Abstract
Recent efforts to design personalized cancer immunotherapies use predicted neoantigens, but most neoantigen prediction strategies do not consider proximal (nearby) variants that alter the peptide sequence and may influence neoantigen binding. We evaluated somatic variants from 430 tumors to understand how proximal somatic and germline alterations change the neoantigenic peptide sequence and also impact neoantigen binding predictions. On average, 241 missense somatic variants were analyzed per sample. Of these somatic variants, 5% had one or more in-phase missense proximal variants. Without incorporating proximal variant correction (PVC) for MHC Class I neoantigen peptides, the overall False Discovery Rate (FDR) (incorrect neoantigens predicted) and the False Negative Rate (FNR) (strong-binding neoantigens missed) across peptides of lengths 8–11 were estimated as 0.069 (6.9%) and 0.026 (2.6%), respectively.
Over the past two decades, approaches to identify and screen for antigens, both self and non-self, have evolved rapidly1,2. This is due in part to advances in sequencing technologies, in the accuracy of algorithmic identification of somatic variants, and in computational modeling to predict the binding affinity of the resulting novel, tumor-specific peptides to major histocompatibility complex (MHC) molecules3. Thus, current immunogenomic approaches can identify somatic variants that give rise to tumor-specific mutant antigens or ‘neo’-antigens and evaluate their ability to bind to MHC Class I and Class II molecules3.
Typically, to evaluate strong-binding neoantigens from genomic sequencing data, the raw sequencing reads from tumor and normal DNA libraries are aligned to the human reference genome, and somatic variants are identified by comparison of tumor to normal read alignments. The resulting somatic variants of interest (SVOI) are then annotated to predict protein sequence changes and to infer possible neoantigenic peptides. Individual neoantigenic peptides are selected by sliding an amino acid window (usually 8–11-mers) across the variant position to consider each possible ‘register’. These peptides are assessed using various algorithms to predict binding affinity to MHC and determine the strongest binding epitopes. These predicted neoantigenic peptides are prioritized as we have previously described4. The cancer vaccine design process, from read alignment to variant calling and neoantigen prediction typically assumes the reference genome sequence surrounding each somatic variant is representative of the patient’s genome sequence.
However, any sequence variant proximal to a SVOI in the patient’s genome that differs from the human reference may alter the amino acid sequence of the resulting peptide (note, proximal is defined here as ‘situated close to’ or ‘nearby’, not the classic genetics meaning of ‘closer to the centromere’). Existing pipelines that are used for computational prediction of neoantigens from sequencing data, such as MuPeXI5 and pVAC-Seq4, do not explicitly incorporate patient-specific nearby germline or somatic variants (collectively referred to as ‘proximal variants’ hereafter) into the peptide sequence considered in neoantigen prediction. Some pipelines such as Vaxrank6 infer the coding sequence from assembly of tumor RNA reads, thus accounting for both somatic and germline variants implicitly, but this is largely dependent on the availability of RNA-Seq data. Failing to account for patient-specific nearby germline or somatic variants (i.e. proximal variants) could impact the efficacy of a vaccine, possibly resulting in immunization with incorrect peptides or failure to identify highly neoantigenic peptides.
To investigate these possibilities, we identified somatic and germline variants proximal to SVOIs in a data set of tumor sequencing studies representing different tissue sites and mutational loads. For this analysis, given that the upper-bound for the length of MHC-binding peptides (accounting for both Class I and Class II) is typically considered to be 30 amino acids7,8 we chose a nucleotide window of 89 bp upstream and downstream of each SVOI in which to identify relevant proximal variants (Methods). We limited our analysis to only include missense proximal variants and SVOIs. We then incorporated these proximal variants in the final peptide sequences (proximal variant correction; PVC) and re-evaluated the resulting peptide set using our neoantigen prediction pipeline (pVAC-Seq)4. Our results suggest that taking individual proximal variation into account can have a significant effect on the accuracy of neoantigen selection, resulting in a more personalized vaccine design.
