Abstract
Objectives
Statins are the most commonly prescribed medications for the treatment of atherosclerotic cardiovascular disease. Statin-associated adverse effects occur in ∼10% of patients and are associated with polymorphisms in several key genes coding for transporters and metabolizing enzymes that affect statin pharmacokinetics. In the present study, we examine the association between cytochrome P450 3A5*3 (CYP3A5*3) T>C (rs776746), COQ G>C (rs4693075), and SLCO1B1 T>C (rs4149056) genetic variants with the risk of myopathy in South Indian patients on statin therapy.
Methods
A total of 202 patients on atorvastatin or rosuvastatin therapy for 12 years were recruited in the study. Genotyping of drug metabolic CYP3A5*3 gene variant and drug transporter genes COQ G>C (rs4693075) and SLCO1B1 T>C (rs4149056) was analyzed by Sanger's sequencing.
Results
In our study subjects, the percentage of patients diagnosed to have statin-induced myopathy was 18%. The majority of the patients were on 10 mg/day dose of either atorvastatin or rosuvastatin. The homozygous nonexpressors genotype CYP3A5*3/3 frequency of the CYP3A5 polymorphism was higher in patients with myopathy. But we could not find association of CYP3A5, COQ, and SLCO1B1 gene polymorphisms with either rosuvastatin or atorvastatin.
Conclusion
Our results clearly demonstrate that the frequency of CYP3A5*3 splicing variant is higher in myopathy group than in the tolerant group. We did not find significant association of genetic polymorphisms in CYP3A5, COQ, and SLCO1B1 with atorvastatin- or rosuvastatin-induced myopathy.
Keywords: Myopathy, Statins, Pharmacogenetics, Dyslipidemia drug transporters
1. Introduction
3-Hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors or statins are widely used drugs in the reduction of cardiovascular events in patients with dyslipidemia, one of the main risk factor for coronary artery disease.1 On the other hand, statin-related severe adverse events are reported in patients treated with statins due to oxidative stress, abnormal mitochondrial function, and imbalance of muscular calcium homeostasis leading to muscular side effects, a common cause of withdrawal and statin discontinuation.2, 3, 4, 5, 6 Statins are associated with adverse clinical effects such as myalgia, myositis, rhabdomyolysis, muscle weakness, muscle cramps, and creatine kinase (CK) elevations after statin withdrawal.4, 5, 6 However, recent data suggest that genetics influences the intolerance and development of statin-induced myopathy, at least for some statins.6
Several candidate genes are proposed to have potential role in statin pharmacokinetics including SLCO1B1 (organic anion transporter family); CYP3A4, CYP3A5, and CYP3C9 (family of cytochrome P450); and ABCB1, ABCC2, and ABCG2 (adenosine triphosphate (ATP)-binding cassette family).7 Single nucleotide polymorphism in these genes may affect the statin transport and metabolism eventually influencing the pharmacokinetics of the drug, which ultimately increases the systemic and intraorganic exposure leading to statin-induced myopathy.6, 7, 8
Solute carrier organic anion transporter family member 1B1 (SLCO1B1) codes for a transporter protein (OATP1B1), which is involved in the transport of various pharmacological compounds including statins, antibiotics, angiotensin converting enzyme (ACE) inhibitors, and xenobiotic compounds.9 The nonsynonymous SLCO1B1 variant T521C, Val174Ala (rs4149056), affects the OATP1B1 protein function by its reduced uptake or transport activity of statins and their metabolites into the liver and hence able to influence variations in plasma concentrations of statins while it also reported to influence the pharmacokinetics of both atorvastatin and rosuvastatin.10, 11 Several studies have shown that SLCO1B1 521C was associated with marginal (<5%) attenuation of the lipid-lowering effect of simvastatin, atorvastatin, lovastatin, and pravastatin.12, 13 In addition, several studies highlighted the association between SLCO1B1 gene c.521T>C polymorphism and statin-related myopathy risk.14, 15 The cytochrome P450 3A5*3 (CYP3A5*3) single-nucleotide polymorphism (SNP) is a splice acceptor variant well studied in CYP3A5 gene, which introduces a frame shift during translation process, resulting in a truncated, nonfunctional protein product.16 The variant allele CYP3A5*3 confers low or undetectable CYP3A5 expression as a result of a single-point mutation within intron 3 of the CYP3A5 gene (c.6986A>G, rs776746).16 COQ2 gene encodes for parahydroxybenzoate–polyprenyltransferase enzyme, and variants within this gene are associated with CoQ10 deficiency and skeletal muscle drug transporters expression leading to statin muscular intolerance.17 However, numerous studies have been proposed from different ethnic groups to demonstrate the risk of statin toxicity.7 The distribution of genotype frequencies vary from one population to another. Studying the impact of pharmacogenetics of statins is necessary to establish pharmacokinetics, efficacy, and tolerability of these drugs in each population. Hence, the present study is aimed to investigate the association of statin-induced myopathy with the CYP3A5*3 T>C (rs776746), COQ G>C (rs4693075), and SLCO1B1 T>C (rs4149056) variants in South-Indian patients on statin therapy.
