Summary
The gut microbiota has been typically viewed as an environmental factor for human health. Twins are well suited for investigating the concordance of their gut microbiomes and decomposing genetic and environmental influences. However, existing twin studies utilizing metagenomic shotgun sequencing have included only a few samples. Here, we sequenced fecal samples from 250 adult twins in the TwinsUK registry and constructed a comprehensive gut microbial reference gene catalog. We demonstrate heritability of many microbial taxa and functional modules in the gut microbiome, including those associated with diseases. Moreover, we identified 8 million SNPs in the gut microbiome and observe a high similarity in microbiome SNPs between twins that slowly decreases after decades of living apart. The results shed new light on the genetic and environmental influences on the composition and function of the gut microbiome that could relate to risk of complex diseases.
Introduction
The gut microbiota is central to host metabolism and immune homeostasis and has been implicated in diseases from colorectal cancer and diabetes to autism spectrum disorders (Clemente et al., 2012; Kamada et al., 2013). The gut microbiota has typically been viewed as an environmental factor, which responds to long-term as well as short-term dietary changes (Cotillard et al., 2013; David et al., 2014; Sommer and Bäckhed, 2013; Wu et al., 2011). On the other hand, gut microbial taxa and genes have been shown to stably associate with an individual (Faith et al., 2013; Li et al., 2014; Schloissnig et al., 2013). Human genes such as Fut2 interact with the gut microbiota and play a role in Crohn’s disease (Goto et al., 2014; Rausch et al., 2011; Wacklin et al., 2011). It remains unclear, however, to what extent the gut microbiome is determined by human genetics.
Twins, with their high genetic and environmental resemblance, are well suited for investigating the role of the gut microbiome and decomposing the genetic and environmental factors. However, existing twin studies were mainly based on 16S rRNA gene amplicon sequencing or included only a few metagenomic shotgun-sequenced samples (Goodrich et al., 2014; Reyes et al., 2010; Smith et al., 2013; Turnbaugh et al., 2009), limiting their interpretation, especially regarding gut microbial species or strains and functional capacity.
A high-quality reference gene catalog is a starting point for metagenomic, metatranscriptomic, and metaproteomic studies, representing both cultured and uncultured entities in the microbiome (Li et al., 2014; Nielsen et al., 2014; Qin et al., 2010, 2012; Sunagawa et al., 2015). The Metagenomics of the Human Intestinal Tract (MetaHIT) project sequenced 760 samples from Denmark and Spain, but important regions such as the UK remain uncharted, and the sampling scheme offered little chance to infer host genetic influences (Li et al., 2014).
SNPs, short insertions or deletions (indels), and copy number variations (CNVs) based on mapping to reference bacterial genomes have been reported in the gut microbiome (Greenblum et al., 2015; Hu et al., 2013; Schloissnig et al., 2013). An early study on isolated strains of the methanogen, Methanobrevibacter smithii, revealed greater sharing of its SNPs between twins than with their mothers or other individuals (Hansen et al., 2011). In general, however, it is not known how such microbiome variations are shared or differ between twins.
Here, we sequenced fecal samples from 250 adult twins in the TwinsUK registry (Goodrich et al., 2014; Moayyeri et al., 2013), leading to a microbial reference gene catalog from 1,517 samples as well as sequenced microbial genomes. We identified SNPs from the gut microbiome and observed a high degree of microbial SNP sharing between twins, especially monozygotic twins. We demonstrated moderate to high heritability of many microbial taxa and functional modules in the gut microbiome, and identified heritable markers previously associated with type 2 diabetes (T2D), rheumatoid arthritis, and colorectal cancer.
Results
Gut Microbiome of the TwinsUK Cohort
We collected fecal samples from 35 monozygotic (MZ) and 92 dizygotic (DZ) female twin pairs from the UK (Goodrich et al., 2014; Moayyeri et al., 2013) and performed metagenomic shotgun-sequencing on 250 samples (one MZ and three DZ samples failed to yield an Illumina HiSeq library and thus could not be sequenced, Table S1A). An average of 74 million high-quality non-human reads were obtained per sample, totaling 1.8 terabyte (TB) sequences (Table S1B). De novo assembly, gene prediction, and annotation (Li et al., 2014) of these sequences led to a total of 5,901,478 non-redundant genes (Figure S1A). We then merged this gene set with a high-quality reference catalog of 9,879,896 gut microbial genes (IGC for Integrated reference Gene Catalog), compiled from 1,267 Danish, Spanish, Chinese, and American samples and 511 sequenced prokaryotic genomes or draft genomes present in the cohort (Li et al., 2014) (Figure S1; Table S1C). The updated reference catalog contained 11,446,577 genes, ensuring a saturated mapping ratio of the sequencing reads to gene-coding regions (on average 80.21%, Table S1B) (Li et al., 2014).