Results
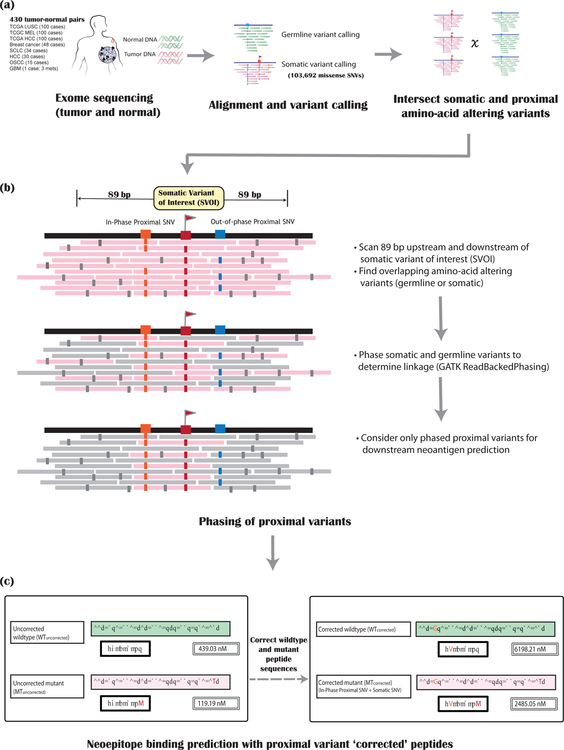
To determine how frequently proximal variants occur within the vicinity of an SVOI, we assessed 430 tumors with varying mutational loads identified from whole genome/exome sequence data of matched normal and tumor tissue (Figure 1,Methods). Specifically, data from 100 cases each of melanoma, hepatocellular carcinoma and lung squamous cell carcinoma were obtained from TCGA. We also evaluated data from 48 cases of HER2+ breast cancer, 34 cases of small cell lung cancer, 30 cases of hepatocellular carcinoma, 15 cases of oral squamous cell carcinoma, and one hypermutated glioblastoma (one primary and two metastatic samples) from in-house studies. After performing alignment and variant calling, we confirmed the linkage of SVOIs and proximal (somatic or germline) variants by phasing the variants using GATK9 [Figure 1b] (Methods). Then, the list of SVOIs from each of the samples was intersected with the respective lists of in-phase amino acid-altering proximal variants to assess their presence within the chosen nucleotide window.
Figure 1: Overview of the pipeline.
The steps required for incorporating and assessing the impact of proximal variants on neoantigen binding prediction are depicted as a flow diagram. There are three main steps. (a) Alignment and variant calling of matched tumor (pink) and normal (green) sequencing data. (b) Phasing of proximal somatic and germline variants: The pink bars represent the tumor sequence reads, with mismatches/sequencing errors shown in small gray rectangles. For a somatic variant of interest (SVOI; labeled with a red flag), we scan 89 bp on either side to assess for proximal germline or somatic SNVs (labeled with blue and orange boxes). These proximal variants are then phased together to determine linkage. Only proximal variants that are in phase (orange box) with the SVOI (red box) are considered for downstream neoantigen analysis. Other (out-of-phase) proximal variants (blue box) are ignored. (c) Neoantigen binding predictions are then assessed after performing proximal variant correction (PVC). The left panel shows the ‘uncorrected’ wildtype and mutant peptides along with their respective binding scores for a single SVOI example. The right panel shows PVC (‘corrected’) peptides and scores for this SVOI.
Missense variants overlap with missense proximal variants
Out of 430 tumor samples analyzed, 380 samples (88.3%) had at least one (range: 1 to 377) missense SVOI in phase with a proximal missense variant. Of a total of 103,673 missense variants identified in these tumors, there were 7,783 SVOIs (7.5%) with a proximal missense variant (somatic or germline) within 89 nucleotides on either side. 5,344 of these missense SVOIs (5.1%) were also in phase with their respective proximal variants. In most cases (93.8%), SVOIs had a single proximal germline or somatic variant in phase, but occasionally multiple (range: two to six) variants were proximal to the SVOI. An average of 241 missense somatic variants were analyzed per sample. Per patient, an average of 6.5% of SVOIs had a proximal missense variant, and 5% had one or more proximal missense variants in phase with the SVOI. On average, 62.2% of these proximal variants were germline missense variants and 37.7% were somatic missense variants. The majority (68.0%) of proximal somatic variants were contributed by Dinucleotide Polymorphisms (DNPs). Most variant callers (including those used for the harmonized analysis of TCGA data in the Genomic Data Commons) report DNPs as two separate SNVs. Excluding the DNPs, on average, 88.4% of the proximal variants were germline missense SNPs, and 11.6% were somatic missense SNVs. Supplementary Table 1 shows, for each sample, the percentage of SVOIs harboring any neighboring variants within the specified 89 bp window and the percentage of the total SVOIs that had any proximal variants in phase. It also shows the breakdown of numbers of somatic versus germline proximal variants for each sample, along with the numbers of variants contributed by DNPs.