2. Materials and methods
2.1. Study subjects
Two hundred and two study subjects with mean average age of 63 years who were on either atorvastatin or rosuvastatin for 12 years of duration were enrolled in the study. The institutional ethics committee approved the study, and informed consent from each patient was obtained. The demographic and clinical details were recorded in a standard format. Patients with comorbid conditions such as hepatic insufficiency; hematologic, kidney, liver, or malignant disease; hypothyroidism; hypovitaminosis D, other drugs that are likely to interact with statins; patients with major bleeding events within 7 days before enrollment; or the use of oral anticoagulant agents were excluded from the study.
2.2. Genetic analysis
Whole blood samples were collected in ethylenediamine tetra acetic acid. Genomic DNA extraction was performed using standard organic phenol-chloroform procedure, followed by checking with NanoDrop instrument for quality assessment (Thermo Fisher Scientific). The reference genomic sequence of CYP3A5*3 (rs776746), COQ (rs4693075), and SLCO1B1 (rs4149056) was retrieved from the Ensembl database. Target-specific primer pairs were designed using Primer3 Web tool version 4. The genotyping of these polymorphisms were determined by using polymerase chain reaction (PCR) method, followed by sequencing. The PCR amplification was done for CYP3A5*3, c.6986A>G (rs776746) polymorphism using the following primer pair: (1) forward, 5′-TCACGTCGGGATCTGTGAT-3′; (2) reverse, 5′-GACTGTGGAGTGCTGTGGAG-3’. PCR conditions for the amplification were as follows: (1) initial denaturation at 94°C for 5 min; (2) followed by 38 cycles of 30 s at 94°C, 30 s at 52.2°C, and 30 s at 72°C for extension; and (3) a final extension of 10 min at 72°C. The PCR amplification of SLCO1B1, rs4149056, Val174Ala was carried out using the following primer pair: (1) forward, 5′-CTTCATCTTCCGCCATGATT-3′ and (2) reverse, 5′-GATCCCAGGGTAAAGCCAAT-3′. The PCR conditions were as follows: (1) initial denaturation at 94°C for 5 min, (2) 94°C for 30 s, (3) 58°C for 30 s, (4) 72°C for 30 s for 38 cycles, and (5) final extension at 72°C for 10 min. The genotyping of COQ, rs4693075, polymorphism was performed using the forward primer 5′-CGTTGTACTGCAATGACCTGTT-3′ and reverse primer 5′-TCCGGCACTTACTTATTTCAG-3′ and the PCR conditions as follows: (1) initial denaturation at 94°C for 5 min, (2) 94°C for 30 s, (3) 50.2°C for 30 s, (4) 72°C for 30 s for 38 cycles, and (5) final extension at 72°C for 10 min.
The PCR was performed in a 10-μl reaction mixture containing 0.15 μl of each primer (10 pmol/μl), 5 μl of Takara EmeraldAmp GT PCR master mix, 2 μl of DNA template (50 ng/μl), and 2.8 μl of double-distilled water using on a thermal cycler (Eppendorf). Qualitative check was carried out for amplified PCR fragments with 2% agarose gel electrophoresis, followed by sequencing of amplicons using the BigDye terminator version 3.1 cycle sequencing kit (Applied Biosystems) on ABI Prism 3730xl Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions. Sequencing chromatograms were visualized with naked eye using FinchTV software, and genotype information was recorded.
2.3. Assessment of statin-induced myopathy
The following mild-to-moderate symptoms of statin-induced myopathy, including fatigue, muscle pain, muscle tenderness, muscle weakness, nocturnal cramping, and tendon pain, were recorded at the time of enrollment. Patients with the abovementioned symptoms with serum CK levels more than five times the normal values were grouped under statin-induced myopathy group, and rest of the patients without the symptoms and normal CK values were grouped as nonmyopathy group or tolerant group18, 19.
2.4. Statistical analysis of data
The differences within demographic between groups were expressed as percentages and were compared using the chi-square test or Fisher's exact test. The correlation between myopathy with clinical history of diabetes, hypertension, and age was evaluated using logistic regression analysis. CYP3A5*3 T>C (rs776746), COQ G>C (rs4693075), and SLCO1B1 T>C (rs4149056) genotype frequencies were tested for Hardy–Weinberg equilibrium using chi-square tests. The association results were estimated using odds ratio (ORs) along with 95% confidence intervals (CI). The distribution of allele and genotype frequencies between the groups was analyzed by Fisher's exact test using 2 × 2 contingency table (www.faculty.vassar.edu/lowry/VassarStats.html). A p < 0.05 was considered statistically significant.
