Abstract
Current antivirals can control but not eliminate hepatitis-B-virus (HBV), because HBV establishes a stable nuclear cccDNA. Interferon-α treatment can clear HBV but is limited by systemic side effects. Here we describe how interferon-α can induce specific degradation of the nuclear viral DNA without hepatotoxicity and propose lymphotoxin-β-receptor activation as a therapeutic alternative. Interferon-α and lymphotoxin-β-receptor activation up-regulated APOBEC3A and 3B cytidinedeaminases, respectively, in HBV-infected cells, primary hepatocytes and human liver-needle biopsies. HBV-core protein mediated the interaction with nuclear cccDNA resulting in cytidine-deamination, apurinic/apyrimidinic site formation and finally cccDNA degradation that prevented HBV-reactivation. Genomic DNA was not affected. Thus, inducing nuclear deaminases - e.g., by lymphotoxin-β-receptor activation - allows development of new therapeutics that combined with existing antivirals may cure hepatitis B.
Hepatitis B virus (HBV) infection remains a major public health threat with more than 350 million humans chronically infected worldwide at risk of developing end-stage liver disease and hepatocellular carcinoma. Each year, more than 600,000 humans die from consequences of chronic HBV infection. A prophylactic vaccine has been available for hepatitis B for almost thirty years, but the overall number of chronic infections remains high.
HBV is a small, enveloped DNA virus replicating via an RNA intermediate. The encapsidated viral genome consists of a 3.2 kb partially double-stranded relaxed circular DNA (rcDNA) molecule. The virus has optimized its life-cycle for long-term persistence in the liver (1). Upon translocation to the nucleus, the rcDNA genome is converted into a covalently closed circular DNA (cccDNA), which serves as the template for viral transcription and secures HBV persistence. Nucleos(t)ide analogs are efficient antivirals but only control and do not cure HBV infection owing to the persistence of HBV cccDNA. Therefore, long-term treatment is required, which is expensive and may lead to concomitant resistance (2). Interferon (IFN)-α is licensed for hepatitis B therapy and treatment with this cytokine can result in virus clearance in a proportion of patients; however, its efficacy is limited and high doses are not tolerated (3). Thus, efficient and nontoxic elimination of cccDNA in hepatocytes is a major goal of HBV research.
Using animal models, it has been shown that HBV replication, and in particular the cccDNA content of the liver, can be affected by noncytopathic mechanisms involving cytokines such as interferons and tumor necrosis factor (TNF), which influence RNA and capsid stability (4–7). Here, we describe an antiviral mechanism that interferes with cccDNA stability and is distinct from influences of antiviral cytokines on cccDNA activity (8).
High-Dose IFN-α Leads to cccDNA Degradation in HBV-Infected Hepatocytes
IFN-α is known to exert transcriptional, post-transcriptional and epigenetic antiviral effects on HBV (8–12). To study the effect of IFN-α on HBV cccDNA, we used HBV-infected, differentiated HepaRG (dHepaRG) cells and primary human hepatocytes (PHH). These are human cell types susceptible to HBV infection (13, 14) and responsive to IFN-α treatment in vitro (fig. S1A). IFN-α treatment did not lead to detectable hepatotoxicity, even at very high doses (fig. S1B). Treating dHepaRG cells with 500 or 1000 IU/ml IFN-α controlled HBV-DNA synthesis as efficiently as 0.5 μM (5-fold EC50) of the nucleoside analog lamivudine (LAM). IFN-α, however, unlike LAM also significantly reduced expression of HBV-RNA and hepatitis B surface (HBsAg) and e (HBeAg) antigens (Fig. 1A and fig. S1C).
Fig. 1. Degradation of cccDNA in IFN-α treated HepaRG cells and primary human hepatocytes.
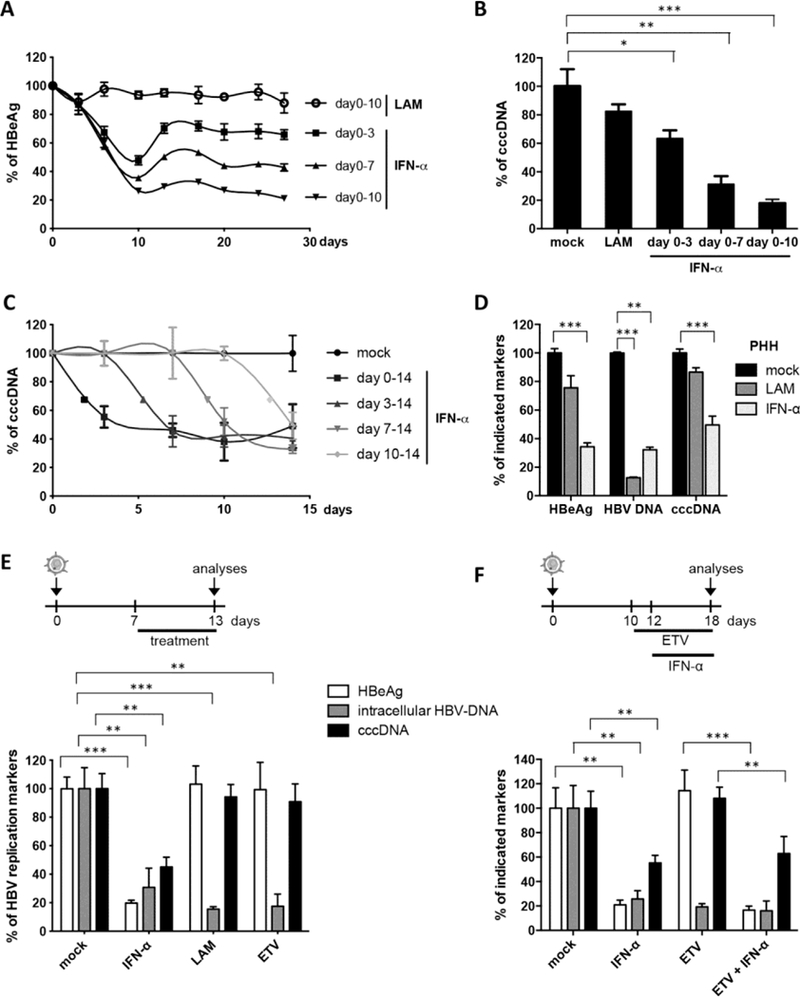
(A, B, C, E, and F) HBV-infected dHepaRG were treated with IFN-α at day 10 post-infection (dpi). Different regimens of treatment were applied as indicated. (D) HBVinfected primary human hepatocyte (PHH) were treated with IFN-α at dpi 3 for 13 days. Levels of HBeAg, total intracellular DNA and cccDNA are given relative to mock treated cells. LAM: lamivudine; ETV: entecavir. Mean values +/− standard deviation of replicates from independent experiments are given; data were analyzed by t test. * p < 0.05, ** p < 0.01 and *** p < 0.001.
