Abstract
In this paper, a fungal strain NT-1, which was screened from orchard soil co-contaminated with acidification and copper, was characterized by morphological, physiological, biochemical and molecular biological techniques. Through application of different concentrations of Cu(II), heavy metal stress was exerted on the strain, and the effects on mycelial biomass, colony diameter and mycelial morphology of NT-1 were studied. Additionally, the impact of different pH values, temperatures, initial Cu(II) concentrations and inoculum doses on the adsorption of Cu(II) by NT-1 was explored. The mechanism of Cu(II) adsorption by NT-1 was studied by adsorption thermodynamics and kinetics, as well as Fourier transform infrared (FTIR) spectroscopy. The results showed that NT-1 belonged to the genus Gibberella, which exhibited high tolerance to both acidic conditions and Cu(II) contamination in the environment. High concentrations of copper stress inhibited the growth of NT-1 to various degrees, leading to the decreases in mycelial biomass and colony diameter, as well as changes in morphology. Temperature, pH and other factors were shown to significantly affect the efficiency of NT-1 with respect to the removal extent of copper from solution. Under optimal conditions (initial copper concentration: 200 mg L−1, temperature: 28°C, pH: 5.0 and inoculum dose: 10%), the maximum copper removal percentage from solution through culture of strain NT-1 within 5 d reached up to 45.5%. The biosorption of Cu(II) by NT-1 conformed to quasi-second order kinetics and Langmuir isothermal adsorption model and was confirmed to be a monolayer adsorption process dominated by surface adsorption. The binding of NT-1 to Cu(II) was mainly achieved by forming polydentate complexes with carboxylate and amide group through covalent interactions and forming Cu-nitrogen-containing heterocyclic complexes via Cu(II)-π interaction. The results of this study provide a new fungal resource and key parameters influencing growth and copper removal capacity of the strain for developing an effective bioremediation strategy for copper contaminated acidic orchard soils.
Keywords: copper resistant fungus, acidic orchard soil, bioremediation, biosorption, Cu(II) removal extent
1. INTRODUCTION
As an essential trace element in organisms, copper (Cu) participates in the synthesis of various types of enzymes. However, excessive copper is highly toxic to organisms and inhibits cell growth, metabolism and other processes. Importantly, the application of a large amount of Bordeaux mixture and other copper-containing fungicides results in a substantial accumulation of copper in the orchard soil (Pietrzak and McPhail 2004). In addition, the improper application of fertilizers and pesticides and the orchard soil acidification caused by long-term planting of fruit trees will further enhance the bioavailability of copper in the soil, which increases the risk for fruit trees to absorb the accumulated copper, which results in reduced fruit yield and quality, thus threatening food security (Gimeno-García et al. 1996). Therefore, it is urgent to develop a green sustainable remediation technique that is economical, efficient and environment-friendly for the acidified orchard soil contaminated with copper.
Currently, among numerous remediation techniques, bioremediation has drawn wide attention due to its low cost, excellent performance, environmental friendliness and other advantages when compared with the traditional physical and chemical remediation techniques (Luo and Tu 2018). Bioremediation includes techniques such as phytoremediation, microbial remediation, as well as their joint remediation. Microorganisms can not only degrade the organic pollutants in the environment, but also immobilize, mobilize or transform the heavy metals in the soil to change their environmental chemical behavior in the soil, thus achieving the purpose of bioremediation. Due to the non-degradability of heavy metal pollutants, the principle of the microbial remediation on heavy-metal-contaminated soil mainly includes biosorption, bio-enrichment and bioconversion to less toxic phases. Microorganisms can either use the negative charges on the cell surface to immobilize heavy metal ions through electrostatic adsorption or complexation, store the absorbed toxic metals in different parts of cells, or bind them on the extracellular matrix (Park and Chon 2016). Furthermore, these microbes can precipitate or chelate these ions on biopolymers, or enrich the heavy metals by interacting with the specific heavy-metal-binding macromolecules, including metal-binding proteins (Gutierrez et al. 2012; Wang et al. 2018). Additional biotransformation can be achieved, which can include redox reactions, methylation and demethylation, as well as the dissolution of heavy metals, organic complexation-coordination degradation, and other processes.
To date, microbial bioremediation of heavy-metal contaminated soils is the most promising sustainable technique, but there are still some limitations in the practical applications, such as how to isolate, screen and acclimatize the microbial strains with high tolerance to the combined pollution of soil acidification and heavy metal-contamination. Moreover, it is equally important to improve the activity, lifespan and environmental safety of these functional microorganisms in soils, while understanding how to promote microbial metabolic activity sensitive to changes in environmental conditions by optimizing nutrients, temperature, humidity, and other key factors.
In this study, a fungal strain NT-1, which was highly resistant to both soil acidification and copper contamination, was screened from an orchard soil with long-term apple cultivation and Bordeaux mixture application history, and this strain was characterized by physiological, biochemical and molecular biological techniques. The response characteristics of strain NT-1 to Cu(II) stress were investigated, and the factors affecting the Cu(II) adsorption by NT-1 were explored, with the mechanism of Cu(II) adsorption by NT-1 studied preliminarily. The aim of this paper was to provide a novel and effective fungal resource and key parameters influencing growth and copper removal by this fungus, which will be beneficial to the research and development of bioremediation technology and mechanism of copper polluted acidic orchard soils by indigenous fungus.
2. MATERIALS AND METHODS
2.1. Screening and Isolation of Copper-resistant Strain
Soil samples for microbial strain screening were collected from the surface soil (0–20 cm) of an orchard with a long-term apple cultivation history in Yantai, Shandong Peninsula. Isolation of copper resistant fungi was set up using gradient dilution culture. Briefly, ten grams of orchard soil was suspended in a flask containing 90 mL of sterile water, subsequently incubated at 28°C on a rotary shaker (150 rpm) for 30 min. A small aliquot was then transferred to a fresh tube for progressive dilution to 10−7. Finally, 100 μL of aliquot of suspension was coated on Luria-Bertani plate containing 100 mg L−1 CuSO4·5H2O (Analytical grade, Sinopharm Chemical Reagent, China). Plates were incubated at 37 °C for 24~48 h. Five colonies with different morphology were isolated and purified, named NT-1 ~ NT-5, respectively. NT-1 is the only fungal strain among the five isolates.