3. Results
3.1. Study cohort and treatment characteristics
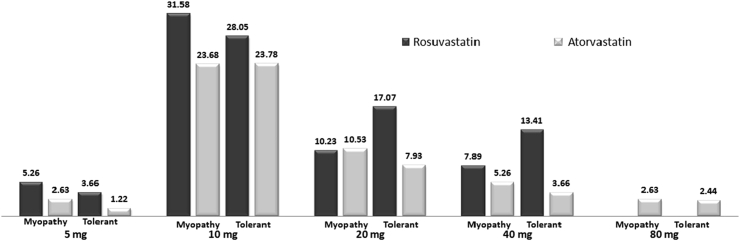
The demographic, clinical parameters of all the patients are shown in Table 1. The percentage of patients with diabetes, hypertension, and smoking were found to be 68.4, 78.94, and 6.57, respectively (Table 1). Of the 202 subjects, 131 (64.85%) were male with a mean age of 63.30 ± 11.05 years and 71 (36.14%) were female with a mean age of 63.36 ± 8.06 years. Among 202, 121 (59.90%) of the patients were on rosuvastatin, whereas 81 (40.09%) were on atorvastatin. With regard to the statin dose, 7% of the patients were on 5 mg, 47% were on 10 mg, 26% on 20 mg, and 20% on 40 mg of rosuvastatin per day compared with 4%, 59%, 21%, 10%, and 6% for these same doses, respectively, for atorvastatin, indicating that majority of the patients were on 10 mg/day dose of either atorvastatin or rosuvastatin. In addition, 6% of the patients were on 80 mg of atorvastatin (Fig. 1). This dose was further stratified into myopathy and nonmyopathy subjects, and results are shown in Fig. 1. Furthermore, multiple logistic regression analysis revealed no association of myopathy with age (p = 0.57), body mass index (p = 0.78), sex (p = 0.48), previous history of type 2 diabetes (p = 0.80), hypertension (p = 0.49), CYP3A5 genotype (p = 0.65), COQ genotypes (p = 0.53), and SLCOB1 genotypes (p = 0.75).
Table 1.
Demographic characteristics of study group.
| Characteristic | Male (n = 131) | Female (n = 71) | P value |
|---|---|---|---|
| Height (cm) | 158.83 ± 15.15 | 152.75 ± 5.15 | ns |
| Weight (kg) | 68.60 ± 10.18 | 65.20 ± 11.98 | ns |
| BMI (kg/m2) | 42.8817 ± 6.46 | 42.95 + 7.59 | ns |
| DM (%) | 84 (64.12) | 52 (73.23) | ns |
| DM duration (years) | 7.267 ± 11.366 | 9.95 ± 8.28 | ns |
| HTN (%) | 100 (76.33) | 63 (88.73) | ns |
| HTN duration (years) | 7.805 ± 7.859 | 9.94 ± 9.02 | ns |
| Myopathy (%) | 22 (16.79) | 16 (22.53) | ns |
| Total cholesterol (mg/dL) | 136.89 ± 37.95 | 138.39 ± 36.80 | ns |
| HDL-C (mg/dL) | 40.09 ± 12.06 | 45.32 ± 11.42 | ns |
| LDL-C (mg/dL) | 70.93 ± 33.24 | 66.66 ± 34.32 | ns |
| VLDL-C (mg/dL) | 31.07 ± 37.36 | 33.22 ± 35.55 | ns |
| Triglycerides (mg/dL) | 135.18 ± 72.24 | 126.78 ± 59.60 | ns |
| Statin at index | |||
| Rosuvastatin, n (%) | 78 (59.54) | 42 (59.15) | ns |
| Atorvastatin, n (%) | 55 (41.98) | 28 (39.43) | ns |
DM, diabetes mellitus; HTN, hypertension; HDL-C- high-density lipoprotein cholesterol; LDL-C- low-density lipoprotein cholesterol; VLDL-C- very low-density lipoprotein cholesterol; ns, Not significant; BMI, body mass index.
Fig. 1.
Distribution of rosuvastatin and atorvastatin dose in the study cohort.