In patients, interruption of LAM treatment results in a rebound of HBV replication (2). Using IFN-α, we observed only a partial or no rebound in HBV-infected dHepaRG cells after treatment cessation (Fig. 1A). Because dHepaRG don’t allow virus spread, reduction of HBeAg and lacking rebound indicated an effect of IFN-α on the established HBV cccDNA transcription template besides the known antiviral effects on viral replication (14). By cccDNA-specific qPCR, we determined an 80% reduction of cccDNA after 10 days of treatment (Fig. 1B). Reduction of cccDNA was confirmed by Southern blot analysis (fig. S1D) and was dose dependent (fig. S1E). cccDNA reduction could be induced at any time point (Fig. 1C) and persisted over time (Fig. 1, A and C). The effect was corroborated in HBV-infected primary human hepatocytes (PHH) (Fig. 1D). In contrast to IFN-α, LAM and even more potent nucleoside analog entecavir (ETV) at very high doses (0.5 μM, 1000-fold IC-50) only inhibited reverse transcription and thus HBV replication, but not viral persistence (Fig. 1E). Pretreatment with ETV did not enhance the effect of IFN-α (Fig. 1F) indicating that IFN-α induces the decay of established HBV cccDNA. Since the doses of IFN-α used to achieve this effect were high, we screened for other cytokines showing similar antiviral effects at moderate doses.
LTβR Activation Controls HBV and Leads to cccDNA Degradation in HBV-Infected Cells
IFN-γ and TNF-α are known to control HBV in a noncytopathic fashion (4, 7), but cannot be used as therapeutics because they cause severe side effects. We tested the effect of lymphotoxin (LT) β receptor (LTβR) activation as an alternative therapeutic option. TNF superfamily members LTα, LTβ and CD258 are the physiological ligands for LTβR and activate several inflammatory, anti-inflammatory, pro- and anti-survival pathways (15). Like hepatocytes (16), dHepaRG (14) and HepG2-H1.3 cells permit HBV replication (17) and express the LTβR (fig. S2, A and B). To activate LTβR, we used a super-agonistic tetravalent bispecific antibody (BS1) and a bivalent anti-LTβR monoclonal antibody (CBE11) (18, 19). As expected, LTβR agonists activated canonical (20) and noncanonical nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathways to trigger p100 cleavage (fig. S2C), RelA phosphorylation (fig. S2D), nuclear RelB and RelA translocation (fig. S2, E and F), and up-regulation of known target genes (fig. S2G) without causing any detectable hepatocytotoxicity (fig. S2H).
To test the effect of LTβR-activation on HBV infection, dHepaRG cells were treated with BS1 for 12 days starting 24 hours prior to HBV infection. LTβR-activation decreased levels of all HBV-markers, including cccDNA by approximately 90% without toxicity (Fig. 2A). The antiviral effect was highly potent with an EC5O of approximately 0.01 μg/mL (fig. S3A). Inhibition of apoptosis did not alter antiviral activity (fig. S4). Neither IFN-β nor classic IFN-stimulated genes were upregulated upon BS1-treatment (fig. S2G) and antiviral activity was independent of IFN-induction (fig. S5).
Fig. 2. LTβR-activation inhibits HBV infection and leads to cccDNA degradation in HepaRG cells and PHH.
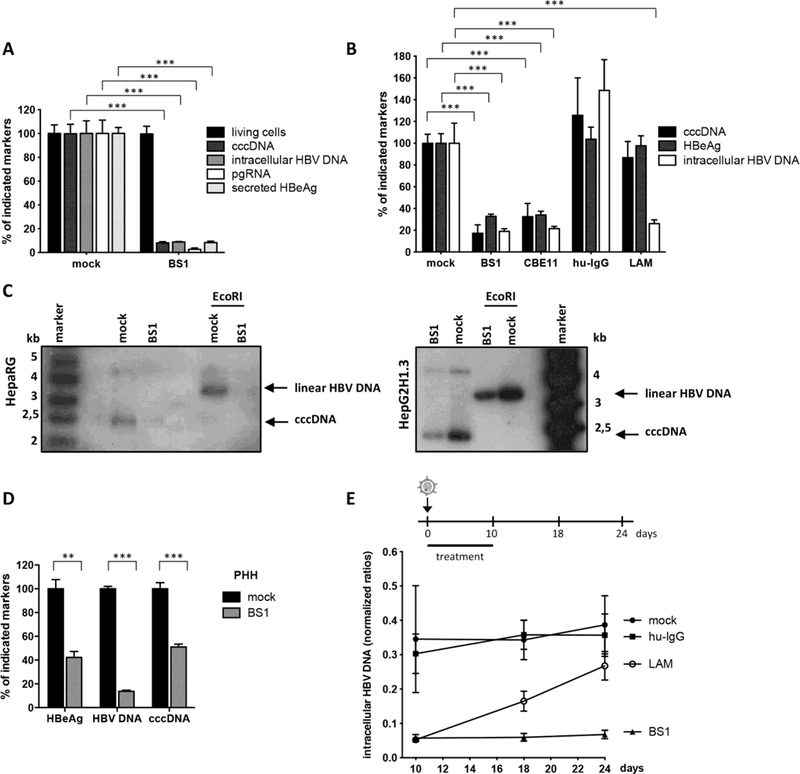
(A and B) HBV-infected dHepaRG were treated with BS1, CBE11, hu-IgG control or lamivudine (LAM). (A) Treatment started 24h before infection for 12 days or (B) at 18 dpi for 10 days. Levels of the indicated HBV markers as well as cell viability are given relative to untreated controls (mock). (C) cccDNA levels were analyzed after 14 days of BS1 treatment by Southern blot in HBV-infected dHepaRG and HBV-replicating HepG2H1.3 cells. Supercoiled cccDNA bands were identified by their expected size and linearization upon EcoRI digestion (3,2 kb). (D) PHH were infected with HBV and treated with BS1 at 7 dpi for 10 days. Levels of the indicated HBV markers were compared to untreated PHH of the same donor (donor 3) (mock). (E) HBV-infected dHepaRG were treated with BS1, hu-IgG control or LAM. Intracellular HBV-DNA was analyzed 8 and 14 days after treatment cessation. Mean values +/− standard deviation of replicates from independent experiments are given; data were analyzed by t test. * p < 0.05, *** p < 0.001.
In vivo, activation of the murine LTβR by systemic application of an agonistic antibody (ACH6) induced RelA and RelB nuclear translocation in hepatocytes of HBV-transgenic mice (fig. S6A), reduced HBV viremia (fig. S6B), HBV RNA (fig. S6C) and HBV core (HBc) protein expression in the liver (fig. S6, D and E). Neither signs of hepatocyte apoptosis (fig. S6F) nor elevation of aminotransferases (ALT) (fig. S6G, right panel) were observed indicating good in vivo tolerability of LTβRactivation. Since HBV-transgenic mice do not establish HBV cccDNA, this indicated additional antiviral effects of LTβR-activation on HBV RNA transcription or stability. Accordingly, discontinuation of LTβRactivation induced an immediate, strong rebound of HBV replication (fig. S6G).