2.2. Morphology and Molecular Identification of Copper Resistant Strains
To allow morphological and molecular characterization of the copper resistant fungus, strain NT-1 was inoculated on potato dextrose agar (PDA) plates and cultivated in a thermostatic incubator at 28°C for 1 week. The growth characteristics of fungal colonies were observed, and the morphology and the mode of mycelial growth and conidia production were examined under microscope using cover-slip culture techniques. The genomic DNA of strain NT-1 was extracted with FastDNA Kit and the conserved sequences in the fungal ribosomal RNA gene were amplified by polymerase chain reaction (PCR) using universal fungal primers ITS1 and ITS4 (White et al. 1990). The amplified product was sent to Shanghai Majorbio Bio-Pharm Technology Co., Ltd. for sequencing. The Basic Local Alignment Search Tool (BLAST) searches in the National Center for Biotechnology Information (NCBI) database were carried out for the obtained sequence and the existing ITS rRNA sequence, and the phylogenetic tree was constructed by MEGA 6.0 software using the neighbor-joining method model.
2.3. Growth Response of Copper-resistant Strain NT-1 to Different Concentrations of Copper Stress
A wire loop of NT-1 mycelium was picked and inoculated on PDA plate with Cu(II) concentrations of 100, 200, 300, 400, 500 and 600 mg L−1, respectively. These concentrations were selected to cover the range of potential soil pore water concentrations that may be encountered in the environment at contaminated or stressed sites. The incubation was carried out in a thermostatic incubator at 28°C for 5 days, after which the growth of mycelium on solid plates was observed, and the colony diameter was measured and recorded.
A wire loop of NT-1 mycelium was also inoculated into the potato dextrose broth with Cu(II) concentrations of 100, 200, 300, 400, 500 and 600 mg L−1, respectively. The incubation was carried out in a thermostatic shaker at a temperature of 28°C and a speed of 150 rpm for 5 days. To monitor strain growth in a liquid medium, fungal culture samples were centrifuged at 5000 rpm for 10 min and dried to constant weight at 80°C, and then recorded as dry mycelial biomass. The mycelial morphologies of the strains in solution at Cu(II) concentrations of 0, 200 and 400 mg L−1 were observed with a scanning electron microscope (Hitachi S-4800 FE-SEM, Hitachi, Japan). SEM samples were prepared according to Ozturk et al. (2009) with appropriate modifications. Briefly, mycelial samples were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) at 4°C for 12 h. After two washes with phosphate buffer, mycelial samples were dehydrated in a graded series of ethanol concentrations from 30% to 100% (v/v). Samples were freeze-dried for 24 h and the dried cells were coated with a thin layer of gold before examined with a scanning electron microscope.
2.4. Effect of Different Factors on the Cu(II) Biosorption by Strain NT-1
In order to investigate the impact of different factors on the concentration of Cu(II) adsorbed by NT-1, 50 mL of potato dextrose broth with 200 mg L−1 Cu(II) was inoculated with 5% (v/v) NT-1 seed culture at logarithmic growth phase, and cultured in a thermostatic shaking incubator at pH 5.0, 28°C, and a speed of 180 r min−1 for 5 d. To test the effect of initial Cu(II) concentration, broth with Cu(II) concentrations of 100, 200, 300, 400, 500 and 600 mg L−1 were also prepared and used for culture. To test the effect of pH, cultures were grown on media with 200 mg L−1 Cu(II) adjusted with 1 mol L−1 HCl(aq) and NaOH(aq) to pH values of 3.0, 4.0, 5.0, 6.0, 7.0, and 8.0; representative of encountered pH values in agronomic soils. The effect of soil temperature was studied by incubating cultures with 200 mg L−1 Cu(II) at 20°C, 28°C and 37°C, while effect of inoculum dose was studied by inoculating 200 mg L−1 Cu(II) media with 2.5, 5.0, and 10.0 mL of NT-1 seed culture.
All of the above experiments were repeated in triplicate. After the cultivation, the fungal solution was centrifuged at 4000 rpm for 15 min and the supernatant was filtered through a 0.45 μm membrane and diluted, with the Cu(II) content in the solution was determined with an atomic absorption spectrophotometer (TAS-990, Beijing Persee Instrument Co. Ltd.). The Cu(II) adsorption removal extent Q (%) (Eq. 1) and the Cu(II) adsorption capacity (qe) (Eq. 2) of the strain were calculated by the following equations,
| (1) |
| (2) |
where C0 and Ce are the initial concentration and final concentration of Cu(II) in the culture medium (mg L−1), qe is the adsorption capacity of adsorbent (mg g−1) at the adsorption equilibrium, V is the broth volume (L), and m is the mass of the adsorbent (g).
2.5. Cu(II) Adsorption Characteristics of the Strain NT-1
Under the conditions of pH 5.0, 25°C, a concentration of 2.0 g L−1 fungal hyphae were agitated at 180 rpm for 24 h, the isothermal adsorption process of the strain NT-1 at different Cu(II) concentrations (50–600 mg L−1) was studied. A 0.01 mol L−1 NaNO3 solution was used as a background solution for different Cu(II) concentrations, and 1 mol L−1 HCl and NaOH solutions were used to adjust the pH. Langmuir and Freundlich adsorption isotherm models were used to simulate the Cu(II) adsorption by NT-1. The Langmuir model is suitable for monolayer adsorption and assumes that the adsorption occurs at specific sites of the adsorbent (Eq. 3), while the Freundlich model is based on the non-uniform adsorption of adsorbent surface and assumes that the adsorption heat decreases exponentially with increasing coverage (Eq. 4), and they were expressed as follows,
| (3) |
| (4) |
where qe is the equilibrium adsorption capacity (mg g−1), Ce is the concentration of Cu(II) in the solution at equilibrium (mg L−1), KL is the Langmuir equilibrium parameter (L mg−1) and is related with the affinity at the binding site, and KF and n are Freundlich constants, representing the adsorption capacity and adsorption strength, respectively.
At an initial Cu(II) concentration of 200 mg L−1, the same above adsorption conditions were maintained and changes in Cu(II) concentrations in solution over time were analyzed, and thus the Cu(II) adsorption by the strain NT-1 at different adsorption times were studied. The kinetic processes for adsorption were fitted to quasi-first-order (Eq. 5) and quasi-second-order kinetic (Eq. 6) models as follows,
| (5) |
| (6) |
where qe is the equilibrium adsorption capacity (mg g−1), and k1 and k2 are the adsorption rate constants for quasi-first order and quasi-second-order kinetic models, respectively.