Predicted binding affinity changes with PVC
To identify neoantigens capable of eliciting an effective anti-tumor T-cell response, it is critical to both determine the correct tumor-specific peptide sequence and assess its ability to bind MHC10. First, we sought to assess how accounting for proximal variants in the neoantigen peptide sequence may influence binding affinity to MHC. In order to evaluate this, we quantified the impact of missing or incorrectly selected strong-binding neoantigens when ignoring proximal variants. We compared binding affinity scores before and after PVC for each patient’s peptides against their respective MHC Class I alleles.
A typical Class I neoantigen binding evaluation and screening is carried out by sliding over shorter sub-peptide registers4. To evaluate strong-binding Class I neoantigens of lengths 8–11-mers, we ideally scan 7–10 amino acids on each side of the mutated amino acid resulting from the SVOI. Even if a proximal variant alters an amino acid in the full peptide window, it may not be included in every register we consider as a candidate neoantigen (Supplementary Figure 1).
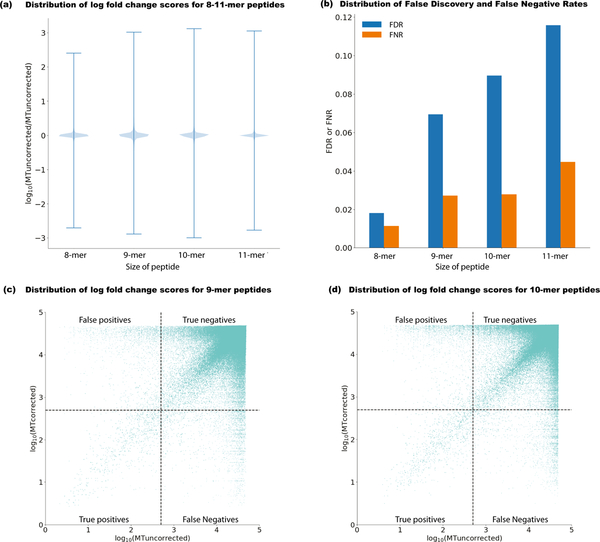
In some rare cases, a proximal variant may translate to the same amino acid sequence as the SVOI, or the SVOI and proximal variant both lead to amino acid changes if considered in isolation, but if they are in phase and considered together, they result in no change to the amino acid sequence. To take into account these cases and accurately assess the effect of amino-acid changes due to proximal variants on binding predictions, we only considered those registers that contained both the proximal variant and the SVOI amino acid changes, when translated together. Across 8–11-mers, on average 45.95% of all neoantigen peptide registers contained both. Figure 2 summarizes the effect of proximal variants on neoantigen binding affinity. Although the effect is less pronounced for 8-mers, the smallest length we examined, we see drastic changes in binding affinity due to PVC across all four peptide lengths (represented as log10 of mutant (MT) epitope fold change (MTuncorrected/MTcorrected), with ranges spanning from −3.0 to 3.1 for 8–11-mers (Figure 2a). Figures 2c–d show the distribution of log(MT fold change) scores for 9-mer and 10-mer peptides, respectively. For both peptide lengths, most weak binders stay within the same range before and after PVC but very few strong binders remain unchanged, after PVC. We chose 500 nM as the binding affinity cutoff for a potential binder, as most known T-cell epitopes have an affinity value of less than 500 nM11. For the binding prediction changes, we only considered a call as erroneous if PVC yielded at least a 10% change in predicted binding affinity.
Figure 2: Mischaracterization of neoantigens before proximal variant correction.