The previously published reference gene catalog already allowed for mapping of an average 77.96% of the reads (Figure 1; Table S1B), confirming the utility of the reference even for samples from a country different from the original reference collection (Li et al., 2014). However, genes from the 250 newly sequenced metagenomes uniquely enabled 1.41%–13.34% mapping in individual samples (3.47% on average, Figure 1; Table S1B), suggesting identification of rare microbial genes that might be specific to this cohort (Li et al., 2014).
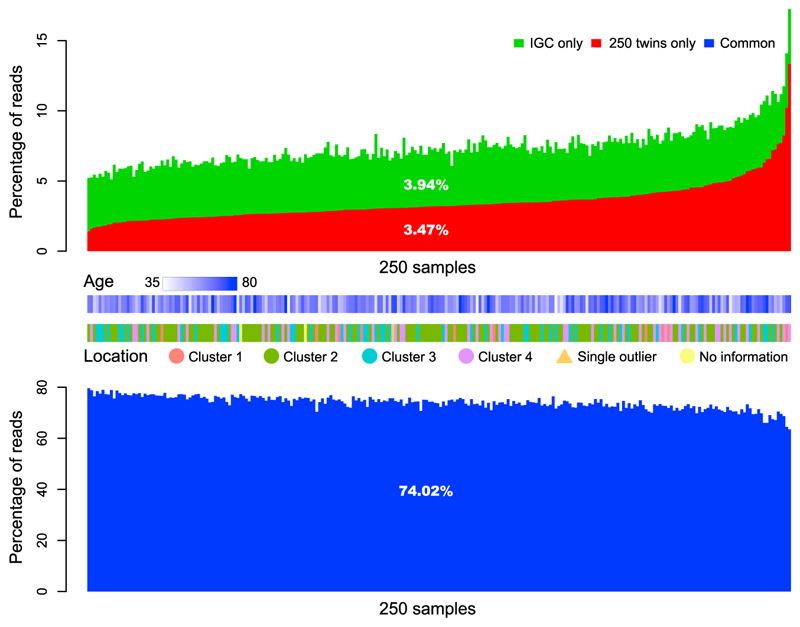
Figure 1. Representation of TwinsUK Samples by the Gene Catalogs.
High-quality non-human metagenomic reads uniquely (green, red) or commonly (blue) aligned to genes from the 250 twins gene catalog and the IGC (Table S1B). The average alignment ratio to each part is shown in the middle. The updated TwinsUK reference gene catalog (1,517 samples + 511 genomes, Table S1C) allows on average 80.21% mapping of the reads (unique + common). This is close to saturation because the percentage of gene-coding regions in all prokaryotic genomes is ~87%, and an estimated 7.25% of sequencing reads with an average length of 77 bp could not be mapped reliably as they only partially overlapped with genes (Li et al., 2014). Age and location of the samples are shown for reference.
15.3% of the 11,446,577 genes were uniquely annotated to a genus, and 42.0% of the genes could be functionally annotated to Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous groups (KOs), similar to previous studies (Arumugam et al., 2011; Li et al., 2014; Qin et al., 2012) (Table S1C).
Consistent with reported differences in the gut microbiota between people from different countries (Li et al., 2014; Human Microbiome Project Consortium, 2012; Tyakht et al., 2013; Yatsunenko et al., 2012), current location (when divided into four main regions across the UK) influenced the abundance profile of gut microbial genes in the cohort (p = 0.0369, Bray-Curtis distance in PERMANOVA, but p = 0.0912 after Benjamini-Hochberg (BH) correction for multiple testing, Figure 2A; Tables S1A and S2A). The gut microbiome between people in the same region was more similar than between people from different regions (p = 1.99e-18, Wilcoxon rank-sum test, Figures 2A–2C). This location effect may have been confounded by host genetics where twins inhabit a similar region; therefore, to reduce this effect (and the influence of common environment before separating), we compared the gut microbiome between twin sisters. Although involving a small number of twin pairs, the results indicated that pairs living in different regions are more different than pairs living in the same region (p = 0.0436, one-tailed Wilcoxon rank-sum test, Figures 2B–2D). There was no major difference between the relative number of MZ and DZ twins living in the same or different regions (p > 0.05, Fisher’s exact test), and many combinations of different regions were covered (Table S1A). While current location appears to be a relevant environmental factor for the gut microbiome in this cohort, we were unable to determine whether this effect was further confounded by non-twin genetics and other geographic differences, for instance, diet, between regions of the UK (Leslie et al., 2015).
