 1836–44. [DOI] [PubMed] [Google Scholar]
- (28).Canis K; McKinnon TA; Nowak A; Haslam SM; Panico M; Morris HR; Laffan MA; Dell A Mapping the N-glycome of human von Willebrand factor. Biochem. J. 2012, 447 (2), 217–28. [DOI] [PubMed] [Google Scholar]
- (29).Samor B; Mazurier C; Goudemand M; Debeire P; Fournet B; Montreuil J Preliminary results on the carbohydrate moiety of factor VIII/von Willebrand factor (FVIII/vWf). Thromb. Res 1982, 25 (1–2), 81–9. [DOI] [PubMed] [Google Scholar]
- (30).Samor B; Michalski JC; Mazurier C; Goudemand M; De Waard P; Vliegenthart JF; Strecker G; Montreuil J Primary structure of the major O-glycosidically linked carbohydrate unit of human von Willebrand factor. Glycoconjugate J. 1989, 6 (3), 263–70. [DOI] [PubMed] [Google Scholar]
- (31).Canis K; McKinnon TA; Nowak A; Panico M; Morris HR; Laffan M; Dell A The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J. Thromb. Haemostasis 2010, 8 (1), 137–45. [DOI] [PubMed] [Google Scholar]
- (32).Solecka BA; Weise C; Laffan MA; Kannicht C Site-specific analysis of von Willebrand factor O-glycosylation. J. Thromb. Haemostasis 2016, 14 (4), 733–46. [DOI] [PubMed] [Google Scholar]
- (33).Lenting PJ; Pegon JN; Christophe OD; Denis CV Factor VIII and von Willebrand factor-too sweet for their own good. Haemophilia 2010, 16 (Suppl 5), 194–9. [DOI] [PubMed] [Google Scholar]
- (34).Ongay S; Boichenko A; Govorukhina N; Bischoff R Glycopeptide enrichment and separation for protein glycosylation analysis. J. Sep Sci 2012, 35 (18), 2341–72. [DOI] [PubMed] [Google Scholar]
- (35).Huang BY; Yang CK; Liu CP; Liu CY Stationary phases for the enrichment of glycoproteins and glycopeptides. Electrophoresis 2014, 35 (15), 2091–107. [DOI] [PubMed] [Google Scholar]
- (36).Fanayan S; Hincapie M; Hancock WS Using lectins to harvest the plasma/serum glycoproteome. Electrophoresis 2012, 33 (12), 1746–54. [DOI] [PubMed] [Google Scholar]
- (37).Jiang J; Tian F; Cai Y; Qian X; Costello CE; Ying W Site-specific qualitative and quantitative analysis of the N- and O-glycoforms in recombinant human erythropoietin. Anal. Bioanal. Chem 2014, 406 (25), 6265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yin H; Zhu J; Wu J; Tan Z; An M; Zhou S; Mechref Y; Lubman DM A procedure for the analysis of site-specific and structure-specific fucosylation in alpha-1-antitrypsin. Electrophoresis 2016, 37 (20), 2624–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kolli V; Schumacher KN; Dodds ED Engaging challenges in glycoproteomics: recent advances in MS-based glycopeptide analysis. Bioanalysis 2015, 7 (1), 113–31. [DOI] [PubMed] [Google Scholar]
- (40).Lee JY; Lee HK; Park GW; Hwang H; Jeong HK; Yun KN; Ji ES; Kim KH; Kim JS; Kim JW; Yun SH; Choi CW; Kim SI; Lim JS; Jeong SK; Paik YK; Lee SY; Park J; Kim SY; Choi YJ; Kim YI; Seo J; Cho JY; Oh MJ; Seo N; An HJ; Kim JY; Yoo JS Characterization of Site-Specific N-Glycopeptide Isoforms of alpha-1-Acid Glycoprotein from an Interlaboratory Study Using LC-MS/MS. J. Proteome Res 2016, 15 (12), 4146–4164. [DOI] [PubMed] [Google Scholar]