2.6. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
After NT-1 strain was cultivated in liquid media with Cu(II) concentrations of 0 and 200 mg L−1, respectively, the fungal cells were collected by centrifugation and washed three times with deionized water. After vacuum freeze-drying, a small amount of lyophilized fungal cells and KBr (spectroscopically pure) were ground and mixed under dry conditions and pressed into thin tablet samples (the weight ratio of fungal cells to KBr: 1%), and a FTIR system (FT/IR-4100, Jasco, Japan) was used to collect and record the spectral characteristics. The IR spectra had a wave number range of 400–4000 cm−1 and a resolution of 4 cm−1.
2.7. Data Processing and Analysis
Origin Pro 8.0 software was used for data processing and plotting. The analysis of variance (ANOVA) of differences among the treatments was carried out with SPSS 20.0 statistical software, and the significance of the difference in the mean values of the treatments was determined by one-way ANOVA.
3. RESULTS AND DISCUSSION
3.1. Screening, Identification and Physiological and Biochemical Characteristics of the Copper-resistant Strains
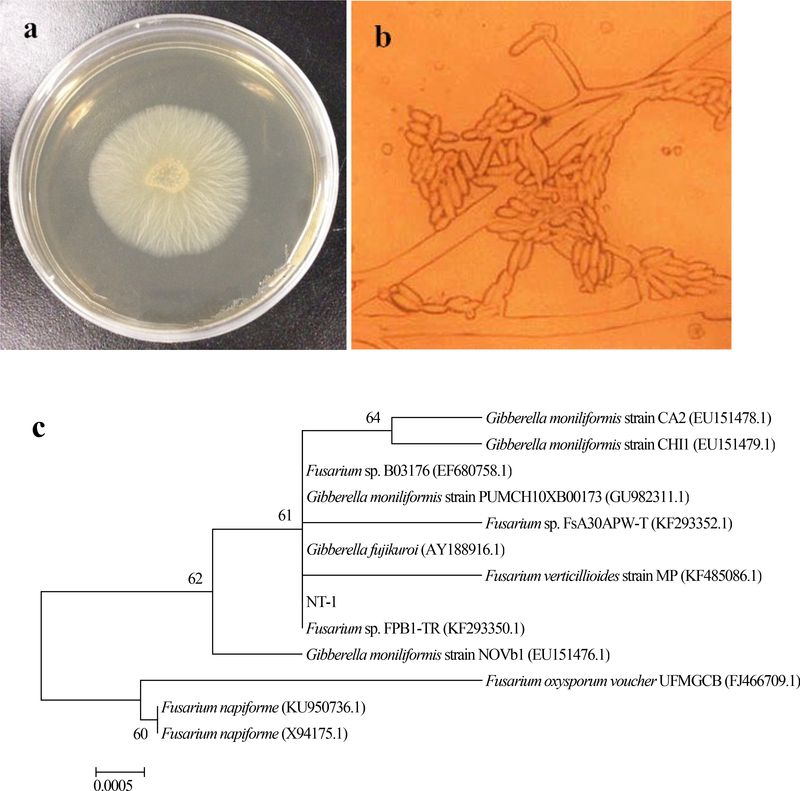
Fungal strain NT-1 grew rapidly on PDA plate, and the colonies were observed to be circular in shape, with a regular edge and a velvety texture, and tightly bound to the medium. After 3 d of cultivation at 28°C, the diameter of the colonies reached 56–58 mm, and the colonies were milky white or yellowish with a slight protrusion in the center (Fig. 1a). Mycelial growth of NT-1 was observed under the optical microscope using cover-slip method, and it was found that mycelium had thin and long branches with an asymmetric distribution, and conidiophores were found in the substrate. Conidia were observed to have two forms. Specifically, smaller conidia were seen to have oval and compact morphology, growing on solitary phialide, and observed to aggregate on top of the phialide, showing deep coloration. Conversely, large conidia were spindle-shaped in large quantities, growing at mycelial branch points (Fig. 1b).
Fig. 1.
Morphological and molecular identification of strain NT-1 (a fungal colony morphology on PDA plate; b fungal mycelium morphology under optical microscope; c phylogenetic tree based on ITS rRNA gene sequence)
The BLAST search of ITS rRNA sequencing results of NT-1 was performed in NCBI database and the phylogenetic tree was plotted. As can be seen from Fig. 1c, strain NT-1 had a 100% similarity with Gibberella moniliformis (GenBank Accession No. JF499676.1). Thus, based on the morphological and molecular characteristics of the strain, NT-1 was preliminarily identified as Gibberella sp. NT-1.
3.2. Physiological Response of Strain NT-1 to Different Concentrations of Cu(II) Stress
The copper-tolerant strain NT-1 was inoculated on PDA plates and liquid media containing different concentrations of Cu(II) and cultivated at 28°C for 5 days, and the colony diameter on the plates and the biomass of the fungal cells in the liquid medium were measured (dry weight basis). On day 5 of cultivation, the growth of NT-1 was observed to be significantly inhibited on the PDA plates with Cu(II) concentrations over 400 mg L−1. With increasing Cu(II) concentrations, the fungal colony diameter was further reduced, and almost no colony was observed on the plates with Cu(II) concentrations of over 400 mg L−1. In the copper-containing liquid medium, the growth of the strain NT-1 was also gradually inhibited as the concentration of Cu(II) increased. When the concentration of Cu(II) reached 600 mg L−1, the growth of the strain was significantly inhibited. However, after 5 days of cultivation, a certain amount of mycelial growth could still be observed, indicating a stronger tolerance of this strain to Cu(II) in liquid culture condition than that in solid culture. This may be because the heavy metal stress in liquid medium can stimulate mycelium to increase the secretion of extracellular mucilaginous material (ECMM). With polysaccharide and protein as the main components, ECMM can effectively protect the fungal cells, reduce the toxic effects of heavy metal ions on the cells, and increase the tolerance of mycelia to Cu ( Paraszkiewicz et al. 2007; Vesentini et al. 2007; Paraszkiewicz et al. 2009; Sun et al. 2009).