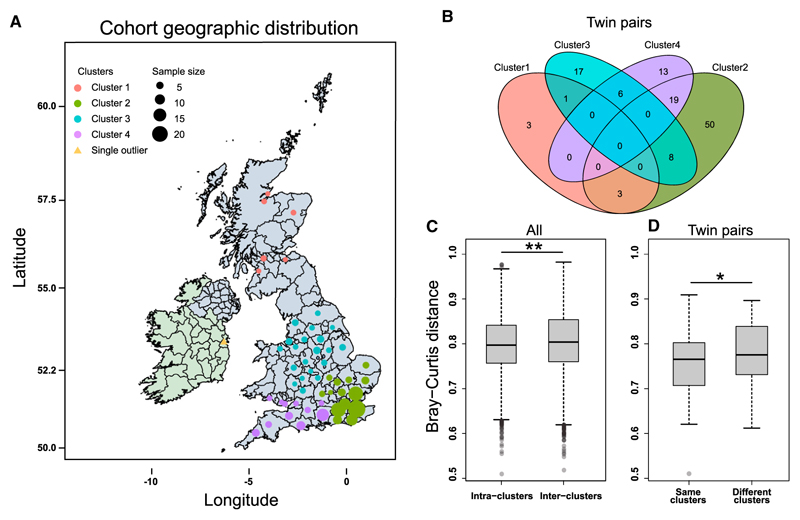
Figure 2. Effect of Current Location on the Gut Microbiome.
(A) Distribution of the subjects among geographic locations (Table S1A). The longitude and latitude of each location were hierarchically clustered to yield four regions (geo-clusters), represented by different colors, except for one sample from Ireland (not shown in B and C). The size of each circle scales with the number of subjects (samples) from that county.
(B) Venn diagram for the number of the twin pairs in the same geo-cluster and in two different geo-clusters.
(C) Bray-Curtis distance of the gut microbial gene profile between any two samples in the same region (intra-clusters), or in different regions (inter-clusters). Plotted are interquartile ranges (IQRs; boxes), medians (dark lines in the boxes), the lowest and highest values within 1.5 times IQR from the first and third quartiles (whiskers above and below the boxes), and outliers beyond the whiskers (circles). p = 1.99e-18, Wilcoxon rank-sum test.
(D) Bray-Curtis distance of the gut microbial gene profile between paired twins in the same or different geo-clusters. p = 0.0363, one-tailed Wilcoxon rank-sum test. As the volunteer in Ireland has a twin sister in geo-cluster 2 (Table S1A), this pair is included in this panel.
Factors including year of birth (p = 0.0348, Bray-Curtis distance in PERMANOVA, p = 0.0912 after BH, Table S2A), age at sampling (p = 0.0429 before and p = 0.0912 after BH), BMI (p = 0.0020 before and p = 0.0170 after BH), and vegetarian or vegan diet (p = 0.0410 before and p = 0.0912 after BH) also impacted the gut microbiome in our cohort, while order of birth, menopause, smoking, drinking, and levels of exercise were not significant (p > 0.05, Bray-Curtis distance in PERMANOVA, Table S2A). None of these factors were nearly as important as twin pair information (p = 0.0001 with 9,999 permutations in PERMANOVA, p = 0.0017 after BH, pseudo-R2 = 0.5426, Table S2A), and the factor that explained the second largest proportion of variances was current location (pseudo-R2 = 0.0141, Table S2A).