- (41).Sanda M; Goldman R Data Independent Analysis of IgG Glycoforms in Samples of Unfractionated Human Plasma. Anal. Chem 2016, 88 (20), 10118–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Huang LJ; Lin JH; Tsai JH; Chu YY; Chen YW; Chen SL; Chen SH Identification of protein O-glycosylation site and corresponding glycans using liquid chromatography-tandem mass spectrometry via mapping accurate mass and retention time shift. Journal of chromatography. A 2014, 1371, 136–45. [DOI] [PubMed] [Google Scholar]
- (43).Windwarder M; Altmann F Site-specific analysis of the O-glycosylation of bovine fetuin by electron-transfer dissociation mass spectrometry. J. Proteomics 2014, 108, 258–68. [DOI] [PubMed] [Google Scholar]
- (44).Wisniewski JR; Zougman A; Nagaraj N; Mann M Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6 (5), 359–62. [DOI] [PubMed] [Google Scholar]
- (45).Morelle W; Michalski J-C Analysis of protein glycosylation by mass spectrometry. Nat. Protoc 2007, 2 (7), 1585–1602. [DOI] [PubMed] [Google Scholar]
- (46).Kaji H; Yamauchi Y; Takahashi N; Isobe T Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protoc 2007, 1 (6), 3019–27. [DOI] [PubMed] [Google Scholar]
- (47).Ma C; Zhao X; Han H; Tong W; Zhang Q; Qin P; Chang C; Peng B; Ying W; Qian X N-linked glycoproteome profiling of human serum using tandem enrichment and multiple fraction concatenation. Electrophoresis 2013, 34 (16), 2440–50. [DOI] [PubMed] [Google Scholar]
- (48).Wang LH; Li DQ; Fu Y; Wang HP; Zhang JF; Yuan ZF; Sun RX; Zeng R; He SM; Gao W pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom 2007, 21 (18), 2985–91. [DOI] [PubMed] [Google Scholar]
- (49).Wilkins MR; Lindskog I; Gasteiger E; Bairoch A; Sanchez JC; Hochstrasser DF; Appel RD Detailed peptide characterization using PEPTIDEMASS–a World-Wide-Web-accessible tool. Electrophoresis 1997, 18 (3–4), 403–8. [DOI] [PubMed] [Google Scholar]
- (50).Doubet S; Bock K; Smith D; Darvill A; Albersheim P The Complex Carbohydrate Structure Database. Trends Biochem. Sci 1989, 14 (12), 475–7. [DOI] [PubMed] [Google Scholar]
- (51).Ceroni A; Maass K; Geyer H; Geyer R; Dell A; Haslam SM GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res 2008, 7 (4), 1650–9. [DOI] [PubMed] [Google Scholar]
- (52).Nowak AA; Canis K; Riddell A; Laffan MA; McKinnon TA O-linked glycosylation of von Willebrand factor modulates the interaction with platelet receptor glycoprotein Ib under static and shear stress conditions. Blood 2012, 120 (1), 214–22. [DOI] [PubMed] [Google Scholar]
- (53).Levery SB; Steentoft C; Halim A; Narimatsu Y; Clausen H; Vakhrushev SY Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta, Gen. Subj 2015, 1850 (1), 33–42. [DOI] [PubMed] [Google Scholar]
- (54).O’Donnell JS; McKinnon TA; Crawley JT; Lane DA; Laffan MA Bombay phenotype is associated with reduced plasma-VWF levels and an increased susceptibility to ADAMTS13 proteolysis. Blood 2005, 106 (6), 1988–91. [DOI] [PubMed] [Google Scholar]
- (55).Frese CK; Altelaar AF; Hennrich ML; Nolting D; Zeller M; Griep-Raming J; Heck AJ Mohammed, S., Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res 2011, 10 (5), 2377–88. [DOI] [PubMed] [Google Scholar]
- (56).Lynch CJ; Lane DA N-linked glycan stabilization of the VWF A2 domain. Blood 2016, 127 (13), 1711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.