3.2. CYP3A5*3, COQ, and SLCO1B1 polymorphisms associated with myopathy
The allele and genotype frequencies of CYP3A5*3, COQ, and SLCO1B1 were shown in Table 2. The allele and genotype frequency distribution was in accordance with Hardy–Weinberg equilibrium (p > 0.05). No significant differences were found for these polymorphisms between male and female study subjects. Genetic data of CYP3A5*3 and COQ polymorphisms were segregated based on the presence and absence of statin-induced myopathy, and results are shown in Table 2. CYP3A5*3 polymorphism showed significant genotype distribution between the myopathy group and tolerant group (TT+TC vs. CC; 52.63% vs. 34.15%) (OR: 2.14, CI 95%: 1.04–4.37, p = 0.04), whereas COQ polymorphism did not show any difference between the myopathy group and tolerant group (TT+TC vs. CC recessive model, OR: 0.98, CI 95%: 0.48–1.99, p = 1.00, and dominant model TT vs TC+TC, OR: 1.68.14, CI 95%: 0.36–7.72, p = 0.54) (Table 2). The genotype frequency of the SLCO1B1 (rs4149056) polymorphism was 93.56% for homozygous wild genotype, 6.43% for heterozygous individuals, and no homozygous mutants were observed in the study cohort. Furthermore, segregation of SLCO1B1 allele and genotype frequencies between the myopathy and tolerant group did not reveal any evidence for statin-induced myopathy (TT vs TC+TC, OR: 1.32.14, CI 95%: 0.34–5.04, P = 0.71). Among 202 patients treated with statin therapy, 38 patients (18.81%) had myopathy, among which 20 (55.26%) patients were on rosuvastatin and 17 (44.73%) patients were on atorvastatin. Furthermore, CYP3A5*3, COQ, and SLCO1B1 allele and genotype frequency data were stratified according to individual statin to find the effect of polymorphism on statin-induced myopathy and could not find association with either rosuvastatin or atorvastatin (Table 3, Table 4).
Table 2.
Genotype and allele frequency distribution of CYP3A5*3, COQ and SLCO1B1 in myopathy and tolerant group.
| Gene (rs number) | Genotype and allele | Myopathy, n = 38 | Tolerant, n = 164 | OR | CI at 95% | P value |
|---|---|---|---|---|---|---|
| CYP3A5*3 (rs776746) | TT | 4 (10.53) | 21 (12.80) | |||
| TC | 14 (36.84) | 87 (53.05) | 1.24 | 0.40–3.87 | 0.79 | |
| CC | 20 (52.63) | 56 (34.15) | 2.14 | 1.04–4.37 | 0.04* | |
| T | 22 (28.95) | 129 (39.33) | ||||
| C | 54 (71.05) | 199 (60.67) | 1.59 | 0.92–2.73 | 0.11 | |
| COQ (rs4693075) | GG | 2 (5.26) | 14 (8.54) | |||
| GC | 16 (42.11) | 63 (38.41) | 1.68 | 0.36–7.72 | 0.54 | |
| CC | 20 (52.63) | 87 (53.05) | 0.98 | 0.48–1.99 | 1 | |
| G | 20 (26.32) | 91 (27.74) | ||||
| C | 56 (73.68) | 237 (72.26) | 1.07 | 0.61–1.89 | 0.88 | |
| SLCO1B1 (rs4149056) | TT | 35 (92.10) | 154 (93.90) | |||
| TC | 3 (7.89) | 10 (6.09) | 1.32 | 0.34–5.04 | 0.71 | |
| CC | 0 (0) | 0 (0) | ||||
| T | 73 (96.55) | 318 (96.95) | 1.30 | 0.35–4.86 | 0.71 | |
| C | 3 (3.44) | 10 (6.09) |
OR, odds ratio; CI, class interval. *P value less than 0.05 is considered as significant.
Table 3.
Genotype and allele frequency distribution of CYP3A5*3 and COQ in myopathy and tolerant group on atorvastatin treatment.
| Gene (rs number) | Genotype and allele | Atorvastatin |
OR (CI at 95%) | |
|---|---|---|---|---|
| Myopathy (n = 17) | Tolerant (n = 64) | |||
| CYP3A5*3 (rs776746) | TT | 0 (0.00) | 7 (10.94) | 0.9581 (0.34–2.67) |
| TC | 9 (52.94) | 26 (40.63) | ||
| CC | 8 (47.06) | 31 (48.44) | ||
| T | 9 (26.47) | 40 (31.25) | 1.26 (0.54–2.9) | |
| C | 25 (73.53) | 88 (68.75) | ||
| COQ (rs4693075) | GG | 0 (0) | 4 (6.25) | 0.87 (0.29–2.55) |
| GC | 8 (47.06) | 24 (37.50) | ||
| CC | 9 (52.94) | 36 (56.25) | ||
| G | 8 (23.53) | 32 (25.00) | ||
| C | 26 (76.47) | 96 (75.00) | 1.08 (0.44–2.63) | |
| SLCO1B1 (rs4149056) | TT | 14 (82.35) | 61 (95.31) | |
| TC | 03 (17.64) | 03 (4.68) | 4.35 (0.79–23.91) | |
| CC | 0 (0) | 0 (0) | ||
| T | 31 (91.17) | 125 (97.65) | 4.0 (0.77–20.95) | |
| C | 03 (8.82) | 03 (2.34) | ||
OR, odds ratio; CI, class interval.
Table 4.