To investigate whether LTβR-activation would affect established HBV cccDNA in the context of a persistent infection and prevent HBV reactivation, dHepaRG cells were treated with LTβR agonists BS1 or CBE11 when a stable, nuclear cccDNA pool had established. All HBV markers, including HBV cccDNA, were reduced upon LTβR-activation in HBV-infected dHepaRG cells (Fig. 2, B and C, and fig. S3) as well as in stably transfected HepG2H1.3 cells containing high levels of cccDNA (Fig. 2C). In HBV-infected primary human hepatocytes (PHH), LTβR agonisation reduced HBV cccDNA, HBeAg secretion and even more pronounced HBV-DNA replication (Fig. 2D). cccDNA degradation was more effective (up to 95%) when treatment was prolonged (fig. S3, C and D). Treatment interruption for 10 days was almost as efficient as continuous treatment (fig. S3C) indicating that LTβR agonists induce a persistent antiviral effect. In contrast to LAM treatment, no rebound of HBV-replication was observed when BS1 treatment stopped (Fig. 2E). Hence, LTβR activation not only suppressed HBV replication but also caused nuclear cccDNA degradation, needed to achieve virus elimination.
LTβR Activation and IFN-α Treatment Induce Deamination and Apurinic/Apyrimidinic (AP) Site Formation in cccDNA
To investigate if cccDNA degradation upon LTβR-activation or IFN-α treatment was a result of DNA damage, we examined cccDNA deamination by differential DNA denaturation PCR (3D-PCR) (21). Low denaturing temperatures were sufficient for cccDNA amplification from HBV-infected dHepaRG cells and for PHH treated with IFN-α or BS1, compared with untreated, LAM- or ETV-treated cells (Fig. 3A and fig. S7, C and D). Using a cocktail of recombinant proteins containing all enzymes necessary for DNA repair (preCR mix), we could reverse the denaturation of cccDNA (Fig. 3A, lower panels). The fact, that denaturation temperatures of mock, LAM and ETV treated cells also shifted, indicated that this modification of HBV cccDNA existed even without exposure to exogenous drugs. Deamination of cccDNA (Fig. 3A, right panel) and a drop in cccDNA levels after treatment with CBE11 (table S1) was confirmed in vivo in human liver chimeric uPA-SCID mice infected with HBV. Sequencing analyses showed G/A transitions occurred under treatment (Fig. 3B and fig. S7, A and B) indicating deamination of cytidines to uridine in the HBV cccDNA minus strand. At lower denaturation temperatures G/A transitions became more obvious (Fig. 3C and fig. S7A). These data showed that both LTβR-activation and IFN-α treatment led to cccDNA deamination in vitro and in vivo, and help to explain the G/A hypermutation observed in patient samples (21).
Fig. 3. Deamination and AP-site formation in cccDNA upon IFN-α treatment and LTβR-activation.
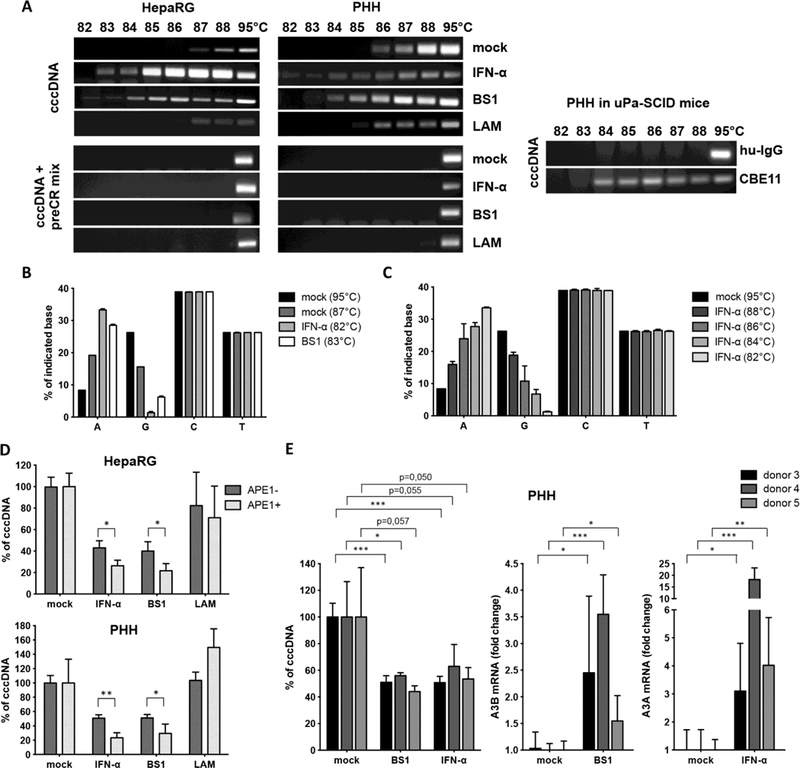
(A) dHepaRG (left) and PHH (middle panel) were infected with HBV and treated with IFN-α, BS1 or LAM. Human chimeric uPA/SCID mice were treated with CBE11 or hu-IgG control (right panel). 3D-PCR analyses were performed on cccDNA left either untreated (upper panels) or treated with a PreCR mix (lower panels). (B and C) 3D-PCR products from HBV-infected dHepaRG cells treated as indicated (IFN-α, BS1 or mock) were cloned and sequenced and mutations were analyzed. (D) Total DNA extracts from HBV-infected cells treated as indicated were digested with APE1, and cccDNA content was compared to mock-treated cells. In (B), (C), and (D), mean values +/− standard deviation of biological triplicates from two independent experiments are given; data were analyzed by t test. * p < 0.05, ** p < 0.01. (E) PHH were infected with HBV and treated with BS1 or IFN-α at 7 dpi for 10 days. Levels of the indicated cccDNA as well as A3A and A3B mRNA expression were compared to untreated PHH (mock) of the same donor.
Importantly, neither deamination nor mutations of genomic DNA were observed by 3D-PCR (fig. S8A) or by deep sequencing of selected housekeeping or IFN- and LTβR-target genes (fig. S8B). This indicated that DNA modifications were specifically targeted to viral cccDNA.
After cytidine deamination, DNA-glycosylases recognize the damaged DNA and cleave N-glycosidic bonds to release the base and create an accessible AP site that can then be cleaved by endonucleases (22). These AP sites can either be repaired, can lead to mutations upon DNA replication or can induce DNA degradation (23). We quantified AP sites created by LTβR-activation or IFN-α treatment. However, no increase of AP sites in total DNA extracts from dHepaRG cells or PHH treated with IFN-α or LTβR-agonists (fig. S8C) was found, reassuring that our treatments did not lead to detectable damage in genomic DNA. Because AP sites in the small (3.2 kb) cccDNA are very likely to be missed by this analysis, we digested total DNA extracts with an AP-endonuclease (APE1) and then amplified cccDNA by qPCR. APE digestion further decreased cccDNA extracted from dHepaRG cells and PHH treated with IFN-α or LTβR-agonists but not with LAM (Fig. 3D). Taken together, our data indicate that both, LTβR-activation or IFN-α treatment induced deamination and AP-site formation in HBV cccDNA leading to its degradation, but did not affect genomic DNA.