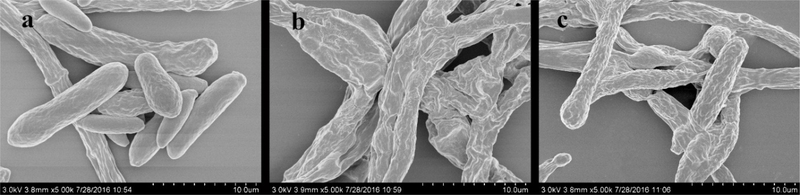
Scanning electron microscopy (SEM) was used to observe the effect of different concentrations of Cu(II) on the mycelial morphology. The results showed that NT-1 grew well in copper-free medium, with healthy extension of mycelia, and spores appearing turgid (Fig. 2a). At a Cu(II) concentration of 200 mg·L−1, the surface of mycelia started to shrink and wrinkle, with obvious protrusions (Fig. 2b). When the Cu(II) concentration was further increased to 400 mg·L−1, the shrinkage and wrinkles of mycelia became even more apparent and the number of spores in the field of view decreased substantially (Fig. 2c). The ability of this fungus to reduce copper concentrations is an important aspect for consideration, as likely related to cell surface adsorption, and could be associated with the changes in cell surface morphology. The chitin and chitosan isolated from the cell wall of Cunninghamella elegans by Franco et al. (2004) was found to adsorb a large amount of Pb and Cu in solution, and Letnik et al. (2017) found that a large amount of copper chelated to the cell surface of Micrococcus luteus treated with 10 mmol L−1 CuSO4 by SEM/EDX (scanning electron microscopy with energy dispersive X-ray spectroscopy) analysis. Cell walls are an important site for filamentous fungi to adsorb and enrich heavy metals, and the polysaccharides, proteins and other organic ligands in cell walls can specifically bind with heavy metals and effectively reduce heavy metal ion toxicity exerted on cells, and thus represent an mechanism for filamentous fungi to cope with heavy metal toxicity (Mullen et al. 1992; Suresh and Subramanyam 1998). As one of the most important components of fungal cell wall, chitin is closely related to fungal growth and spore production ability (Klis et al. 2002). Therefore, under high concentration of copper stress, the shrinkage and wrinkles of mycelia and a decrease in the number of spores could be closely related to the synthesis and degradation of chitin.
Fig. 2.
Scanning electron microscope profiles of NT-1 mycelium morphology of NT-1 under different Cu(II) stress. (a 0 mg L−1; b 200 mg L−1; c 400 mg L−1)
3.3. Effect of Different Factors on the Copper Biosorption by Strain NT-1
3.3.1. Effect of pH on the copper biosorption
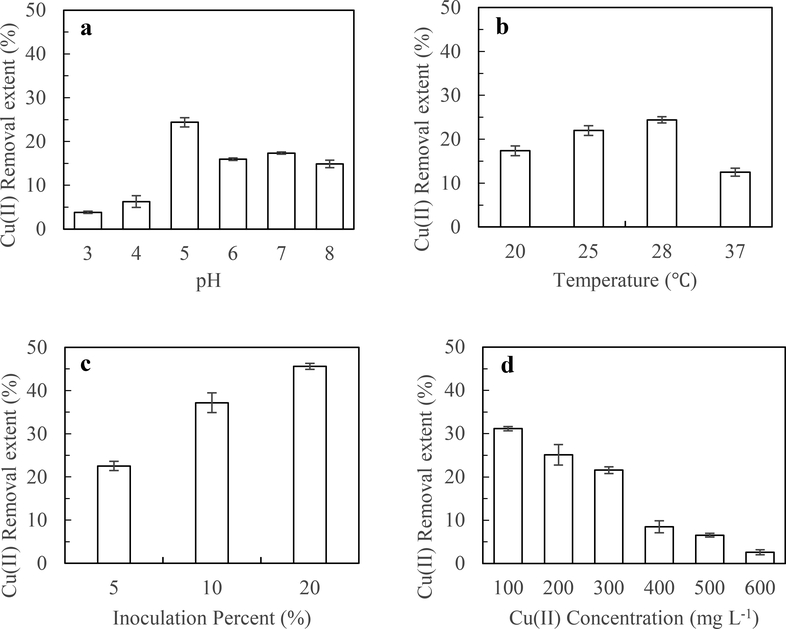
As can be seen from Fig. 3a, strain NT-1 was observed to grow within a broad pH range of 3.0–8.0, indicating that the strain has strong adaptability to the acidification stress. When the initial pH of the solution was in the range of 3.0–5.0, the Cu(II) removal extent of the strain increased significantly with increasing pH and reached a peak value of 24.4% at pH = 5.0. When the initial pH of the solution was in the range of 5.0–8.0, Cu(II) removal extent by strain NT-1 decreased with increasing pH, suggesting that strain NT-1 has strong H+ tolerance, which is also consistent with the fact that strain NT-1 was screened and isolated from orchard soil co-contaminated with acidification and copper. The pH is an important factor influencing the adsorption effect by affecting the valence state of the metal ions, changing the adsorption sites of the biofilm, and influencing the permeability of the cell membrane and the activity of biological functional groups. At low pH (i.e. pH < 3.0), competitive adsorption exists between a large number of hydronium ions and Cu(II), and H+ occupies numerous adsorption sites, thus reducing the Cu(II) adsorption to the active site. Furthermore, low pH values (i.e. a pH of 3.0) can also affect enzyme activity and inhibit the growth of fungi, resulting in decreased removal capability (Sun et al. 2014). When pH was higher than 8.0, Cu(II) began to react with OH− to form precipitates, which were suspended in solution and removed. The Cu(II) adsorbed on the surface of the fungal cells was prone to precipitation with hydroxide and was attached on the cell surface, thus affecting the transport of cellular enzymes and other carriers and reducing copper removal capability (Yahaya et al. 2009). Our result is consistent with these previous researches.
Fig. 3.
Effect of different elements factors on the removal extent of copper. a pH; b Temperature; c Inoculation amounts; d Initial Cu(II) concentration
3.3.2. Effect of temperature on the copper biosorption
As can be seen from Fig. 3b, within the temperature range of 20–28°C, copper removal extent by strain NT-1 increased with increasing temperature and reached a peak value (24.4%) at 28°C. After that, as the temperature increased, the extent of copper removal achieved by NT-1 gradually decreased. The results suggest that strain NT-1 has a wide range of adaptability to growth temperature and can grow in the temperature range of 20–37°C. Excessively low or high temperatures were unfavorable to the growth of the strain, which would likely reduce microbial metabolic rates, and would affect synthesis of metal reductase, metal transport protein and other active substances in fungal cells (Cooksey 1994; Puig et al. 2002), thus reducing the Cu(II) removal by NT-1.
