Letter to the Editor
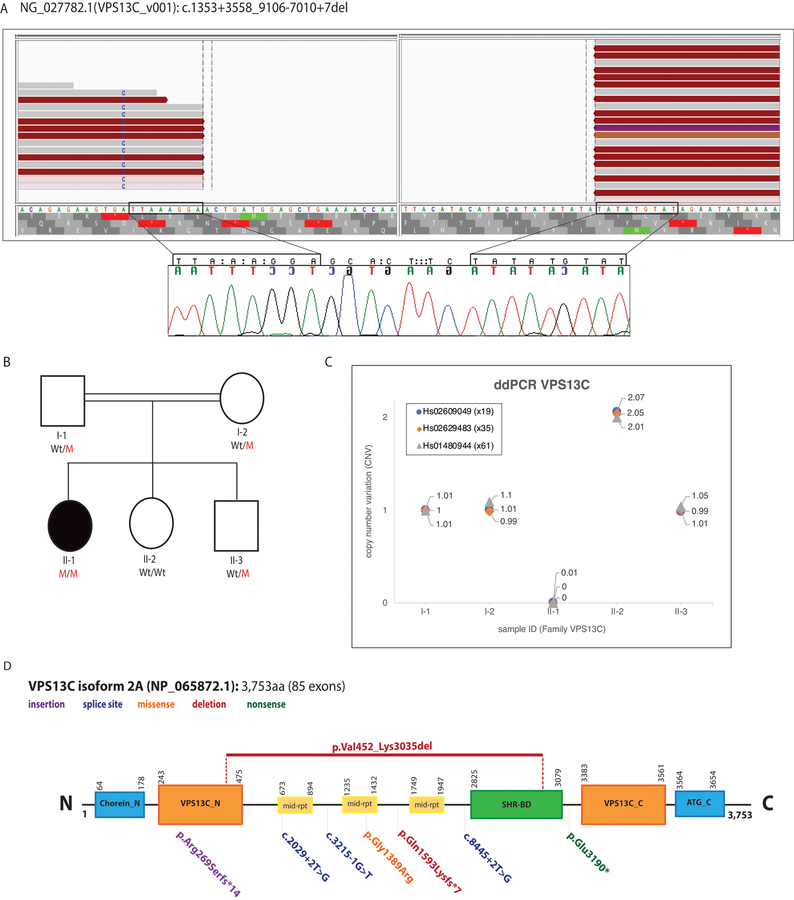
We report the clinical symptoms and the causal genetic defect of an isolated patient with parkinsonism and sensorimotor polyneuropathy that was born to healthy parents (Supplementary material, Fig. 1A). Whole genome sequencing (WGS), performed and processed as described in the Supplementary Material, led us to the identification of novel and rare genetic variations, including 606 non-synonymous, 69 splice-site, 24 stop-gained, stop- and start-lost, and 111 frame-shift mutations, as well as 63 different genomic deletions/duplications. Because recessive alleles may be present in normal population, all homozygous and compound-heterozygous variants were explored and included in the analysis, but only 11 homozygous missense mutations were found to be present in our patient (Supplementary Table 2). Among the identified deletions/duplications, a large homozygous genomic deletion, comprising 50 coding exons (from exon 17 to exon 66) of the Vacuolar Sorting Associated Protein 13 (VPS13C) gene, which was previously identified mutated in families with asymmetric akinetic rigidity and cognitive impairment (Supplementary Table 3),1, 2 was found in the patient’s genome. This large deletion consisted of c.1353+3558_9106–7010del and resulted in p.Val452_Lys3035del, (NM_020821/NM_001018088). The deletion breakpoints (chr15: 62,189,609–62,289,207) and its segregation with disease status were later confirmed through Sanger sequencing (Figs. 1A, 1B), which also revealed a 7-bp insertion (GCACTTC) (Fig. 1A). Three VPS13C exons (exons 19, 35, 61) expanding the deletion breakpoints were also subject to copy number variation (CNV) quantification by performing droplet digital PCR (ddPCR; QX100 system, Bio-rad, USA) as previously described.3 Only the affected member showed no copies of the implicated exons, further confirming this large VPS13C deletion as the causal genetic defect in our reported patient(Fig. 1C).
Fig 1.