Genotype and allele frequency distribution of COQ and SLCO1B1 in myopathy and tolerant groups on rosuvastatin treatment.
| Gene (rs number) | Genotype and allele | Rosuvastatin |
OR (CI at 95%) | |
|---|---|---|---|---|
| Myopathy (n = 21) | Tolerant (n = 102) | |||
| COQ(rs4693075) | GG | 2 (9.52) | 11 (10.78) | 1.14 (0.23–5.60) |
| GC | 8 (38.09) | 39 (38.24) | 1.05 (0.41–2.7) | |
| CC | 11 (55.00) | 52 (50.98) | ||
| G | 12 (28.57) | 61 (29.90) | ||
| C | 30 (71.42) | 143 (70.10) | 1.06 (0.51–2.2) | |
| SLCO1B1(rs4149056) | TT | 21 (100) | 95 (93.13) | |
| TC | 0 (0) | 07 (6.86) | NA | |
| CC | 0 (0) | 0 (0) | ||
| T | 42 (0) | 197 (96.56) | NA | |
| C | 0 (0) | 07 (3.43) | ||
OR, odds ratio; CI, class interval; NA, not available.
4. Discussion
The interindividual variability in response to HMG-CoA reductase inhibitors, or statins, with regard to both efficacy and safety has been well documented. Earlier studies from India have investigated the efficacy of statins in the reduction of lipid parameters20, 21; however, no studies have been reported on the influence of pharmacogenetics on statin-induced myopathy. To the best of our knowledge, this the first study to investigate the role of pharmacogenetics on statin-induced myopathy in South-Indian patients. Statin therapy is limited with skeletal muscle toxicity associated with elevated systemic drug exposure, and up to 10% of statin-treated individuals will experience muscle pain or weakness, in rare cases.3, 12, 22 Studies have shown that muscle-related symptoms include muscle weakness (∼3%),23, 24 myalgia (2%–22%),25, 26 and life-threatening rhabdomyolysis (0.0004%),27 which tend to occur several months to years after statin initiation. In a randomized (JUPITER) clinical trial with rosuvastatin 20 mg daily or placebo and with an average follow-up of 17 months, they found that muscle symptoms (pain, stiffness, or weakness) was noted in 16.0% of the rosuvastatin group and 15.4% of the placebo group.10 In the present study, we found 18.81% of patients diagnosed to have statin-induced myopathy, which is higher than that reported in literature. This could be due to the inclusion of patients who are on chronic treatment of atorvastatin or rosuvastatin. Hence, we found myopathy-related symptoms even at lower doses of atorvastatin or rosuvastatin. Wu et al. studied 26 cases of statin-associated autoimmune myopathies and found myalgias in 38% of cases, muscle weakness in 100% of cases, and rhabdomyolysis in 12% of confirmed cases of statin-associated myopathies.28 Specific genetic polymorphisms have been identified as potential contributors to variability in statin absorption, systemic distribution, metabolism, and elimination eventually leading to statin-induced myopathy in individuals with higher stain dose.29
Pharmacokinetic case–control study on patients receiving atorvastatin showed 2.4-fold and 3.1-fold higher systemic exposures of atorvastatin metabolites lactone and p-hydroxyatorvastatin, respectively.30 Furthermore, statin lactones are potent inhibitors of the enzymatic activity of complex III of respiratory chain–inducing cytotoxicity and reducing respiration and mitochondrial ATP production in statin-induced myopathy patients.31 SLCO1B1 T521C polymorphism results in decreased statin transport into liver cells and, therefore, theoretically should confer a decreased risk of statin-associated liver toxicity due to attenuated hepatic statin exposure. More importantly, SLCO1B1 521C carriers have increased systemic statin exposure and increased risk of statin myopathy.14, 32, 33 The Statin Response Examined by Genetic Haplotype Markers (STRENGTH) pharmacogenetic study on safety and efficacy of atorvastatin, simvastatin, and pravastatin revealed that composite adverse event (CAE) was associated with SLCO1B1*5 genotype (CAE in individuals with TT, TC, and CC alleles: 19%, 27%, and 50%, respectively) and female gender.34 A genome-wide scan revealed a strong association of myopathy with the SLCO1B1 SNP rs4363657 and strong linkage disequilibrium with another nonsynonymous SNP rs4149056 (r2 = 0.97). Carriers of SLCO1B1 T>C allele had 4.5 times increased risk with TC genotype and 16.9 times higher risk with CC than TT homozygotes and further more than 60% of myopathy cases reported with C variant allele.14 Dosing recommendations were prescribed for simvastatin, based on rs4149056 genotype, because of intermediate myopathy risk with “TC” genotype and high myopathy risk with “CC” genotype.35 SLCO1B1 variants do not have clinical importance in atorvastatin therapy because only fewer studies have shown positive association,34 but majority of the studies found no association with SLCO1B1 with atorvastatin-induced myopathy.24, 36, 37 In the present study, the genotype frequency of the SLCO1B1 (rs4149056) polymorphism was 93.56% for homozygous wild “TT” genotype and 6.43% for heterozygous “TC” individuals, and no homozygous “CC” mutants were observed in the study subjects. Furthermore, segregation of SLCO1B1 allele and genotype frequencies between the myopathy and tolerant group did not reveal evidence for statin-induced myopathy. In atorvastatin group, we observed the absence of homozygous CC variant and presence of TC of 17.64% in the myopathy group and 4.68% in the tolerant group; though the frequency is high in the myopathy group, we did not find a significant association with atorvastatin-induced myopathy.