3.3.3. Effect of inoculum dose on the copper biosorption by strain NT-1
As can be seen from Fig. 3c, the copper removal extent by NT-1 was increased substantially as the inoculum dose increased. This is because when the mass concentration of Cu(II) is constant and the adsorption sites on the cell surface are not saturated, a higher inoculum dose of fungal cells will result in more active sites provided by the fungal cells for adsorbing heavy metal ions, providing more chances for the fungal cell surface to contact and bind with metal ions, achieving a higher removal capacity of copper ions (Amirnia et al. 2015). However, when the inoculum dose was increased from 10% to 20%, the removal extent of copper in the solution also increased significantly, but not proportionally. This observation demonstrates that when the fungal cell concentration exceeds a certain critical point, with further increasing fungal cell concentration, the group interactions are enhanced due to the mutual adsorption and aggregation between fungal cells, and some of the effective binding sites are occupied (Li and Yu 2014; Li et al. 2017), resulting in a decrease in the utilization rate of the binding sites. Therefore, to achieve the most efficient copper removal rate in liquid media in this study, the most effective inoculum dose is 10%.
3.3.4. Effect of initial Cu(II) concentration on copper biosorption
As shown in Fig. 3d, the Cu(II) removal extent by strain NT-1 exhibited an apparent downward trend as the Cu(II) concentration in the system gradually increased. When Cu(II) concentration was in the range of 100–300 mg L−1, Cu(II) removal extent by NT-1 reached more than 20%. When Cu(II) concentration was higher than 300 mg L−1, the Cu(II) removal extent by NT-1 was decreased substantially, revealing that high Cu(II) concentration has a strong toxic effect on cell growth and cell membrane function, and that the growth of strain NT-1 and its ability to adsorb Cu(II) were inhibited. Andreazza et al. (2010) and Kiran et al. (2017) reported that with the increasing of initial metal concentration, extracellular secretions including polysaccharide and enzymes secreted by microorganisms were decreased significantly, thus result in the decreasing of metal removal extents.
3.4. Study on the Mechanism of Cu(II) Biosorption by Strain NT-1
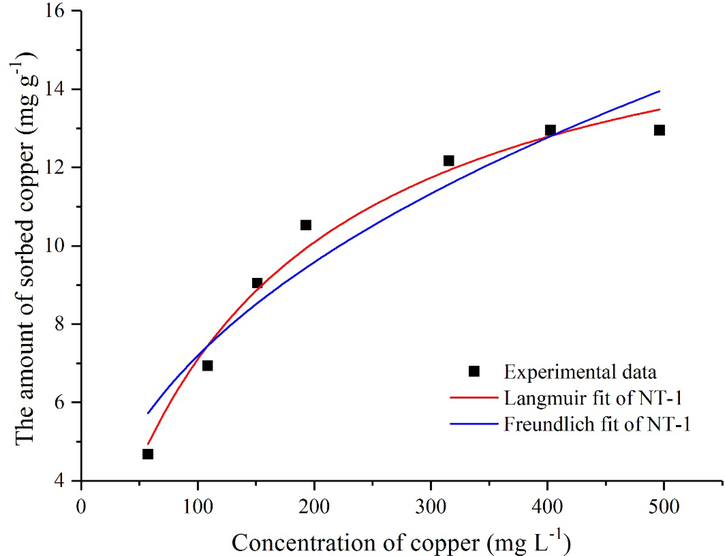
At a certain temperature, when the adsorption reaches equilibrium, the equilibrium adsorption capacity of the adsorbent qe and the equilibrium concentration of metal ions in solution Ce can be described by an isothermal adsorption equation, and the type of adsorption isotherm equation can be used to understand the interaction between adsorbents and metal ions and the surface properties of adsorbents. In order to predict biosorption behavior and calculate the biosorption capacity, Langmuir and Freundlich adsorption isotherm models were used to simulate the adsorption of Cu(II) by strain NT-1 in this study. Fig. 4 displays the adsorption isotherm for strain NT-1. The results show that with increasing Cu(II) concentration, the copper adsorption capacity of NT-1 demonstrated an apparent upward trend before reaching the equilibrium in a saturated state. This can be attributed to an increase in the probability of interaction between metal ions and mycelia (Öztürk et al. 2004). In addition, a higher initial metal ion concentration provides more driving force to overcome the mass transfer resistance of metal ions between the aqueous phase and the solid phase and increase the probability of collision between the metal ions and the mycelia (Ozdemir et al. 2004). According to the equilibrium adsorption capacity data and the solution equilibrium concentration data in Fig. 4, the maximum adsorption capacity from the fitting using Langmuir equation was 17.43 mg g−1. The comparison of the correlation coefficients of the two adsorption models suggested that the adsorption process was showed good fit to the Langmuir equation, indicating that the Cu(II) adsorption by NT-1 is a monolayer adsorption process dominated by surface adsorption.
Fig. 4.
Cu(II) adsorption isotherms of NT-1
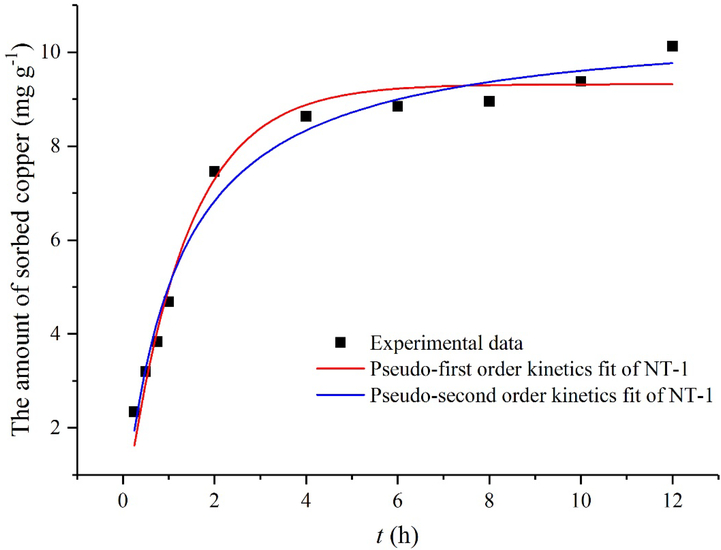
Biosorption kinetics are one of the most important characteristics in describing the adsorption rate and determining the rate-controlling step. In order to elucidate the kinetics of Cu(II) adsorption by NT-1, the experimental data were fitted with pseudo-first order kinetics and pseudo-second order kinetics (Fig. 5). The kinetics of Cu(II) adsorption by NT-1 fit well with pseudo-second-order kinetic model (R2 = 0.981), and the theoretical adsorption capacity (10.69 mg g−1) was highly consistent with the actually measured value (10.12 mg g−1). Therefore, a quasi-second-order kinetic adsorption model is more suitable for describing the dynamic Cu(II) adsorption by NT-1. These results also showed that the adsorption of Cu(II) in the solution by NT-1 is controlled by chemisorption mechanism.