LTβR Activation and IFN-α Treatment Up-Regulate Expression of Nuclear APOBEC3 Deaminases
IFN-α is known to induce several cytidine deaminases (23, 24). We performed genome-wide expression profiling of HBV-infected dHepaRG cells after LTβR-activation (fig. S9A) and classified regulated genes according to their activity and properties (fig. S9B). Hereby, APOBEC3B (A3B) was identified to be the most up-regulated gene with nucleic acid binding properties (fig. S9C).
Analysis of all APOBEC3 family members showed that LTβRactivation leads to strong up-regulation of A3B and to minor extent A3G in HBV-infected dHepaRG and PHH, and after systemic application in human liver chimeric uPA-SCID mice (fig. S10A). A3B expression was induces by LTβR-activation in a dose-dependent manner and expression levels steadily increased during continuous treatment (fig. S11) correlating with a concomitant increase in treatment efficacy over time (fig. S3C). Treatment of PHH isolated from different donors with LTβRagonist BS1 resulted in cccDNA degradation at different levels (Fig. 3E and fig. S10B), which could neither be explained by the level of A3B upregulation (Fig. 3E) nor by detection of a previously described (25) genomic deletion of the A3B allele, which seems to correlate with HBV persistence in infected patients (fig. S10, B and C).
In contrast to LTβR-activation, IFN-α treatment induced mainly A3A, but also A3F and A3G expression in HBV-infected dHepaRG cells and PHH (fig. S12A), and A3D expression in isolated PHH. By systemic IFN treatment of chimpanzees (26), A3A was strongly upregulated in liver needle biopsies (fig. S12B). Activation of A3A, A3F and A3G after IFN-α treatment was dose- and time-dependent, and decreased after an initial peak despite continuous treatment indicating that cells become refractory to IFN-α (fig. S13). In patients treated with subcutaneous pegylated IFN-α, needle biopsies obtained at different time points confirmed a rapid, strong upregulation of A3A and to a lower extend of A3G in the liver peaking at 16 hours post treatment (fig. S12C). Expression levels declined after this time point and remained low until day 6 post treatment confirming a fast but only transient induction of A3A by IFN-α treatment. Interestingly, the level of A3B or A3A induction in BS-1 and IFN-α treated PHH, respectively, did not directly correlate with the level of cccDNA degradation (Fig. 3E). The fact that IFN-α only induces a transient A3A induction and cells rapidly become refractory to IFN-α may account for the limited effect of IFN-α treatment in HBV-infected patients (3).
APOBEC3A or APOBEC3B Activity Is Essential to Induce cccDNA
Degradation
Among the APOBEC3 family members up-regulated in our experiments, only A3A and A3B located to the nucleus (fig. S14) where they can gain access to cccDNA. To verify that they are indeed responsible for the induction of cccDNA degradation, we overexpressed the HIV-Vif protein (known to promote the degradation of all APOBEC3 proteins except A3B (27, 28)) in dHepaRG cells in a tetracycline-regulated fashion. Expression of HIV-Vif reduced A3A, A3F and A3G expression (fig. S15A), reverted IFN-α-induced cccDNA deamination and prevented cccDNA degradation induced by IFN-α treatment (Fig. 4A). However, expression of HIV-Vif did not alter A3B levels (fig. S15B) and had no impact on cccDNA degradation by LTβR-activation (fig. S15C). To specifically address the role of A3A or A3B in cccDNA degradation we further knocked down A3A and A3B in dHepaRG cells under IFN-α or LTβR-agonist treatment, respectively, and observed reduced cccDNA deamination (Fig. 4, B and C, left panels). A3A as well as A3B knock-down completely reverted cccDNA degradation, but could not rescue the additional effect of IFN-α or LTβR-activation on HBV replication (Fig. 4, B and C, right panels).
Fig. 4. Analysis of cccDNA deamination and degradation.
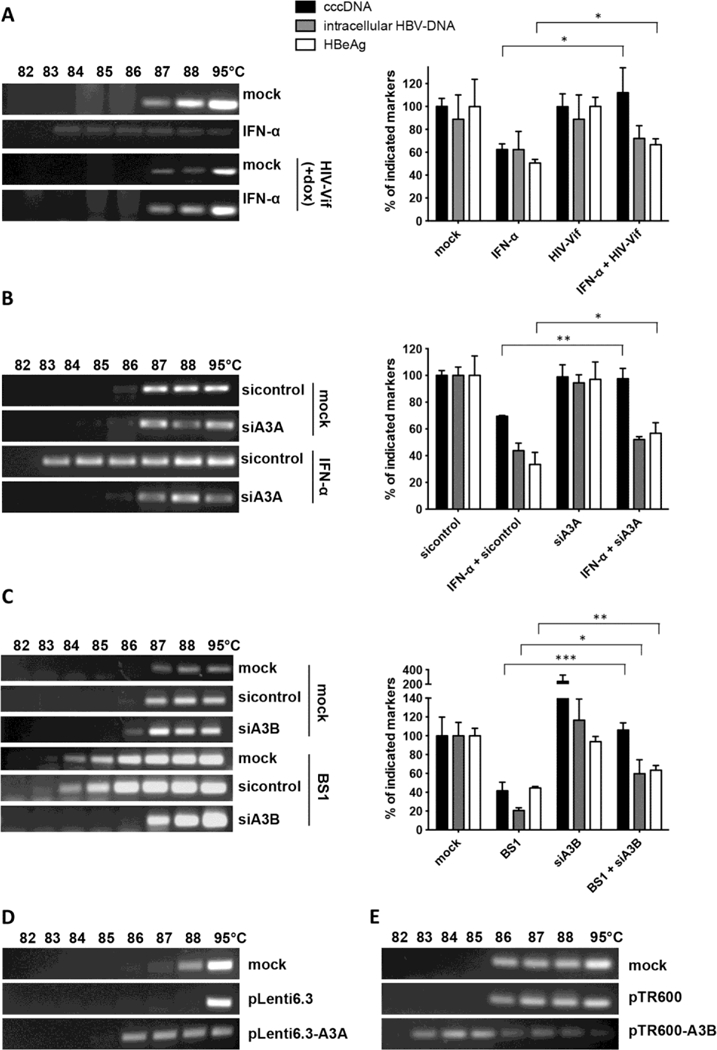
(A to C) cccDNA denaturation was analyzed by 3D-PCR (left panels); levels of HBeAg, total intracellular DNA and cccDNA are given relative to mock treated cells (right panels). (A) dHepaRG-tA-Vif cells treated with IFN-α for 10 days with and without doxycycline (dox)-induced HIV-Vif expression. HBV-infected dHepaRG cells treated with (B) IFN-α or (C) BS1 transfected with siRNA against A3A or A3B, respectively, or sequence nonspecific siRNA (sicontrol). Mean values +/− standard deviation of independent replicates and experiments are given; data were analyzed by t test. * p < 0.05, ** p < 0.01 and *** p < 0.001. (D) cccDNA denaturation analysis by 3D-PCR in HepG2-H1.3 cells overexpressing A3A or (E) A3B from lentiviral vector plasmid pLenti6.3 or pTR600, respectively, for 5 days.