Cytochrome P450 3A (CYP3A) subfamily members are important drug-metabolizing enzymes involved in atorvastatin, lovastatin, simvastatin, and pravastatin metabolism.7 The most frequent and commonly studied CYP3A5 polymorphism is the loss of function of CYP3A5*3 (rs776746) allele.38 The carriers of homozygous CYP3A5*3 demonstrated higher serum CK levels with greater degree of atorvastatin-induced muscle damage, whereas no association was found with efficacy or tolerability of simvastatin.38, 39 Kivistö et al reported that the mean serum low-density lipoprotein cholesterol concentration was 24% higher in subjects possessing the CYP3A5*1 allele than in subjects homozygous for the CYP3A5*3 allele, and lovastatin, simvastatin, and atorvastatin were significantly less effective in CYP3A5 expressors than in nonexpressors indicating interindividual variations in response to statins.40
In the present study, the percentage of patients with myopathy and positive for CYP3A5*3 was 52.5%, whereas in the tolerant group, it was 34.1% indicating that the number of patients with myopathy with CYP3A5*3 is significantly higher than that in the tolerant group. On the contrary, another study did not find any significant associations among CYP3A4*1B and CYP3A5*3C and the effect of simvastatin on low-density lipoprotein cholesterol levels after two and six months of treatment.16 Similarly, we did not find any significant association of atorvastatin-induced myopathy with the CYP3A5, COQ, and SLCO1B1 genetic variants and of rosuvastatin-induced myopathy with COQ and SLCO1B1 genetic variants.
A study on 133 subjects who developed myopathy on statin therapy and 158 in the tolerated group showed significant association of nonsynonymous polymorphism in exon 5 (rs6535454) and noncoding polymorphism in intron 4 (rs4693075) with increased risk of statin intolerance among homozygotes mutant carriers (OR,:2.42 and 2.33, respectively).41 An observational case–control study on 76 cases of muscular intolerance showed significant association with intolerance in rosuvastatin-treated subjects with the rs4693075 variant of COQ2 gene.22 Ruaño et al. has evaluated 31 candidate genes based on the previous literature in 793 patients (377 with myalgia and 416 without myalgia) and found significant association with COQ2 (rs4693570).42 In the present study, the COQ polymorphism did not show any difference between the myopathy group and tolerant group both in recessive and dominant models. Further studies are required to study the other genotypes and also to correlate the plasma concentrations of the drug in patients who are on statin therapy. The major limitations of the present study include small sample size and inclusion of patients with chronic treatment of atorvastatin or rosuvastatin. Further studies are warranted with increased sample size with inclusion of newly diagnosed dyslipidemic patients on statin treatment with regular follow-up to assess the association of statin-induced myopathy with the genotype.
5. Conclusion
In conclusion, our present study shows that the homozygous nonexpressors genotype of CYP3A5 gene is associated with myopathy in South-Indian population. We did not find significant association of genetic variants in CYP3A5, COQ, and SLCO1B1 with atorvastatin- or rosuvastatin-induced myopathy.
Authors' contributions
N.R. performed clinical investigations and contributed samples. V.K.K. provided research material. S.K.K. and B.I. conducted experiments and analyzed the data. S.K.K., B.I., and V.K.K. wrote and corrected the manuscript.
Conflict of interest
All authors have none to declare.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgments
The authors are thankful to the patients for participation in the study. They also express thanks to M/s Bioserve Bio-Technologies Pvt. Ltd., Hyderabad, India, for their support for sequencing services and oligo synthesis.