The effect of accounting for proximal variants in neoantigen selection is summarized in several ways (n = 380 biologically independent samples with at least one proximal variant). (a) Violin plot (distribution of all data in blue and whiskers indicating max/min values) showing the change in uncorrected neoantigen binding using the existing approach (MTuncorrected) versus PVC (MTcorrected), represented as log10 MT fold change (MTuncorrected / MTcorrected) across 8–11-mers for all variants in phase with the somatic variant of interest. (b) For 8–11-mer peptides, the False Negative Rate (FNR) (shown as orange bars) represents the number of instances when a truly strong-binding peptide was mistaken as a weak-binding peptide (MTuncorrected > 500 nM, and MT fold-change < 1.1 ). The False Discovery Rate (FDR) (shown in blue bars) represents the number of instances where a strong-binder before PVC (MTuncorrected < 500nM) is determined to have an incorrect peptide sequence as a result of a proximal variant. (c) Log10 scaled comparison of corrected versus uncorrected binding scores for 9-mer peptides considering patient-specific MHC Class I alleles. Dotted lines demarcate the binding affinity threshold of 500 nM. (d) Log10 scaled comparison of corrected versus uncorrected binding scores for 10-mer peptides considering patient-specific MHC Class I alleles.
Impact of PVC on False Discovery and False Negative Rates
In addition to the effect a proximal amino acid substitution may have on a neoantigen’s binding potential, it is also important to consider whether the peptide sequence of the selected neoantigen is correct and representative of the sequence in the tumor. Failure to do so may affect the immunogenic potential of the neoantigen being selected, as the uncorrected neoantigen will not produce tumor-specific T-cells, even if it binds well and is presented by the MHC.
To determine how many neoantigens were being erroneously predicted, and the effect that mischaracterization of neoantigens due to proximal variants would have on candidate selection, we calculated the False Negative Rate (FNR) and False Discovery Rate (FDR) after applying PVC. The FNR and FDR represent probabilities of potential MHC binders (binding affinity < 500 nM) being discarded (false negatives) and of erroneous peptides being mistaken for potential binders (false positives), respectively.
An average of 9 SVOI and 10 neoantigenic peptides were mischaracterized per case. As a consequence, 1,165 potential binders (MTcorrected < 500 nM) were erroneously rejected, and 3,305 peptides which were strong binders before PVC were misidentified across all 430 patients investigated here. Overall, FNR and FDR across lengths 8–11 were 0.026 and 0.069, respectively (Figure 2b).
As a representative example, Supplementary Figure 2 illustrates data from one of the TCGA melanoma samples with a heterozygous missense SNV in the reverse strand gene MARCH10 that overlaps an in-phase heterozygous germline single nucleotide polymorphism (SNP), 21 nucleotides upstream. When translated, this germline SNP results in S357F (NP_001275708.1:p.Phe357Ser) alteration that is 7 amino acids downstream to the missense somatic variant F350S (NP_001275708.1:p.Ser350Phe). This variant directly affects the final neoantigen sequence for a peptide of any length (> 8-mer). To evaluate the effect of this germline SNP on the binding affinity of the neoantigen peptide, we calculated the binding affinity of the uncorrected versus the PVC neoantigenic peptides. The binding affinity of the best register for a 10-mer peptide using the uncorrected approach (MTuncorrected = 55.44 nM) is within the range for a good binder (< 500 nM). However, after including this patient’s proximal germline variant, the binding affinity for the same register decreases almost 70-fold (MTcorrected = 3766.72 nM), thus predicting a very weak binder. Using the uncorrected analysis approach, one might have selected this neoantigenic peptide for a vaccine but after PVC, the candidate peptide is unsuitable. This result illustrates the importance of using the individual variation of the germline genome while selecting and designing neoantigens for personalized immunotherapy.
Discussion
There are some caveats/limitations of our approach. Firstly, the analysis was restricted only to single nucleotide changes (i.e. missense somatic SNVs that are near another germline or somatic SNV), and did not seek to evaluate whether other, potentially relevant types of variants were found nearby. These include insertions and deletions (both somatic and germline)12 and different types of structural variants that often have a more significant impact on peptide sequences but also are rarer than SNVs. Phasing of indels and structural variants is also not currently handled by software such as GATK’s ReadBackedPhasing. Secondly, our analysis ‘window’ (89 bp) was defined in genomic coordinates. It is substantially more complicated to consider this window size in the context of transcriptome coordinates, since intronic coordinates must be ignored when scanning upstream and downstream. This is further complicated in genes with alternative transcripts and hence multiple introns and exons to consider. Our ability to determine phase for variants separated by an intron would be limited in WGS or exome data (although could be evaluated in RNA-seq data with sufficient read lengths). Lastly, for this study, we only considered neoantigen binding predictions to MHC Class I molecules. MHC Class II peptides are much longer due to an open binding groove and hence, the subsequent impact of proximal variants on the peptide sequence would be even more pronounced. Due to these limitations, our results are likely an underestimation of the impact of PVC.