To confirm the impact of A3A and A3B on cccDNA deamination, we overexpressed A3A and A3B, respectively, in HBV-replicating HepG2-H1.3 (Fig. 4, D and E). Cytidine-deamination of nuclear cccDNA by A3A and A3B is in accordance with other studies showing that both localize to the nucleus (29) and may be involved in the elimination of foreign DNA (23).
APOBEC3A Interacts with the HBV Core Protein and Binds to cccDNA
APOBECs have evolved to restrict retroviral replication (30) as well as DNA transfer into cells. They are able to clear foreign nuclear DNA (23, 31), but it remains unclear how HBV cccDNA DNA was recognized and whether it was specifically targeted in our experiments. To assess specificity, we generated cell lines replicating a mammalian replicon plasmid pEpi containing a linear HBV 1.3-fold overlength sequence. From the linear HBV-genome, HBV replication was initiated and in addition to the pEpi-H1.3 replicon HBV cccDNA was established in the nucleus. Treatment with either IFN-α or LTβR-agonist BS1 inhibited HBV replication and resulted in deamination and degradation of HBV cccDNA, but not of the HBV-sequence containing replicon (fig. S16). This indicated that deamination and subsequent degradation induced by both treatments is HBV cccDNA specific.
HBV core protein associates with A3G (32) and HBV cccDNA (33) and thus was a candidate to mediate the targeting of A3 deaminases to HBV cccDNA. Confocal microscopy indicated a co-localization of A3A and A3B with HBV core in different cell lines and PHH (Fig. 5 and fig. S17). Chromatin immunoprecipitation (ChIP) experiments using stably (fig. S18A) or transiently transfected HepG2H1.3 cells or HBV-infected and IFN-α treated dHepaRG cells, showed that HBV core protein and A3A both bind to the cccDNA minichromosome (Fig. 6A). Supporting the possibility that a guardian protein prevents A3A direct binding to DNA (34), we could not detect A3A binding to genomic DNA (fig. S18B) even in the presence of HBV core, which has been reported to also bind to cellular DNA (35).
Fig. 5. Co-localization of A3A and A3B with HBV core protein (HBc).
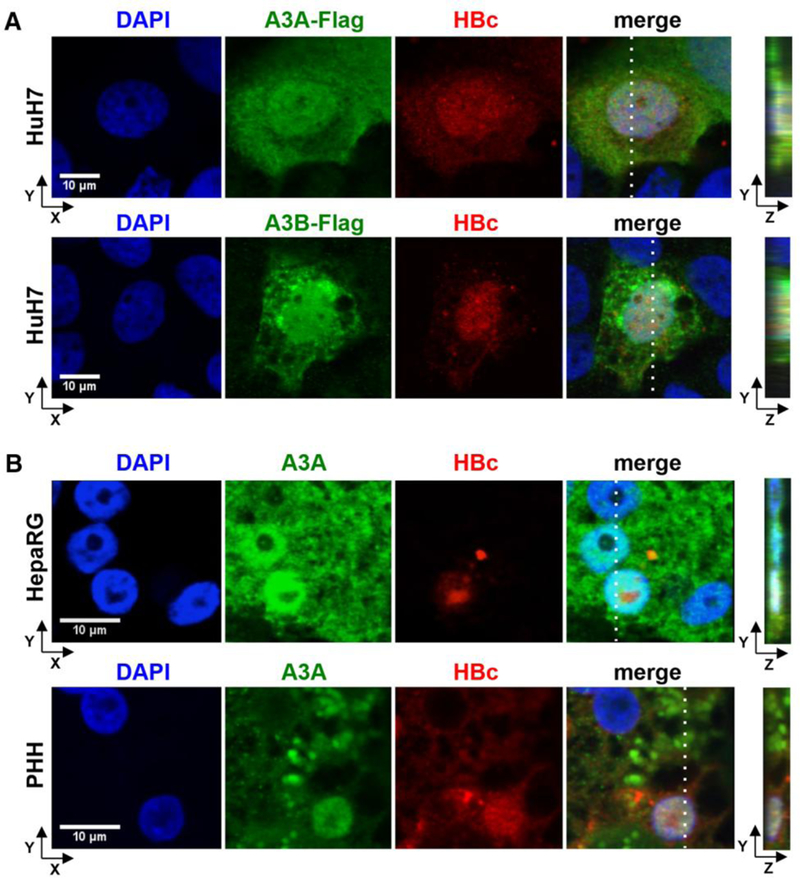
(A) HuH7 cells were co-transfected with an HBV1.1-fold genome and A3A-Flag or A3B-Flag expressing plasmids and stained using DAPI, anti-HBc and anti-FLAG antibodies. (B) HBV-infected dHepaRG and PHH were treated with IFN-α at day 7 post infection for 3 days. A3A and HBc were analyzed by immunofluorescence staining. Right panels indicate z stacks taken at the dotted lines.
Fig. 6. Interaction of A3A, HBV core protein (HBc) and cccDNA.
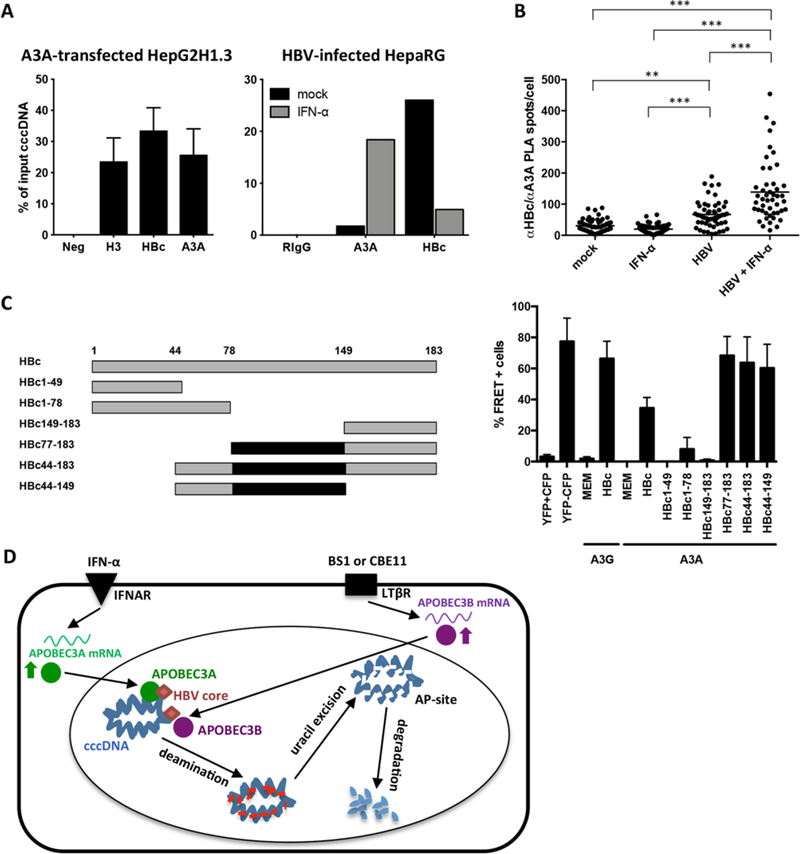
(A) Chromatin immunoprecipitation (ChIP) was performed using lysates of HepG2H1.3 cells transfected with A3Aexpressing plasmid, or HBV-infected dHepaRG cells treated with IFN-α for 3 days. IPs using antibodies against histone H3, A3A, HBc and control rabbit IgG (RIgG) were analyzed by qPCR for cccDNA. (B) Interaction between HBc and A3A was assessed by proximity ligation assay (PLA) in HBV-infected, IFN-α treated dHepaRG. PLA-spots were quantified in single cells by software-based spot-counting. Data were analyzed by one-way ANOVA. ** p < 0.01 and *** p < 0.001. (C) Serial HBV core-deletion mutants (left panel) were fused to CFP and interaction with A3A-YFP was assessed by FACS-FRET in HuH7.5 hepatoma cells (right panel). Cells cotransfected with CFP and YFP served as controls to exclude false positive FRET and subtract background signals. A CFP-YFP fusion construct was used as positive control. Mean values ± standard deviation of FRET-positive cells from 3–4 independent experiments are given. Black boxes indicate shared regions of HBc mutants giving a FRET signal. (D) Model of cccDNA degradation induced by IFN-α treatment or LTβR-activation.