Fig. 5.
Cu(II) adsorption kinetics fit of NT-1
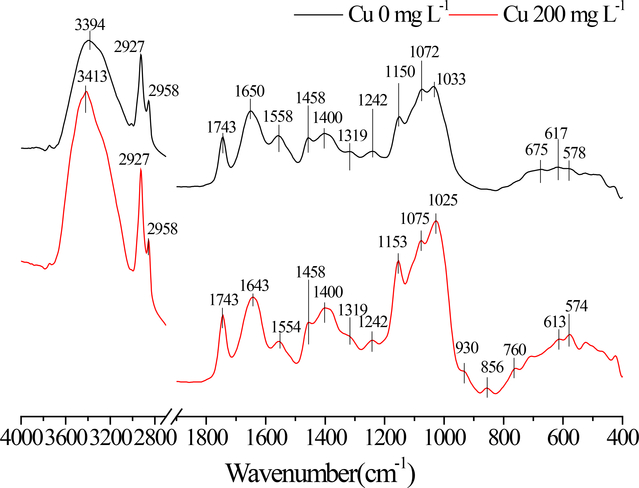
The IR spectra were used to characterize the changes in the surface functional groups before and after the Cu(II) biosorption by strain NT-1, and the IR spectra are shown in Fig. 6. The IR absorption peaks were assigned according to the literature (Abdolali et al. 2015; Yang et al. 2015; Chen and Wang 2016). The cell surface of NT-1 contains a large amount of carboxyl, amide, and hydroxyl groups as well as other organic functional groups, providing numerous binding sites for Cu(II). The absorption peaks between 3600–3200 cm−1 are mainly caused by the stretching vibration of O-H arising from carbohydrates in the cells, which participate in hydrogen bonding interactions. Under the impact of Cu(II), these peaks were observed to become sharper and shift towards higher frequencies, indicating that hydrogen bonding interaction involving O-H were weakened, possibly leading to the unstable structures of polypeptide or protein on the cell surface. The absorption peak at 1743 cm−1 is due to the vibration of C=O in carboxylic acids, and the absorption peaks at 1650 cm−1 and 1400 cm−1 correspond to the asymmetric and symmetric stretching vibrations of the carboxylate ions, respectively. The ratios of absorption peak height at 1650 ± 10 cm−1 and 1743 cm−1 before and after addition of Cu(II) (i.e. COO−/COOH ratios), were 0.928 and 0.967, respectively, probably due to an increase in pH of the solution after the cell surface binding of Cu(II), which represents an increase from 5.00 to 7.90 and an increase in the degree of COOH deprotonation. When Cu(II) was not added, the interval between two peaks at 1650 cm−1 and 1400 cm−1 was 250 cm−1. After the interaction with Cu(II), the asymmetric stretching frequency of carboxylate ions decreased to 1643 cm−1 and the peak interval was reduced to 243 cm−1, indicating that copper as a transition metal is covalently bonded to the carboxylate groups on the cell surface to form polydentate complexes (Evangelou et al. 2002).
Fig. 6.
The FTIR spectra of NT-1 before and after Cu(II) adsorption
The absorption peak at 1550 ± 10 cm−1 in Fig. 6 was caused by the deformation of CON-H of the protein amide II, which shifts to a lower frequency after addition of Cu(II), indicating that copper can also bind with amide groups on the fungal cell surface through covalent interaction. Khan et al. (2016) has proposed that the mechanism of heavy metal Cd immobilization on the cell surface of Pichia hampshirensis 4Aer is mainly achieved by the coordination binding of Cd with hydroxyl and amide groups in chitin, as the major component of most fungal cell walls is chitin (Dhillon et al. 2013). In this study, it was determined that amide groups are important sites for strain NT-1 to bind with Cu. In addition, nucleic acid and other substances in the fungal cells contain aromatic heterocyclic compounds such as purine and pyrimidine. These aromatic heterocyclic compounds can provide the necessary π-electrons, while having strong coordination ability, and can form stable complexes with various metal ions. After the interaction between the strain and Cu(II), the frequency of the deformation vibration of CH in aromatic heterocyclic compounds at 860–750 cm−1 was increased. Therefore, it is speculated that the Cu(II)-π interaction occurs during the reaction to form Cu-nitrogen-containing heterocyclic complexes. Wang et al. (2015) also found a similar phenomenon in studies involving Pb adsorption by biochar materials rich in aromatic heterocyclic compounds, such as furan and pyridine. It is noteworthy that the intensity of the absorption peak in the range of 1150–930 cm−1 increased considerably after the addition of Cu. This is probably caused by the changes in the physiological characteristics of NT-1 under the copper stress condition and an increase in the amounts of produced polysaccharides and phosphate esters on the cell surface. This is consistent with the result that the cell surface characteristics observed using SEM started to change in the culture medium with a Cu(II) concentration of 200 mg L−1.
4. CONCLUSION
In this paper, a copper-resistant fungal strain NT-1, which was isolated and preserved from copper-contaminated orchard soil, was identified as Gibberella sp. NT-1 based on the morphological, physiological, biochemical and molecular biological characteristics. The strain could grow in a wide range of relevant soil pH and Cu(II) concentrations; however a high concentration of Cu(II) could inhibit the growth of NT-1, resulting in the decreases in biomass and colony diameter, the shrinkage and wrinkles on the fungal cell surface, and other physiological responses. It was found that temperature, pH, initial Cu(II) concentration, inoculum dose and other factors could substantially affect Cu(II) removal extent by NT-1. The Cu(II) adsorption by NT-1 followed a quasi-second-order kinetic equation and Langmuir isothermal adsorption model, and was a monolayer adsorption process dominated by surface adsorption. The binding of Cu(II) to NT-1 was mainly achieved by forming polydentate complexes with carboxylate and amide groups and forming Cu-nitrogen-containing heterocyclic complexes through Cu(II)-π interaction. In the future, key parameters and mechanisms obtained from this study will be adopted to guide the implementation soil microcosm experiment and field trial of fungal bioremediation of copper-contaminated acidic soil.