Moreover, even with seemingly small false discovery and false negative rates, the importance of accounting for the effect of proximal variants is clear when we consider clinical vaccine design scenarios. For example, 10 or fewer peptides are usually selected for the final vaccine from a larger number of initial candidates. Given this scenario, we calculated the probability of choosing at least 1 weak binder or of omitting 1 strong binder in the final vaccine, without PVC. For the first probability, we calculated 1 - (1-FDR)10 = 0.513 and for the second, we calculated 1 - (1-FNR)10 = 0.228. The probability that at least one of these errors occurs for each patient evaluated, is 1 - (1-FDR)10*(1-FNR)10 = 0.624. Thus, for neoantigen identification in 100 patients, we can expect that approximately 51 patients would receive a suboptimal vaccine specifically due to receiving a neoantigen with an incorrect peptide sequence, 23 would receive a suboptimal vaccine specifically due to missing a strong-binding neoantigen, and 62 would receive a suboptimal vaccine due to at least one of these causes.
Design of personalized cancer vaccines is complex, time consuming, and expensive. Previous work has shown that only about 16–43% of the predicted neoantigenic peptides included in a vaccine formulation yield CD8+ T-cell response13–16. Our study demonstrates the importance of ensuring the selected neoantigens correctly represent the individual’s genome and therefore maximize the likelihood of eliciting an immune response. PVC based on the patient’s genome can eliminate errors during neoantigen candidate selection, potentially increasing the efficacy of personalized vaccines. Further studies may also demonstrate the importance of considering proximal variants when using neoantigen load to predict response to checkpoint blockade inhibition therapies.
Online Methods
Sequence data alignment and variant calling
To investigate the prevalence of proximal variants (germline SNPs or somatic variants), we analyzed publicly available sequencing data from the TCGA as well as datasets generated in-house, altogether representing seven different tissue sites. These data sets were chosen to adequately represent low, medium and high mutational burden tumors.
Analysis of in-house whole genome/exome sequencing datasets was performed as previously described4,17,18. Briefly, raw sequencing reads from both the tumor and normal were aligned to the human reference genome sequence (either GRCh37 or GRCh38) using BWA19, then merged and deduplicated using Picard (see URLs). A combination of three or four different variant callers was used to identify somatic variants by comparison of tumor and normal variant calls: Samtools20, Sniper21, Strelka22, and VarScan23,24. These variants were filtered as previously described25,26 and then manually reviewed using IGV per the standard operating procedures27 to obtain a list of high confidence variant calls. On average, 80% of the filtered variants passed manual review. Germline variant analysis of the normal samples was performed using Samtools.
For the TCGA data, aligned tumor and normal BAMs from BWA (version 0.7.12-r1039) as well as somatic variant calls from VarScan2 (in VCF format) were downloaded from the Genomic Data Commons (GDC). We restricted our analysis to only consider ‘PASS’ variants in these VCFs that are higher confidence than the raw set. Since TCGA does not provide germline variants, we used GATK’s HaplotypeCaller to perform germline variant calling using default parameters. These calls were refined using VariantRecalibrator in accordance with GATK Best Practices28.
For this study, we restricted the variant calls to only include missense SNVs, in both- TCGA as well as in-house datasets.
Phasing of variants to assess linkage
Somatic and germline missense variant calls from each sample were combined using GATK’s CombineVariants, and the variants were subsequently phased using GATK’s ReadBackedPhasing algorithm.
In silico HLA-typing
OptiType29 was used to perform in silico HLA typing for the in-house samples. For the datasets downloaded from TCGA, existing in silico HLA typing information was obtained from The Cancer Immunome Atlas (TCIA30) database.