HBV core protein co-immunoprecipitated A3A in HepG2H1.3 cells and transfected HuH7 cells indicating physical interaction with A3A (fig. S19). Direct interaction of HBV core expressed after HBV infection and A3A induced by IFN-α was confirmed by proximity ligation assay (PLA) (Fig. 6B and fig. S20) and fluorescence resonance energy transfer (FRET) analysis (Fig. 6C). By deletion analysis, we determined that the central region of HBc (aa 77 to 149) is involved in the interaction with A3A (Fig. 6C and fig. S21).
These data suggest that A3A is targeted to cccDNA by interaction with HBV core. No such targeting to genomic DNA has been described so far. Since APOBEC3 deaminases are thought to act on single stranded DNA (36), one possibility is that A3A and A3B act on cccDNA when it is transiently rendered single-stranded by RNA polymerase II before transcription initiation.
We suggest, therefore, the following mechanism of APOBECdependent degradation of HBV cccDNA (Fig. 6D). High dose IFN-α treatment or LTβR-activation up-regulate the expression of A3A and A3B, respectively, which subsequently co-localize or directly interact with HBV core in infected hepatocytes, translocate to the nucleus, where they are brought into close contact with cccDNA by HBV core. Now, APOBECs can deaminate cccDNA that is transiently rendered singlestranded during transcription. Uracils in HBV cccDNA are recognized and excised by cellular DNA glycosylases leading to formation of AP sites, which are then recognized by cellular AP endonculeases (23) leading to cccDNA digestion. Why cccDNA is degraded instead of being repaired by the cellular DNA repair machinery remains elusive so far. Using a mixture of various enzymes, we were able to repair deaminated cccDNA in tubo (Fig. 3A) suggesting induction of an additional factor promoting DNA degradation or an impaired function of the repair machinery rather than a lack of recognition by the repair machinery. Thus, we can only speculate that either the number of AP sites introduced after treatment is too high and exceeds the capacity of the cellular repair machinery or that IFN-α treatment or LTβR-activation or even HBV itself (37) modulate the repair machinery. This may shift the equilibrium from cccDNA repair (38) to degradation.
Ideally, a cure for HBV infection needs to eliminate cccDNA. Therefore, cytokines or cytokine-receptor agonists that can trigger HBV cccDNA deamination and its degradation are interesting antiviral candidates. Antivirals that induce A3A/B activity should be combined with nucleos(t)ide analogs to avoid the replenishment of nuclear cccDNA after degradation. LTβR-agonists were active at low doses and we did not observe any toxicity in vitro or in vivo nor did we detect any modification of genomic DNA. Constitutive overexpression of LTα/β for more than one year has been associated with inflammatory liver disease and hepatocellular carcinoma (16). As antivirals, however, LTβR-agonists would only be used for a limited period of time minimizing the risk of side effects. Moreover, LTβR-activation was already explored as a cancer treatment (18).
A recent study has shown a significantly higher frequency of an A3B deletion allele in persistent HBV carriers and hepatocellular carcinoma patients compared with healthy controls (25). This finding was further supported by the moderate deamination of cccDNA even in absence of treatment, and by the observation that knockdown of A3B in the absence of any treatment increased cccDNA levels. Although deregulated expression of A3A and A3B has been shown to correlate with genomic DNA mutations (39, 40), we did not detect any alterations of genomic DNA using analyses of AP sites, 3D-PCR analysis and deep sequencing of a set of human genes.
Our data indicate that cccDNA degradation is possible and can be induced without side-effects on the infected host cell. An important task will be testing of combinations of nucleos(t)ide analogs with novel antiviral strategies (e.g., LTβR agonists or adoptive T-cell therapy (41)) to activate A3A or A3B to cure hepatitis B.
Supplementary Material
Acknowledgments:
We would like to thank Romina Bester, Theresa Asen, Kerstin Ackermann, Kathrin Kappes, Martin Feuerherd, Robert Baier, Ruth Hillermann, Ute Finkel, Aikatherini Krikoni and Fang Zhang for their technical support, Prof. Luigi Terracciano for analysis of acute hepatitis patients, Prof. Frank Chisari for providing HBV transgenic mice (HBV 1.3.32), Prof. Thorsten Buch and Olivia Prazeres da Costa for help with array analysis and data discussions, Lena Allweiss and Anne Groth for help by generating and treating humanized uPA/SCID mice, as well as Siemens Healthcare Diagnostics for providing reagents. The study was supported by grants from FCC (Fédération belge Contre le Cancer) to ED, an ERC Starting grant (LiverCancerMechanism) to MH, the German Research Foundation (SFB 841 to MD, SFB TR 36 to MH, SFB TR 22), the Peter-Hans Hofschneider foundation and the Helmholtz Alliances HAIT (to UP) and PCCC (to MH). We acknowledge the support of the nonprofit foundation HTCR, which holds human tissue on trust, making it broadly available for research on an ethical and legal basis. Patent application EP12006XXX filed at the European patent office: ‘Lymphotoxin signaling activation and its downstream mediators eliminate HBV ccc DNA’. Microarray data have been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo/) and have the accession number GSE46667. Human liver-chimeric UPA/SCID mice were handled in accordance with protocols approved by the Ethical Committee of the city and state of Hamburg (permission number G12/015). Experiments with HBV-transgenic mice were performed in accordance to the German legislation governing animal studies and the Principles of Laboratory Animal Care guidelines, NIH (55.1–1-54–2531.3–27-08). The study protocol for the experiment with Chimpanzee was approved at the Southwest Foundation for Biomedical Research, San Antonio, TX (IACUC 869 PT, approved in 2004).