Table 1.
Langmuir and Freundlich model parameters for adsorption of Cu(II) by NT-1
| Langmuir | Freundlich | |||||
|---|---|---|---|---|---|---|
| qm (mg g−1) | KL (L mg−1) | R2 | KF | n | R2 | |
| NT-1 | 17.43 | 0.006 | 0.979 | 1.071 | 2.421 | 0.917 |
Table 2.
The major peaks and assignment in FTIR spectra of NT-1 before and after Cu(II) adsorption
| Position of absorption peak (cm−1) | Peak assignment | Type of group |
|---|---|---|
| 3600−3200 | O−H vibration | OH forming hydrogen bonds |
| 2927, 2958 | Asymmetric and symmetric stretching vibrations of C−H | Aliphatic CH, CH2, and CH3 |
| 1743 | C=O vibration | COOH |
| 1650 ± 10 | C=O vibration; Asymmetric stretching vibration of COO− | Protein amide I Carboxylate ion |
| 1550 ± 10 | CON−H deformation | Protein amide II; aromatic heterocyclic compounds |
| 1458 | C=C vibration; C−H2, C−H3 bending deformation | Aromatic carbon Aliphatic CH2 and CH3 |
| 1400 | COO− symmetric stretching vibration | Carboxylate ion |
| 1319 | O−H in-plane bending deformation | Phenolic hydroxyl group |
| 1242 | C−N vibration; −P, −S | Protein amide III; phosphates and sulfates on the biofilm surface |
| 1150 ± 10 | C−O vibration | C-O in esters; aliphatic OH |
| 1072 ± 10 | C−O−C vibration | Polysaccharide ring |
| 1030 ± 20 | P−OCH3 vibration | Phosphate esters |
| 930 ± 10 | P−O−P vibration | Polyphosphate esters |
| 860−750 | C−H out-of-plate bending deformation | CH of aromatic heterocyclic compounds |
| 615 ± 10 | O−H out-of-plate bending deformation | OH of polysaccharide ring |
Acknowledgement
The current research was funded by the National Key R&D Program of China (2016YFE0106400), and the National High Technology Research and Development Program (2012AA06A204–4, 2013AA06A211–4). Although EPA contributed to this article, the research presented was not performed by or funded by EPA and was not subject to EPA’s quality system requirements. Consequently, the views, interpretations, and conclusions expressed in this article are solely those of the authors and do not necessarily reflect or represent EPA’s views or policies.
References
- Abdolali A et al. (2015) Characterization of a multi-metal binding biosorbent: chemical modification and desorption studies. Bioresource technology 193:477–487. 10.1016/j.biortech.2015.06.123 [DOI] [PubMed] [Google Scholar]
- Amirnia S, Ray MB, Margaritis A (2015) Heavy metals removal from aqueous solutions using Saccharomyces cerevisiae in a novel continuous bioreactor–biosorption system. Chemical Engineering Journal 264:863–872. 10.1016/j.cej.2014.12.016 [DOI] [Google Scholar]
- Andreazza R, Pieniz S, Wolf L, Lee M-K, Camargo FA, Okeke BC (2010) Characterization of copper bioreduction and biosorption by a highly copper resistant bacterium isolated from copper-contaminated vineyard soil. Science of the Total Environment 408:1501–1507. 10.1016/j.scitotenv.2009.12.017 [DOI] [PubMed] [Google Scholar]
- Chen C, Wang J (2016) Uranium removal by novel graphene oxide-immobilized Saccharomyces cerevisiae gel beads. Journal of environmental radioactivity 162:134–145. 10.1016/j.jenvrad.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Cooksey DA (1994) Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS microbiology reviews 14:381–386. [DOI] [PubMed] [Google Scholar]
- Dhillon GS, Kaur S, Brar SK, Verma M (2013) Green synthesis approach: extraction of chitosan from fungus mycelia. Critical reviews in biotechnology 33:379–403. 10.3109/07388551.2012.717217 [DOI] [PubMed] [Google Scholar]
- Evangelou V, Marsi M, Chappell M (2002) Potentiometric–spectroscopic evaluation of metal-ion complexes by humic fractions extracted from corn tissue Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 58:2159–2175. 10.1016/S1386-1425(01)00690-4 [DOI] [PubMed] [Google Scholar]
- Franco LdO, Maia RdCC, Porto ALF, Messias AS, Fukushima K, Campos-Takaki GMd (2004) Heavy metal biosorption by chitin and chitosan isolated from Cunninghamella elegans (IFM 46109) Brazilian. Journal of Microbiology 35:243–247. 10.1590/S1517-83822004000200013 [DOI] [Google Scholar]
- Gimeno-García E, Andreu V, Boluda R (1996) Heavy metals incidence in the application of inorganic fertilizers and pesticides to rice farming soils. Environmental pollution 92:19–25. 10.1016/0269-7491(95)00090-9 [DOI] [PubMed] [Google Scholar]
- Gutierrez T, Biller DV, Shimmield T, Green DH (2012) Metal binding properties of the EPS produced by Halomonas sp. TG39 and its potential in enhancing trace element bioavailability to eukaryotic phytoplankton. Biometals 25:1185–1194. 10.1007/s10534-012-9581-3 [DOI] [PubMed] [Google Scholar]
- Khan Z, Rehman A, Hussain SZ (2016) Resistance and uptake of cadmium by yeast, Pichia hampshirensis 4Aer, isolated from industrial effluent and its potential use in decontamination of wastewater. Chemosphere 159:32–43. 10.1016/j.chemosphere.2016.05.076 [DOI] [PubMed] [Google Scholar]
- Kiran MG, Pakshirajan K, Das G (2017) Heavy metal removal from multicomponent system by sulfate reducing bacteria: mechanism and cell surface characterization. Journal of hazardous materials 324:62–70. 10.1016/j.jhazmat.2015.12.042 [DOI] [PubMed] [Google Scholar]
- Klis FM, Mol P, Hellingwerf K, Brul S (2002) Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS microbiology reviews 26:239–256. 10.1016/S0168-6445(02)00087-6 [DOI] [PubMed] [Google Scholar]