(A) WGS reads (Top) and Sanger chromatograms (Bottom) illustrating the large homozygous VPS13C deletion (Blank area, no reads) identified in an isolated patient with parkinsonism (https://databases.lovd.nl/shared/variants/0000368891). The WGS reads were visualized using the IGV tool. The Sanger chromatograms show the evidence of a 7-bp insertion between the deletion breakpoints (nucleotides between rectangles). The breakpoints of the deletion are highlighted with rectangles. (B) Pedigree of the family with parkinsonism and sensorimotor polyneuropathy. Wt/M indicates heterozygous carrier for the VPS13C large deletion; M/M indicates homozygous carrier; and Wt/Wt indicates non-carrier. Affected patient is represented with a black circle. (C) CNV plots of VPS13C exons 19, 35, and 61 obtained through ddPCR (QX100 system, Bio-rad) and corresponding taqman assays (Applied Biosystems) are shown. Homozygous Wt alleles are represented with a CNV score of 2 or close to 2, meaning both alleles carry a copy of the exon under investigation; heterozygous mutant alleles are represented with a CNV score of 1 or close to 1, meaning only one allele carry a copy of the exon while the exon is absent in the other allele; homozygous mutant alleles are represented with a CNV score of 0, meaning the exon under investigation is absent in both alleles. Only subject II-1 is homozygous carrier for the identified VPS13C deletion. (D): VPS13C protein (NP_065872.1) structure located at chromosome 15q22.2 and predicted by SMART (http://smart.embl-heidelberg.de). Functional domains along with reported VPS13C mutations are shown. The VPS13C mutation identified in this study is shown at the top of the protein. Chorein_N: N terminal region of Chorein, VPS13C_N: N terminal domain, mid-rpt: VPS13_mid_repeats, SHR-BD: SHR binding domain, VPS13C_C: C terminal domain, ATG_C: Autophagy related protein C terminal domain.
Subsequently, the proximal and distal sequences (~2kb) to the breakpoints were examined to determine their homology (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the presence of repeated sequences (http://www.roadmapepigenomics.org/), as these might help to elucidate its generating mechanisms.4 Short homologies and two long interspersed elements (LINE) of 6kb (L1PA3: chr15: 62,188,436–62,194,473) and 1.3kb (L2a: chr15: 62,289,333–62,290,611), respectively, were found to be located nearby the breakpoints, suggesting that this deletion was originated through transposable element insertions (TEI).5 Both proximal and distal regions showed weak transcription and were found to be enriched for the following chromatin marks: H3K4me3, H3K4me1, HeK9me3, H3K27me3.
Because the performed electrophysiological studies revealed a demyelinating sensory-motor polyneuropathy in favor of Charcot-Marie-Tooth (CMT) disease, we examined all genetic variation identified in the known CMT genes but only found previously reported benign variants (Supplementary Material and Table 4), suggesting that the identified VPS13C deletion might contribute to the CMT phenotype seen in our patient, which might help with the diagnosis of prospective VPS13C patients.
In summary, we report the identification and characterization of a large L1 retrotransposition-mediated VPS13C deletion in a patient with parkinsonism, further confirming the role of the VPS13C in the pathogenesis of PD. Besides the strong effect of the identified mutation on the protein, our patient showed normal cognitive status and a milder clinical picture than that observed in previously reported patients. Lastly, the hypothesis that the identified VPS13C deletion might be responsible for the CMT phenotype seen in our patient needs to be further explored as it also might be caused by mutations in an unknown CMT gene.
Supplementary Material
Acknowledgments
We thank the patient, the relatives, and other participants for their cooperation in this study.
Funding source:
This work was in part supported by the Shahid Beheshti University of Medical Sciences, the American Parkinson Disease Association (APDA; CPR), and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (R01NS079388; CPR).
Footnotes
Financial disclosure/conflict of interest:
The authors declare they have no conflict of interest.
References
- 1.Schormair B, Kemlink D, Mollenhauer B, et al. Diagnostic exome sequencing in early-onset Parkinson’s disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson’s disease. Clin Genet 2018;93(3):603–612. [DOI] [PubMed] [Google Scholar]
- 2.Lesage S, Drouet V, Majounie E, et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am J Hum Genet 2016;98(3):500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez E, Darvish H, Mesias R, et al. Identification of a Large DNAJB2 Deletion in a Family with Spinal Muscular Atrophy and Parkinsonism. Hum Mutat 2016;37(11):1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaer K, Speek M. Retroelements in human disease. Gene 2013;518(2):231–241. [DOI] [PubMed] [Google Scholar]
- 5.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 2005;1(6):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.