Concordance of the Twins’ Gut Microbiome
The difference within MZ or DZ twin pairs was significantly smaller than between unrelated samples, according to gut microbial gene abundance profiles (Figure S2). The MZ twins were slightly more similar than the DZ twins (Figure S2), consistent with previous results from 16S rRNA gene amplicon sequencing (Goodrich et al., 2014; Turnbaugh et al., 2009)
We then explored environmental factors that might contribute to the divergence between twins. The Bray-Curtis distance between twin pairs did not associate with age (p > 0.05 for Pearson’s, Spearman’s, and Kendall’s correlation, Table S2B). This distance did, however, negatively correlate with the age the twins started living apart (no longer share a household, p = 0.0029, 0.0208, 0.0212 for Pearson’s, Spearman’s, and Kendall’s correlation, respectively), and to a lesser extent positively correlated with years they lived apart (p = 0.0629 and correlation coefficient [cc] = 0.168 for Pearson’s correlation, but p > 0.05 for Spearman’s and Kendall’s correlation, Table S2B). As most of the twins lived separately since they were 16–24 years old (Table S1A), the results suggest that adolescence and early adulthood may be critical periods for establishing or maintaining the similarity between the twins’ gut microbiome.
Heritability of the Gut Microbiome
To explore genetic contributions to a person’s gut microbiome, we assessed the correlation between gut microbiota compositions in twin pairs. Of the 14 phyla and 109 genera detected in at least 50% of the samples, 11 phyla and 64 genera (79% and 59%) displayed an intraclass correlation coefficient (ICC) higher in MZ than in DZ pairs (Figure 3A; Tables S3A and S3B); i.e., their abundances were more similar in MZ than in DZ twins.
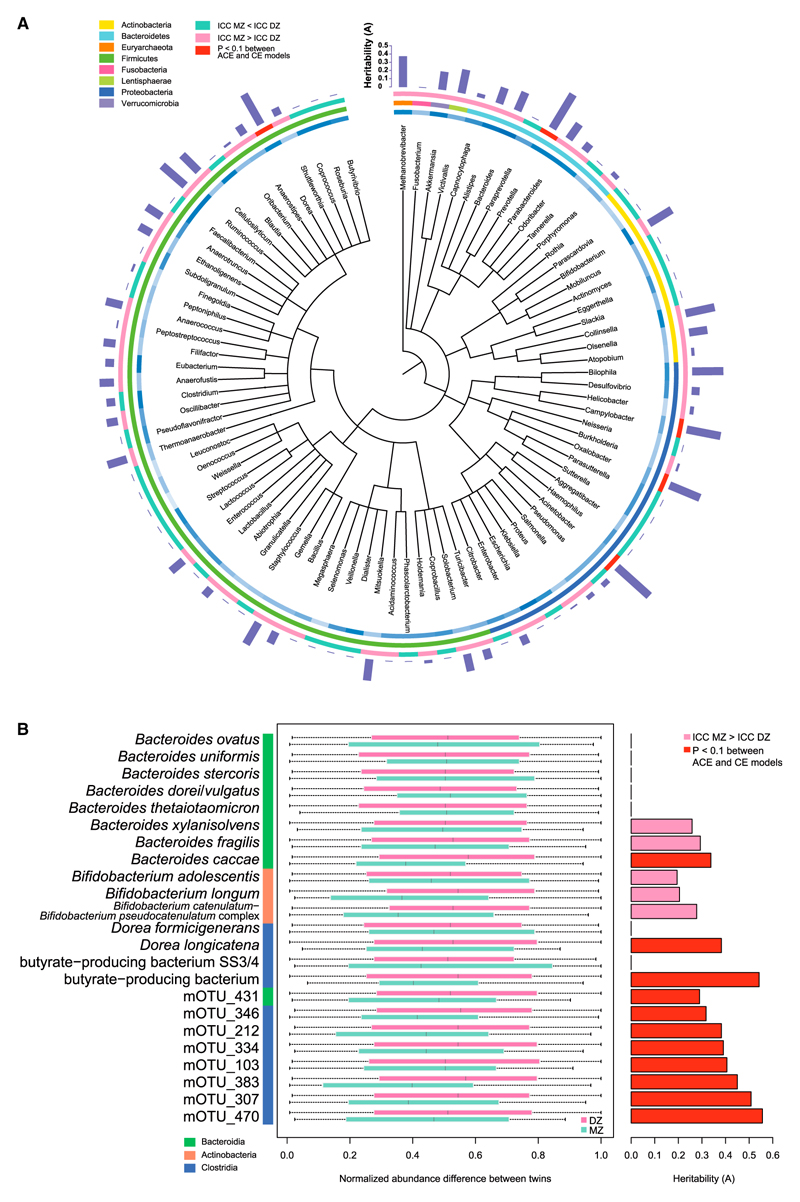
Figure 3. Heritability of Gut Microbial Taxa.