- Letnik I, Avrahami R, Port R, Greiner A, Zussman E, Rokem JS, Greenblatt C (2017) Biosorption of copper from aqueous environments by Micrococcus luteus in cell suspension and when encapsulated. International Biodeterioration & Biodegradation 116:64–72. 10.1016/j.ibiod.2016.09.029 [DOI] [Google Scholar]
- Li D, Xu X, Yu H, Han X (2017) Characterization of Pb2+ biosorption by psychrotrophic strain Pseudomonas sp. I3 isolated from permafrost soil of Mohe wetland in Northeast China. Journal of environmental management 196:8–15. 10.1016/j.jenvman.2017.02.076 [DOI] [PubMed] [Google Scholar]
- Li W-W, Yu H-Q (2014) Insight into the roles of microbial extracellular polymer substances in metal biosorption. Bioresource technology 160:15–23. 10.1016/j.biortech.2013.11.074 [DOI] [PubMed] [Google Scholar]
- Luo Y, Tu C (2018) Twenty Years of Research and Development on Soil Pollution and Remediation in China. Springer, Singapore: 10.1007/978-981-10-6029-8 [DOI] [Google Scholar]
- Mullen M, Wolf D, Beveridge T, Bailey G (1992) Sorption of heavy metals by the soil fungi Aspergillus niger and Mucor rouxii. Soil Biology and Biochemistry 24:129–135. 10.1016/0038-0717(92)90268-3 [DOI] [Google Scholar]
- Ozdemir G, Ceyhan N, Ozturk T, Akirmak F, Cosar T (2004) Biosorption of chromium (VI), cadmium (II) and copper (II) by Pantoea sp. TEM18. Chemical Engineering Journal 102:249–253. 10.1016/j.cej.2004.01.032 [DOI] [Google Scholar]
- Öztürk A, Artan T, Ayar A (2004) Biosorption of nickel (II) and copper (II) ions from aqueous solution by Streptomyces coelicolor A3 (2). Colloids and Surfaces B: Biointerfaces 34:105–111. 10.1016/j.colsurfb.2003.11.008 [DOI] [PubMed] [Google Scholar]
- Ozturk S, Aslim B, Suludere Z (2009) Evaluation of chromium (VI) removal behaviour by two isolates of Synechocystis sp. in terms of exopolysaccharide (EPS) production and monomer composition. Bioresource Technology 100:5588–5593. 10.1016/j.biortech.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Paraszkiewicz K, Bernat P, Długoński J (2009) Effect of nickel, copper, and zinc on emulsifier production and saturation of cellular fatty acids in the filamentous fungus Curvularia lunata. International Biodeterioration & Biodegradation 63:100–105. 10.1016/j.ibiod.2008.03.015 [DOI] [Google Scholar]
- Paraszkiewicz K, Frycie A, Słaba M, Długoński J (2007) Enhancement of emulsifier production by Curvularia lunata in cadmium, zinc and lead presence. Biometals 20:797–805. 10.1007/s10534-006-9043-x [DOI] [PubMed] [Google Scholar]
- Park JH, Chon H-T (2016) Characterization of cadmium biosorption by Exiguobacterium sp. isolated from farmland soil near Cu-Pb-Zn mine. Environmental Science and Pollution Research 23:11814–11822. 10.1007/s11356-016-6335-8 [DOI] [PubMed] [Google Scholar]
- Pietrzak U, McPhail D (2004) Copper accumulation, distribution and fractionation in vineyard soils of Victoria, Australia. Geoderma 122:151–166. 10.1016/j.geoderma.2004.01.005 [DOI] [Google Scholar]
- Puig S, Lee J, Lau M, Thiele DJ (2002) Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. Journal of Biological Chemistry 277:26021–26030. 10.1074/jbc.M202547200 [DOI] [PubMed] [Google Scholar]
- Sun F, Yan Y, Liao H, Bai Y, Xing B, Wu F (2014) Biosorption of antimony (V) by freshwater cyanobacteria Microcystis from Lake Taihu, China: effects of pH and competitive ions. Environmental Science and Pollution Research 21:5836–5848. 10.1007/s11356-014-2522-7 [DOI] [PubMed] [Google Scholar]
- Sun X-F, Wang S-G, Zhang X-M, Chen JP, Li X-M, Gao B-Y, Ma Y (2009) Spectroscopic study of Zn2+ and Co2+ binding to extracellular polymeric substances (EPS) from aerobic granules. Journal of colloid and interface science 335:11–17. 10.1016/j.jcis.2009.03.088 [DOI] [PubMed] [Google Scholar]
- Suresh K, Subramanyam C (1998) Polyphenols are involved in copper binding to cell walls of Neurospora crassa. Journal of inorganic biochemistry 69:209–215. 10.1016/S0162-0134(97)10001-0 [DOI] [Google Scholar]
- Vesentini D, Dickinson DJ, Murphy RJ (2007) The protective role of the extracellular mucilaginous material (ECMM) from two wood-rotting basidiomycetes against copper toxicity. International biodeterioration & biodegradation 60:1–7. 10.1016/j.ibiod.2006.11.006 [DOI] [Google Scholar]
- Wang T, Yao J, Yuan Z, Zhao Y, Wang F, Chen H (2018) Isolation of lead-resistant Arthrobactor strain GQ-9 and its biosorption mechanism. Environmental Science and Pollution Research 25:3527–3538. 10.1007/s11356-017-0694-7 [DOI] [PubMed] [Google Scholar]
- Wang Z et al. (2015) Investigating the mechanisms of biochar’s removal of lead from solution. Bioresource technology 177:308–317. 10.1016/j.biortech.2014.11.077 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics PCR protocols: a guide to methods and applications 18:315–322. [Google Scholar]
- Yahaya YA, Don MM, Bhatia S (2009) Biosorption of copper (II) onto immobilized cells of Pycnoporus sanguineus from aqueous solution: Equilibrium and kinetic studies. Journal of Hazardous Materials 161:189–195. 10.1016/j.jhazmat.2008.03.104 [DOI] [PubMed] [Google Scholar]
- Yang J, Wei W, Pi S, Ma F, Li A, Wu D, Xing J (2015) Competitive adsorption of heavy metals by extracellular polymeric substances extracted from Klebsiella sp. J1. Bioresource technology 196:533–539. 10.1016/j.biortech.2015.08.011 [DOI] [PubMed] [Google Scholar]