Abstract
There is perhaps no more important time in the history of placebos to consider their role in clinical trials and in medicine. Increasingly well-designed pharmaceutical and academic clinical trials testing promising and established drug and surgical interventions have failed to “beat” the placebo response. The collateral damage resulting from these failures is staggering - novel treatments, many with compelling mechanisms of action and promising Phase 2 trial results, never reach the patient, adversely affecting small and large pharma alike. Recent evidence suggests that variability in placebo response may be attributed in part to genetic variation. Thus, having a better understanding of the genomic underpinnings of the placebo response, the “placebome”, may pave the way to innovatively and more effectively use placebos in drug development.
The “powerful placebo”
Placebos are inert treatments, given historically to enlist the imagination in pleasing difficult, and appeasing untreatable, patients.1 The use of placebos in exposing “fake” treatments resulted in their taking on the negative stigma of the charlatans they were enlisted to discredit. When clinical trials were introduced after World War II to accelerate drug discovery, placebo controls, along with randomization and double-blinding, were implemented to ensure the objective determination of efficacy and safety of any new therapy. By 1955, when Henry Beecher published his landmark paper on placebos,2 it was understood that although the power of placebos was significant - Beecher estimated that approximately 30% of patients responded positively to placebo - drug and placebo responses were additive and, hence, the drug effect could be easily determined by subtraction. For the next half century, placebos would become the benchmark by which new drugs were tested, a veritable gold-standard for drug development. What Beecher did not foresee, however, was how placebos would wreak havoc on clinical trials and come to be a stumbling block for future drug development.
At what cost, failing to beat the placebo response?
In the last decade, a surprising and unexplained trend has emerged. Increasingly, drugs are failing to beat the placebo response. One possibility is that these new drugs are simply not efficacious. Although there are no comprehensive estimates of the extent of this problem, reports in schizophrenia, ADHD, neuropathic pain, clinical depression, and Parkinson disease suggest that while treatment efficacy has remained unchanged, the placebo response rate has crept up. These increased placebo response rates appear unpredictable, and while the sources of variation are not well understood, some studies point to variations in disease severity, non-stringent inclusion criteria, regression to the mean, natural history, treatment adherence, or psychological factors like openness to experience or extraversion, as contributors. Still, without methodology to control the vagaries of the placebo response, whole research areas remain at risk as evidenced by recent cutbacks in the neuroscience divisions of several large pharmaceutical companies.
What does neuroimaging tell us about the placebo response?
At the turn of this century, neuroimaging studies shifted our understanding of placebos from the domain of imagination to physiology and the brain. Brain regions activated in response to placebo treatment, including the ventromedial prefrontal cortex, insula, amygdala, hypothalamus, and periaqueductal gray, revealed that there are regionally distinct pain, autonomic, and immune placebo response pathways. With evidence that signaling and downstream effectors included neurotransmitters (dopamine), hormones (norepinephrine), and cytokines (IL2), the research question turned to whether genetic variation in the molecules mediating these signaling pathways could influence variability in placebo response. Our group recently coined the term “placebome”3 to circumscribe the potential network of genome-related factors that influence the placebo response.4
The Placebome - Towards a genetics of the placebo response
Although placebos have been an integral part of clinical trials throughout the genomics era, there are no genome-wide association studies (GWAS) of the placebo response, and no formal estimates of heritability. To assess what is known about placebo genetics, we started with a comprehensive review of the literature. Using the search terms ([placebo] and [gene] and [SNP]), we identified 28 genes from 42 studies associated with significant variation in outcomes in the placebo arm of a trial. These studies were all small (N<257), and included a mix of clinical trials and experimental paradigms in healthy volunteers. Only two studies included a no-treatment control, the requisite control for placebo research, and most of the studies conducted candidate analyses of genes hypothesized to be associated with the disease or treatment of interest. Given the limitations in this set of genes, we turned to the human interactome (the network of all known human physical protein-protein interactions) to identify other genes/proteins potentially associated with the placebo response. We developed a seed-connector algorithm to connect the original set of placebo response genes (seeds) to each other by adding as few extra nodes (proteins from the interactome) as possible. In this way, we created a placebome module consisting of 54 proteins. These 54 proteins were enriched for neurotransmitters, hormones, and cytokines that could readily be mapped to the pain (i.e., opioid: OPRM1 and OPRD1), autonomic (i.e., catecholamine: ADRA1B and COMT), and immune (i.e., cytokines: IL6 and TNF) placebo response pathways (Figure).
Figure.