References and Notes
- 1.Protzer U, Maini MK, Knolle PA, Living in the liver: hepatic infections. Nat. Rev. Immunol 12, 201–213 (2012). doi: 10.1038/nri3169 Medline [DOI] [PubMed] [Google Scholar]
- 2.Zoulim F, Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int 31, (Suppl 1), 111–116 (2011). doi: 10.1111/j.1478-3231.2010.02399.x Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, Fischer C, Currie G, Brosgart C, Petersen J, Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 44, 675–684 (2006). doi: 10.1002/hep.21282 Medline [DOI] [PubMed] [Google Scholar]
- 4.Guidotti LG, Ando K, Hobbs MV, Ishikawa T, Runkel L, Schreiber RD, Chisari FV, Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. U.S.A 91, 3764–3768 (1994). doi: 10.1073/pnas.91.9.3764 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV, Viral clearance without destruction of infected cells during acute HBV infection. Science 284, 825–829 (1999). doi: 10.1126/science.284.5415.825 Medline [DOI] [PubMed] [Google Scholar]
- 6.Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV, Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc. Natl. Acad. Sci. U.S.A 101, 2129–2134 (2004). doi: 10.1073/pnas.0308478100 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClary H, Koch R, Chisari FV, Guidotti LG, Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol 74, 2255–2264 (2000). doi: 10.1128/JVI.74.5.2255-2264.2000 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M, IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest 122, 529–537 (2012). doi: 10.1172/JCI58847 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rang A, Günther S, Will H, Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J. Hepatol 31, 791–799 (1999). doi: 10.1016/S0168-8278(99)80279-7 Medline [DOI] [PubMed] [Google Scholar]
- 10.Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV, Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol 76, 5646–5653 (2002). doi: 10.1128/JVI.76.11.5646-5653.2002 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieland SF, Guidotti LG, Chisari FV, Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol 74, 4165–4173 (2000). doi: 10.1128/JVI.74.9.4165-4173.2000 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uprichard SL, Wieland SF, Althage A, Chisari FV, Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc. Natl. Acad. Sci. U.S.A 100, 1310–1315 (2003). doi: 10.1073/pnas.252773599 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gripon P, Diot C, Thézé N, Fourel I, Loreal O, Brechot C, GuguenGuillouzo C, Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol 62, 4136–4143 (1988). Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C, Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U.S.A 99, 15655–15660 (2002). doi: 10.1073/pnas.232137699 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf MJ, Seleznik GM, Zeller N, Heikenwalder M, The unexpected role of lymphotoxin beta receptor signaling in carcinogenesis: from lymphoid tissue formation to liver and prostate cancer development. Oncogene 29, 5006–5018 (2010). doi: 10.1038/onc.2010.260 Medline [DOI] [PubMed] [Google Scholar]
- 16.Haybaeck J, Zeller N, Wolf MJ, Weber A, Wagner U, Kurrer MO, Bremer J, Iezzi G, Graf R, Clavien PA, Thimme R, Blum H, Nedospasov SA, Zatloukal K, Ramzan M, Ciesek S, Pietschmann T, Marche PN, Karin M, Kopf M, Browning JL, Aguzzi A, Heikenwalder M, A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell 16, 295–308 (2009). doi: 10.1016/j.ccr.2009.08.021 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jost S, Turelli P, Mangeat B, Protzer U, Trono D, Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J. Virol 81, 10588–10596 (2007). doi: 10.1128/JVI.02489-06 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukashev M, LePage D, Wilson C, Bailly V, Garber E, Lukashin A, Ngam-ek A, Zeng W, Allaire N, Perrin S, Xu X, Szeliga K, Wortham K, Kelly R, Bottiglio C, Ding J, Griffith L, Heaney G, Silverio E, Yang W, Jarpe M, Fawell S, Reff M, Carmillo A, Miatkowski K, Amatucci J, Crowell T, Prentice H, Meier W, Violette SM, Mackay F, Yang D, Hoffman R, Browning JL, Targeting the lymphotoxin-beta receptor with agonist antibodies as a potential cancer therapy. Cancer Res 66, 9617–9624 (2006). doi: 10.1158/0008-5472.CAN-06-0217 Medline [DOI] [PubMed] [Google Scholar]
- 19.Hu X, Zimmerman MA, Bardhan K, Yang D, Waller JL, Liles GB, Lee JR, Pollock R, Lev D, Ware CF, Garber E, Bailly V, Browning JL, Liu K, Lymphotoxin β receptor mediates caspase-dependent tumor cell apoptosis in vitro and tumor suppression in vivo despite induction of NF-κB activation. Carcinogenesis 34, 1105–1114 (2013). doi: 10.1093/carcin/bgt014 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR, The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 17, 525–535 (2002). doi: 10.1016/S1074-7613(02)00423-5 Medline [DOI] [PubMed] [Google Scholar]
- 21.Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP, Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A 102, 8321–8326 (2005). doi: 10.1073/pnas.0408223102 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman JI, Stivers JT, Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 49, 4957–4967 (2010). doi: 10.1021/bi100593a Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS, APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol 17, 222–229 (2010). doi: 10.1038/nsmb.1744 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J, Interferoninducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43, 1364–1374 (2006). doi: 10.1002/hep.21187 Medline [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Cai J, Chang J, Yu D, Wu C, Yan T, Zhai K, Bi X, Zhao H, Xu J, Tan W, Qu C, Lin D, Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum. Mol. Genet 22, 1262–1269 (2013). doi: 10.1093/hmg/dds513 Medline [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Feld JJ, Sapp RK, Nanda S, Lin JH, Blatt LM, Fried MW, Murthy K, Liang TJ, Defective hepatic response to interferon and activation of suppressor of cytokine signaling 3 in chronic hepatitis C. Gastroenterology 132, 733–744 (2007). doi: 10.1053/j.gastro.2006.11.045 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doehle BP, Schäfer A, Cullen BR, Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339, 281–288 (2005). doi: 10.1016/j.virol.2005.06.005 Medline [DOI] [PubMed] [Google Scholar]
- 28.Berger G, Turpin J, Cordeil S, Tartour K, Nguyen XN, Mahieux R, Cimarelli A, Functional analysis of the relationship between Vpx and the restriction factor SAMHD1. J. Biol. Chem 287, 41210–41217 (2012). doi: 10.1074/jbc.M112.403816 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C, APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem 281, 22161–22172 (2006). doi: 10.1074/jbc.M601716200 Medline [DOI] [PubMed] [Google Scholar]
- 30.Münk C, Willemsen A, Bravo IG, An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol 12, 71 (2012). doi: 10.1186/1471-2148-12-71 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL, Harris RS, Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem 287, 34801–34808 (2012). doi: 10.1074/jbc.M112.385161 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turelli P, Mangeat B, Jost S, Vianin S, Trono D, Inhibition of hepatitis B virus replication by APOBEC3G. Science 303, 1829 (2004). doi: 10.1126/science.1092066 Medline [DOI] [PubMed] [Google Scholar]