Choosing an appropriate window for neoantigen analysis
Due to the absence of patient-specific HLA Class II typing information, we limited our neoantigen binding prediction analysis to MHC Class I, though we believe that the Class II peptides are also important in contributing to immunogenicity. Hence, our nucleotide window was chosen such that it encompasses both Class I and Class II MHC peptide lengths, to demonstrate the prevalence of proximal variants within that genomic region. Most strong-binding Class I MHC peptides are around 8–11 amino acids in length. There is no length restriction on Class II MHC peptides due to an open binding groove, and longer peptide lengths are much more common, typically 13–25-mers31 but peptides as long as 30-mer have been reported7,8. The majority (99.2%) of human linear T-cell epitopes with MHC class II restriction currently reported in IEDB32are 8–30-mers. To identify the best binding 30-mer around a missense variant of interest, one would ideally scan 29 amino acids upstream and downstream of the mutant (MT) amino acid, hence a window of 59 amino acids. At the nucleotide level, this corresponds to 87 nucleotides. Given that the frame of the missense mutation is not always known, we allow for 2 extra bases leading to a window size of 89 nucleotides on each side of the SVOI.
The appropriate nucleotide window for any peptide length can be calculated using this formula: ((peptide length −1)*3)+2.
Corrected neoantigen binding prediction using pVACtools
For each sample, the phased variant calls as well as the somatic variant calls were annotated using Variant Effect Predictor (VEP33), specifically using the Downstream plugin as well as the custom Wildtype plugin, available via pVACtools (see URLs). To evaluate the effect of relevant nearby variants on neoantigen identification, we re-assessed the binding affinities of the neoantigens with the corrected mutant peptide sequence (Figure 1c), using NetMHCv4.034,35 via an updated version of the pVACtools software. This version takes as input the VEP-annotated phased VCF file of somatic and germline variants, in addition to the existing VEP-annotated somatic VCF.
Calculating False Discovery and False Negative Rates
To calculate FNR and FDR, we first determined the number of weak binders before PVC that were falsely omitted (‘false negatives’ or ‘FN’), as well as the number of peptides that were identified as strong binders before PVC but whose sequence was altered due to a proximal variant, and thus were incorrectly considered during neoantigen selection (‘false positives’ or ‘FP’). We also calculated the number of peptides which were strong before correction and remained unaltered by proximal variants (‘true positives’ or ‘TP’).
The False Negative Rate (FNR) is then defined as the number of false negatives divided by the number of false negatives plus true positives. FNR represents the chance that a strong binder was mischaracterized as a weak binder before PVC and thus was falsely omitted. The False Discovery Rate (FDR) is defined as the number of false positives divided by the number of all positive calls, including both true positives and false positives. FDR represents the chance that a candidate peptide was identified as a strong binder before PVC but whose sequence was altered due to a proximal variant, and thus was incorrectly considered during neoantigen selection (‘false positives’ or ‘FP’), normalized using the number of peptides which were strong before correction and remained unaltered by proximal variants (‘true positives’ or ‘TP’).
Code availability
The proximal variant analysis code has now been added to the proximal_variants branch of the pVACtools GitHub repository (https://github.com/griffithlab/pVACtools/tree/proximal_variants). We have also packaged this branch and uploaded the package as an alpha release to TestPyPi. The alpha release can be installed by running `pip install -f https://test.pypi.org/project/pvactools/1.0.8/ pvactools==1.0.8` on the command line. The feature will be released with the main pVACtools package as part of the next software release cycle (version 1.1.0).
Data availability
Several of the in-house sequencing datasets used in the study have been previously published and deposited in various databases. All sequence data for the HER2+ breast cancer samples can be accessed via the Database of Genotypes and Phenotypes (dbGAP; study accession: phs001291)36. Data for oral squamous cell carcinoma project and hepatocellular carcinoma samples are part of other manuscripts currently in preparation, and can be accessed under dbGAP study accession phs001623 and phs001106, respectively. Results for the glioblastoma case37 and small cell lung cancer26 have been published and can be accessed under dbGAP study accessions phs001663 and phs001049, respectively. TCGA data can be accessed under dbGaP study accession phs000178.