References
- 1.Jellinger P.S., Smith D.A., Mehta A.E. AACE task force for management of dyslipidemia and prevention of atherosclerosis. American association of clinical endocrinologists' guidelines for management of dyslipidemia and prevention of atherosclerosis. Endocr Pract. 2012 Mar–Apr;18(Suppl 1):1–78. doi: 10.4158/ep.18.s1.1. [DOI] [PubMed] [Google Scholar]
- 2.Guyton J.R. Benefit versus risk in statin treatment. Am J Cardiol. 2006 Apr 17;97(8A):95C–97C. doi: 10.1016/j.amjcard.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Bruckert E., Hayem G., Dejager S., Yau C., Bégaud B. Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients--the PRIMO study. Cardiovasc Drugs Ther. 2005 Dec;19(6):403–414. doi: 10.1007/s10557-005-5686-z. [DOI] [PubMed] [Google Scholar]
- 4.Ghatak A., Faheem O., Thompson P.D. The genetics of statin-induced myopathy. Atherosclerosis. 2010 Jun;210(2):337–343. doi: 10.1016/j.atherosclerosis.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 5.Ruaño G., Thompson P.D., Windemuth A. Physiogenomic analysis links serum creatine kinase activities during statin therapy to vascular smooth muscle homeostasis. Pharmacogenomics. 2005 Dec;6(8):865–872. doi: 10.2217/14622416.6.8.865. [DOI] [PubMed] [Google Scholar]
- 6.Kitzmiller J.P., Mikulik E.B., Dauki A.M., Murkherjee C., Luzum J.A. Pharmacogenomics of statins: understanding susceptibility to adverse effects. Pharmgenomics Pers Med. 2016 Oct 3;9:97–106. doi: 10.2147/PGPM.S86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talameh J.A., Kitzmiller J.P. Pharmacogenetics of statin-induced myopathy: a focused review of the clinical translation of pharmacokinetic genetic variants. J Pharmacogenomics Pharmacoproteomics. 2014 Apr 23;5(2) doi: 10.4172/2153-0645.1000128. pii: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrigoni E., Del Re M., Fidilio L., Fogli S., Danesi R., Di Paolo A. Pharmacogenetic foundations of therapeutic efficacy and adverse events of statins. Int J Mol Sci. 2017 Jan 6;18(1) doi: 10.3390/ijms18010104. pii: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kivistö K.T., Niemi M. Influence of drug transporter polymorphisms on pravastatin pharmacokinetics in humans. Pharm Res. 2007 Feb;24(2):239–247. doi: 10.1007/s11095-006-9159-2. [DOI] [PubMed] [Google Scholar]
- 10.Chasman D.I., Giulianini F., MacFadyen J., Barratt B.J., Nyberg F., Ridker P.M. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: the justification for the use of statins in prevention: an intervention trial evaluating rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet. 2012 Apr 1;5(2):257–264. doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues A.C. Efflux and uptake transporters as determinants of statin response. Expet Opin Drug Metabol Toxicol. 2010 May;6(5):621–632. doi: 10.1517/17425251003713519. [DOI] [PubMed] [Google Scholar]
- 12.Thompson P.D., Clarkson P.M., Rosenson R.S. National lipid association statin safety task force muscle safety expert panel. An assessment of statin safety by muscle experts. Am J Cardiol. 2006 Apr 17;97(8A):69C–76C. doi: 10.1016/j.amjcard.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Peters B.J., Rodin A.S., Klungel O.H. Pharmacogenetic interactions between ABCB1 and SLCO1B1 tagging SNPs and the effectiveness of statins in the prevention of myocardial infarction. Pharmacogenomics. 2010 Aug;11(8):1065–1076. doi: 10.2217/pgs.10.81. [DOI] [PubMed] [Google Scholar]
- 14.SEARCH Collaborative Group. Link E., Parish S., Armitage J. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008 Aug 21;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 15.Hou Q., Li S., Li L., Li Y., Sun X., Tian H. Association between SLCO1B1 gene T521C polymorphism and statin-related myopathy risk: a meta-analysis of case-control studies. Medicine (Baltim) 2015 Sep;94(37):e1268. doi: 10.1097/MD.0000000000001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hustert E., Haberl M., Burk O. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001 Dec;11(9):773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Quinzii C., Naini A., Salviati L. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006 Feb;78(2):345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry A.R., Beach J.E., Pearson G.J. Prevention and management of statin adverse effects: a practical approach for pharmacists. Can Pharm J (Ott) 2018 Apr 4;151(3):179–188. doi: 10.1177/1715163518768534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenson R.S., Baker S.K., Jacobson T.A., Kopecky S.L., Parker B.A. The national lipid association's muscle safety expert panel. An assessment by the statin muscle safety task force: 2014 update. J Clin Lipidol. 2014 May–Jun;8(3 Suppl):S58–S71. doi: 10.1016/j.jacl.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Jaywant S.V., Singh A.K., Prabhu M.S., Ranjan R. Statin therapy/lipid lowering therapy among Indian adults with first acute coronary event: the dyslipidemia residual and mixed abnormalities in spite of statin therapy (REMAINS) study. Indian Heart J. 2016 Sep–Oct;68(5):646–654. doi: 10.1016/j.ihj.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grover A., Rehan H.S., Gupta L.K., Yadav M. Correlation of compliance to statin therapy with lipid profile and serum HMGCoA reductase levels in dyslipidemic patients. Indian Heart J. 2017 Jan–Feb;69(1):6–10. doi: 10.1016/j.ihj.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joy T.R., Hegele R.A. Narrative review: statin-related myopathy. Ann Intern Med. 2009 Jun 16;150(12):858–868. doi: 10.7326/0003-4819-150-12-200906160-00009. [DOI] [PubMed] [Google Scholar]