- 33.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H, Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol 307, 183–196 (2001). doi: 10.1006/jmbi.2000.4481 Medline [DOI] [PubMed] [Google Scholar]
- 34.Aynaud MM, Suspène R, Vidalain PO, Mussil B, Guétard D, Tangy F, Wain-Hobson S, Vartanian JP, Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A. J. Biol. Chem 287, 39182–39192 (2012). doi: 10.1074/jbc.M112.372722 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, Yan Z, Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC Genomics 13, 563 (2012). doi: 10.1186/1471-2164-13-563 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM, Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol 23, 258–268 (2012). doi: 10.1016/j.semcdb.2011.10.004 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee TH, Elledge SJ, Butel JS, Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol 69, 1107–1114 (1995). Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura K, Wang Z, Chowdhury S, Simadu M, Koura M, Muramatsu M, Uracil DNA glycosylase counteracts APOBEC3G-induced hypermutation of hepatitis B viral genomes: excision repair of covalently closed circular DNA. PLoS Pathog 9, e1003361 (2013). doi: 10.1371/journal.ppat.1003361 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry S, Narvaiza I, Linfesty DC, Weitzman MD, APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 12, 444–450 (2011). doi: 10.1038/embor.2011.46 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS, APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013). doi: 10.1038/nature11881 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs K, Böttinger N, Huang LR, Chmielewski M, Arzberger S, Gasteiger G, Jäger C, Schmitt E, Bohne F, Aichler M, Uckert W, Abken H, Heikenwalder M, Knolle P, Protzer U, T cells expressing a chimeric antigen receptor that binds hepatitis B virus envelope proteins control virus replication in mice. Gastroenterology 145, 456–465 (2013). doi: 10.1053/j.gastro.2013.04.047 Medline [DOI] [PubMed] [Google Scholar]
- 42.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U, Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol 55, 996–1003 (2011). doi: 10.1016/j.jhep.2011.02.015 Medline [DOI] [PubMed] [Google Scholar]
- 43.Schulze-Bergkamen H, Untergasser A, Dax A, Vogel H, Büchler P, Klar E, Lehnert T, Friess H, Büchler MW, Kirschfink M, Stremmel W, Krammer PH, Müller M, Protzer U, Primary human hepatocytes—a valuable tool for investigation of apoptosis and hepatitis B virus infection. J. Hepatol 38, 736–744 (2003). doi: 10.1016/S0168-8278(03)00120-X Medline [DOI] [PubMed] [Google Scholar]
- 44.Lee SM, Schelcher C, Demmel M, Hauner M, Thasler WE, Isolation of human hepatocytes by a two-step collagenase perfusion procedure. J. Vis. Exp (79): (2013). Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thasler WE, Weiss TS, Schillhorn K, Stoll PT, Irrgang B, Jauch KW, Charitable State-Controlled Foundation Human Tissue and Cell Research: Ethic and Legal Aspects in the Supply of Surgically Removed Human Tissue For Research in the Academic and Commercial Sector in Germany. Cell Tissue Bank 4, 49–56 (2003). doi: 10.1023/A:1026392429112 Medline [DOI] [PubMed] [Google Scholar]
- 46.Hantz O, Parent R, Durantel D, Gripon P, Guguen-Guillouzo C, Zoulim F, Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J. Gen. Virol 90, 127–135 (2009). doi: 10.1099/vir.0.004861-0 Medline [DOI] [PubMed] [Google Scholar]
- 47.Quasdorff M, Hösel M, Odenthal M, Zedler U, Bohne F, Gripon P, Dienes HP, Drebber U, Stippel D, Goeser T, Protzer U, A concerted action of HNF4alpha and HNF1alpha links hepatitis B virus replication to hepatocyte differentiation. Cell. Microbiol 10, 1478–1490 (2008). doi: 10.1111/j.1462-5822.2008.01141.x Medline [DOI] [PubMed] [Google Scholar]
- 48.Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hösel M, Schirmacher P, Tiegs G, Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology 133, 1156–1165 (2007). doi: 10.1053/j.gastro.2007.07.021 Medline [DOI] [PubMed] [Google Scholar]
- 49.Untergasser A, Zedler U, Langenkamp A, Hösel M, Quasdorff M, Esser K, Dienes HP, Tappertzhofen B, Kolanus W, Protzer U, Dendritic cells take up viral antigens but do not support the early steps of hepatitis B virus infection. Hepatology 43, 539–547 (2006). doi: 10.1002/hep.21048 Medline [DOI] [PubMed] [Google Scholar]
- 50.Summers J, Smith PM, Horwich AL, Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol 64, 2819–2824 (1990). Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao W, Hu J, Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol 81, 6164–6174 (2007). doi: 10.1128/JVI.02721-06 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smyth GK, Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol 3, Article3 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Banning C, Votteler J, Hoffmann D, Koppensteiner H, Warmer M, Reimer R, Kirchhoff F, Schubert U, Hauber J, Schindler M, A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS ONE 5, e9344 (2010). doi: 10.1371/journal.pone.0009344 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnold K, Bordoli L, Kopp J, Schwede T, The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). doi: 10.1093/bioinformatics/bti770 Medline [DOI] [PubMed] [Google Scholar]
- 55.Wynne SA, Crowther RA, Leslie AG, The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3, 771–780 (1999). doi: 10.1016/S1097-2765(01)80009-5 Medline [DOI] [PubMed] [Google Scholar]
- 56.Byeon IJ et al. , NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat. Commun 4, 1890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghoorah AW, Devignes MD, Smaïl-Tabbone M, Ritchie DW, Protein docking using case-based reasoning. Proteins 81, 2150–2158 (2013). doi: 10.1002/prot.24433 Medline [DOI] [PubMed] [Google Scholar]
- 58.Sayle RA, Milner-White EJ, RASMOL: biomolecular graphics for all. Trends Biochem. Sci 20, 374–376 (1995). doi: 10.1016/S0968-0004(00)89080-5 Medline [DOI] [PubMed] [Google Scholar]
- 59.Guidotti LG, Matzke B, Schaller H, Chisari FV, High-level hepatitis B virus replication in transgenic mice. J. Virol 69, 6158–6169 (1995). Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dandri M, Burda MR, Török E, Pollok JM, Iwanska A, Sommer G, Rogiers X, Rogler CE, Gupta S, Will H, Greten H, Petersen J, Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology 33, 981–988 (2001). doi: 10.1053/jhep.2001.23314 Medline [DOI] [PubMed] [Google Scholar]
- 61.Lütgehetmann M, Bornscheuer T, Volz T, Allweiss L, Bockmann JH, Pollok JM, Lohse AW, Petersen J, Dandri M, Hepatitis B virus limits response of human hepatocytes to interferon-α in chimeric mice. Gastroenterology 140, 2074–2083, e1–e2 (2011). doi: 10.1053/j.gastro.2011.02.057 Medline [DOI] [PubMed] [Google Scholar]
- 62.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH, Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. U.S.A 105, 7034–7039 (2008). doi: 10.1073/pnas.0707882105 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