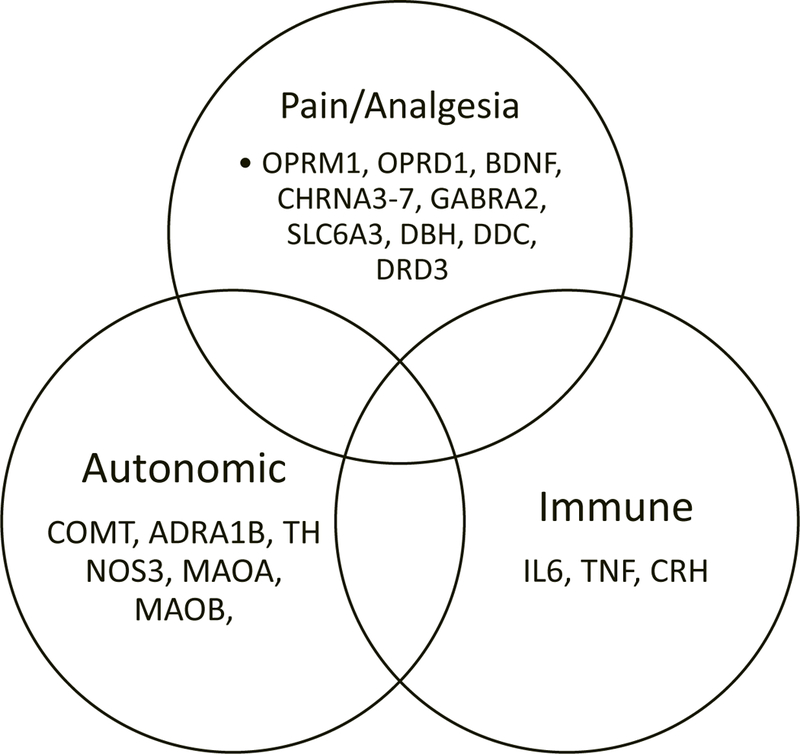
A subset of the placebome genes associated with the pain, autonomic and immune placebo response pathways.
We hypothesized that the placebome, a distinct and novel module (or subnetwork) in the interactome, would be located proximal to modules (subnetworks) for specific diseases known to have a high placebo response. Using bench-mark conditions like anxiety, depression, and migraine disorders known to have a high placebo response, we confirmed that they were, indeed, proximal to the placebome. In contrast, conditions like viremia, uremia, and pneumothorax known to have low-to-no placebo responses were distal. We then compared the distance between an exhaustive set of 859 diseases and the placebome module, and found that there were 252 modules including central nervous system diseases (i.e., epilepsy and Parkinson disease) and substance abuse disorders that were significantly proximal to the placebome. Interestingly, metabolic diseases and some cancers also mapped close to the placebome. Although cancer and metabolic diseases are not known to have high placebo responses, the possibility remains that placebo response in some diseases could be initiated but limited in efficacy. We also mapped symptoms associated with high placebo response (pain, nausea, headache, fatigue, and hot flashes) and found them to be significantly proximal to the placebome compared to low-placebo symptoms like fever.
Additivity revisited
Strikingly, we found that 26 of the 28 placebo seed genes and 40 of the 54 genes in the placebome module were known drug targets. Further, when we examined the proximity of drug targets to the placebome module, we found 15 drug categories, including analgesics, appetite suppressants, and anti-depressives, that mapped significantly closely to the placebome. These observations raise two related questions. First, is the placebo response a ‘druggable’ target? And second, have we all along been developing drugs that target the placebo response? Certainly, naloxone and proglumide have been shown to disrupt the placebo response. What remains to be determined is how genetic variation in the placebo response pathway might affect our interpretation of clinical trial results. Could gene-drug/placebo interactions be an unexplored confounder of observed outcomes? Our findings of differential outcomes in the placebo and drug treatment arms as a function of a placebome gene, catechol-O-methyltransferase (COMT), in two trials, NorCAPITAL5 and the Women’s Health Study6 support this hypothesis and suggest that the placebome can be used to improve precision in drug efficacy determination.
Future directions
The concept of the placebome raises a series of interesting and challenging questions and has the potential to revolutionize our understanding of the placebo response in clinical trial design. Despite the tremendous body of work elucidating the neurological, behavioral, and now genetic underpinnings of the placebo response, placebos have not shaken the stigma of the charlatans they were enlisted to discredit. Any placebo researcher will tell you that scientists and clinicians are still disquieted by discussions about the implications of placebo effects. Because of this unease (a topic in and of itself), half of the data from thousands of clinical trials have been ignored for over half-a-century. This blind spot has cost us and will continue to do so unless and until we turn our scientific rigor towards this problem and address it with precision.
Acknowledgments
Funding: KTH is funded by a Harvard Catalyst faculty fellowship and NIH grant 1K01HL130625.
References
- 1.Finniss DG; Kaptchuk TJ; Miller F; Benedetti F, Biological, clinical, and ethical advances of placebo effects. Lancet 2010, 375 (9715), 686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beecher HK, The powerful placebo. Journal of the American Medical Association 1955, 159 (17), 1602–6. [DOI] [PubMed] [Google Scholar]
- 3.Hall KT; Loscalzo J; Kaptchuk TJ, Genetics and the placebo effect: the placebome. Trends in molecular medicine 2015, 21 (5), 285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang RS; Hall KT; Giulianini F; Passow D; Kaptchuk TJ; Loscalzo J, Network analysis of the genomic basis of the placebo effect. JCI insight 2017, 2 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall KT; Kossowsky J; Oberlander TF; Kaptchuk TJ; Saul JP; Wyller VB; Fagermoen E; Sulheim D; Gjerstad J; Winger A; Mukamal KJ, Genetic variation in catechol-O-methyltransferase modifies effects of clonidine treatment in chronic fatigue syndrome. The pharmacogenomics journal 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall KT; Nelson CP; Davis RB; Buring JE; Kirsch I; Mittleman MA; Loscalzo J; Samani NJ; Ridker PM; Kaptchuk TJ; Chasman DI, Polymorphisms in catechol-O-methyltransferase modify treatment effects of aspirin on risk of cardiovascular disease. Arterioscler Thromb Vasc Biol 2014, 34 (9), 2160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