- 23.Phillips P.S., Haas R.H., Bannykh S., Scripps Mercy Clinical Research Center Statin-associated myopathy with normal creatine kinase levels. Ann Intern Med. 2002;137:581–585. doi: 10.7326/0003-4819-137-7-200210010-00009. [DOI] [PubMed] [Google Scholar]
- 24.Agostini J.V., Tinetti M.E., Han L., McAvay G., Foody J.M., Concato J. Effects of statin use on muscle strength, cognition, and depressive symptoms in older adults. J Am Geriatr Soc. 2007;55:420–425. doi: 10.1111/j.1532-5415.2007.01071.x. [DOI] [PubMed] [Google Scholar]
- 25.Ucar M., Mjorndal T., Dahlqvist R. HMG-CoA reductase inhibitors and myotoxicity. Drug Saf. 2000;22:441–457. doi: 10.2165/00002018-200022060-00003. [DOI] [PubMed] [Google Scholar]
- 26.Graham D.J., Staffa J.A., Shatin D. Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. J Am Med Assoc. 2004;292:2585–2590. doi: 10.1001/jama.292.21.2585. [DOI] [PubMed] [Google Scholar]
- 27.Nichols G.A., Koro C.E. Does statin therapy initiation increase the risk for myopathy? An observational study of 32,225 diabetic and nondiabetic patients. Clin Therapeut. 2007;29:1761–1770. doi: 10.1016/j.clinthera.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Lach B., Provias J.P., Tarnopolsky M.A., Baker S.K. Statin-associated autoimmune myopathies: a pathophysiologic spectrum. Can J Neurol Sci. 2014 Sep;41(5):638–647. doi: 10.1017/cjn.2014.22. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell W.D., Ramsey L.B., Johnson S.G. Impact of pharmacogenetics on efficacy and safety of statin therapy for dyslipidemia. Pharmacotherapy. 2017 Sep;37(9):1172–1190. doi: 10.1002/phar.1981. [DOI] [PubMed] [Google Scholar]
- 30.Hermann M., Bogsrud M.P., Molden E. Exposure of atorvastatin is unchanged but lactone and acid metabolites are increased several-fold in patients with atorvastatin-induced myopathy. Clin Pharmacol Ther. 2006 Jun;79(6):532–539. doi: 10.1016/j.clpt.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Schirris T.J., Renkema G.H., Ritschel T. Statin-induced myopathy is associated with mitochondrial complex III inhibition. Cell Metabol. 2015 Sep 1;22(3):399–407. doi: 10.1016/j.cmet.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Wilke R.A., Ramsey L.B., Johnson S.G., Clinical Pharmacogenomics Implementation Consortium (CPIC) The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012 Jul;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsey L.B., Johnson S.G., Caudle K.E. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014 Oct;96(4):423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voora D., Shah S.H., Spasojevic I. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009 Oct 20;54(17):1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puccetti L., Ciani F., Auteri A. Genetic involvement in statins induced myopathy. Preliminary data from an observational case-control study. Atherosclerosis. 2010 Jul;211(1):28–29. doi: 10.1016/j.atherosclerosis.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 36.Carr D.F., O'Meara H., Jorgensen A.L. SLCO1B1 genetic variant associated with statin-induced myopathy: a proof of- concept study using the clinical practice research data link. Clin Pharmacol Ther. 2013;94:695–701. doi: 10.1038/clpt.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunham L.R., Lansberg P.J., Zhang L. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J. 2012;12:233–237. doi: 10.1038/tpj.2010.92. [DOI] [PubMed] [Google Scholar]
- 38.Fiegenbaum M., da Silveira F.R., Van der Sand C.R. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin Pharmacol Ther. 2005 Nov;78(5):551–558. doi: 10.1016/j.clpt.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Wilke R.A., Moore J.H., Burmester J.K. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenetics Genom. 2005 Jun;15(6):415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Kivistö K.T., Niemi M., Schaeffeler E. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenetics. 2004 Aug;14(8):523–525. doi: 10.1097/01.fpc.0000114762.78957.a5. [DOI] [PubMed] [Google Scholar]
- 41.Oh J., Ban M.R., Miskie B.A., Pollex R.L., Hegele R.A. Genetic determinants of statin intolerance. Lipids Health Dis. 2007 Mar 21;6:7. doi: 10.1186/1476-511X-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruaño G., Windemuth A., Wu A.H. Mechanisms of statin-induced myalgia assessed by physiognomic associations. Atherosclerosis. 2011 Oct;218(2):451–456. doi: 10.1016/j.atherosclerosis.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]