Supplementary Material
Acknowledgements
We are grateful to the patients and their families without whom this study would not be possible. We would like to thank G Dunn for early access to raw data for the published glioblastoma hypermutator case included in our analysis. We would also like to thank R Schreiber and B Carreno for the initial discussions that inspired the study, and for their expertise and guidance during the study. S Swamidass was supported by the NIH National Library of Medicine (R01LM012222 and R01LM012482). O Griffith was supported by the NIH NCI (K22CA188163 and U01CA209936). M Griffith was supported by the National Institutes of Health (NIH), National Human Genome Research Institute (NHGRI; award number R00HG007940) and the NIH National Cancer Institute (NCI) (U01CA209936).
Footnotes
URLs
pVACtools: http://pvactools.org/
Github repository for proximal variant analysis code: https://github.com/griffithlab/pVACtools/tree/proximal_variants
Competing financial interests
The authors declare that they have no competing interests.
Editorial summary:
Including patient-specific information about nearby somatic and germline alterations improves the accuracy of neoantigen prediction, potentially impacting cancer vaccine design.
References
- 1.Hackl H, Charoentong P, Finotello F & Trajanoski Z Computational genomics tools for dissecting tumour–immune cell interactions. Nat. Rev. Genet 17, 441–458 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Liu XS, Shirley Liu X & Mardis ER Applications of Immunogenomics to Cancer. Cell 168, 600–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hundal J et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 8, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjerregaard A-M, Nielsen M, Hadrup SR, Szallasi Z & Eklund AC MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol. Immunother. (2017). doi: 10.1007/s00262-017-2001-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinsteyn A, Hodes I, Kodysh J & Hammerbacher J Vaxrank: A Computational Tool For Designing Personalized Cancer Vaccines. (2017). doi: 10.1101/142919 [DOI] [Google Scholar]
- 7.Meydan C, Otu HH & Sezerman OU Prediction of peptides binding to MHC class I and II alleles by temporal motif mining. BMC Bioinformatics 14 Suppl 2, S13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rammensee HG, Friede T & Stevanoviíc S MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178–228 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Poplin R et al. Scaling accurate genetic variant discovery to tens of thousands of samples. (2017). doi: 10.1101/201178 [DOI] [Google Scholar]
- 10.Łuksza M et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sette A et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol 153, 5586–5592 (1994). [PubMed] [Google Scholar]
- 12.Turajlic S et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol (2017). doi: 10.1016/S1470-2045(17)30516-8 [DOI] [PubMed] [Google Scholar]
- 13.Carreno BM et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahin U et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linette GP & Carreno BM Neoantigen Vaccines Pass the Immunogenicity Test. Trends Mol. Med 23, 869–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 17.Griffith M et al. Genome Modeling System: A Knowledge Management Platform for Genomics. PLoS Comput. Biol 11, e1004274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith M et al. Optimizing cancer genome sequencing and analysis. Cell Syst 1, 210–223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H & Durbin R Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson DE et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Koboldt DC et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith M et al. Comprehensive genomic analysis reveals FLT3 activation and a therapeutic strategy for a patient with relapsed adult B-lymphoblastic leukemia. Exp. Hematol 44, 603–613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner AH et al. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat. Commun 9, 3787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnell EK et al. Standard operating procedure for somatic variant refinement of tumor sequencing data. (2018). doi: 10.1101/266262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Auwera GA et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szolek A et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charoentong P et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Chicz RM et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 358, 764–768 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Vita R et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43, D405–D412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaren W et al. The Ensembl Variant Effect Predictor. (2016). doi: 10.1101/042374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreatta M & Nielsen M Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen M et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12, 1007–1017 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesurf R et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann. Oncol 28, 1070–1077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johanns TM et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov 6, 1230–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Several of the in-house sequencing datasets used in the study have been previously published and deposited in various databases. All sequence data for the HER2+ breast cancer samples can be accessed via the Database of Genotypes and Phenotypes (dbGAP; study accession: phs001291)36. Data for oral squamous cell carcinoma project and hepatocellular carcinoma samples are part of other manuscripts currently in preparation, and can be accessed under dbGAP study accession phs001623 and phs001106, respectively. Results for the glioblastoma case37 and small cell lung cancer26 have been published and can be accessed under dbGAP study accessions phs001663 and phs001049, respectively. TCGA data can be accessed under dbGaP study accession phs000178.