(A) A phylogenetic tree was drawn for the 90 genera seen in at least 50% of the samples. The heritability (A component in the ACE model) was plotted as a bar for each genus. Outer circle, green, ICC MZ < DZ; pink, ICC MZ > DZ; red, ICC MZ > DZ and p < 0.1 between ACE and CE models. Middle circle, colored according to phyla; inner circle, light to dark blue according to mean relative abundance of each genus. Genera that contained less than ten genes in 97% of the samples were not plotted. More detailed data are available in Table S3B for genera and Table S3A for phyla.
(B) Heritability of mOTUs. The rank of relative abundance difference between each MZ or DZ twin pair, normalized to be between zero and 1, was shown as boxplots for Bacteroides, Bifidobacterium, Dorea, butyrate-producing bacterium, and unnamed mOTUs. Class information for all the plotted mOTUs was shown as colored bar on the left. Heritability of these mOTUs according to the ACE model were plotted to the right and color coded: pink, ICC MZ > DZ; red, ICC MZ > DZ and p < 0.1 between ACE and CE models. Detailed results for all mOTUs with more than 50% occurrence are available in Table S3C.
We then estimated heritability of these gut microbial taxa by variance components analysis using the ACE model, which partitions the total variance into genetic effects (A), common environment (C), and unique environment (E) effects (Eaves et al., 1978; Goodrich et al., 2014; Neale and Maes, 1992). After Box-Cox transformation, correction for sequencing amount, age, and age started living apart, and filtering according to ICC (detailed in STAR Methods), 11 of the genera unequivocally showed heritability, i.e., p < 0.1 between ACE and CE models (Figure 3A; Table S3B). Genera in the class Clostridia, especially Dorea displayed a heritability A = 0.422, i.e., 42.2% of its abundance variations could be explained by host genetic effects (90% confidence interval [CI] = 0.094 ~0.583). In contrast, the C component (common environmental effect) was zero for these genera. Other notable genera with host genetic effects included the common Bacteroidetes, Prevotella, and the oxalate-degrading Proteobacteria, Oxalobacter (Liebman and Al-Wahsh, 2011). The major gut commensal, Bifidobacterium, showed a modest genetic component but the lower bound of confidence limits was close to zero (A = 0.309, 90% CI = 0 ~0.486) (Figure 3A; Table S3B). This was also true for the mucin-degrading bacterium, Akkermansia (A = 0.223, 90% CI = 0 ~0.394), the inflammatory bowel disease-associated bacterium, Bilophila (A = 0.378, 90% CI = 0 ~0.531), and the most common archaeaon in the human gut, Methanobrevibacter (A = 0.377, 90% CI = 0 ~0.527). The family Christensenellaceae reported to be the most heritable taxon from a 16S rRNA gene study of the twins (A = 0.39, 95% CI = 0.21 ~0.49) (Goodrich et al., 2014) was not identified here, because no genome is currently available in this family for metagenomic annotation. Although the sample size of the current study is smaller and the lower bound of 90% CI for the heritability estimates is often close to zero, in general, the heritabilities of the gut microbiome were greater than those previously reported using 16S profiles, suggesting that greater power can be achieved with the resolution made possible by the metagenomic analyses. In conclusion, the abundances of a sizable portion of the human gut microbial taxa are likely to be influenced by host genetics.
To better delineate the heritable bacteria or archaea species, we used the metagenomic Operational Taxonomic Units (mOTU) method that is based on universal single-copy marker genes (Mende et al., 2013; Sunagawa et al., 2013). Of the 143 mOTUs present in at least 50% of the samples, 91 mOTUs (64%) showed a higher ICC correlation in MZ than in DZ twins, and 11 mOTUs unequivocally showed heritability (p < 0.1 between ACE and CE models, Figure 3B; Table S3C). These heritable mOTUs included Bacteroides caccae involved in celiac disease (Viitasalo et al., 2014), Dorea longicatena that could metabolize amygdalin, aesculin, inulin, sorbital, etc. (Taras et al., 2002), a butyrate-producing bacterium from Lachnospiraceae and eight more unnamed species in the classes Clostridia and Bacteroidia. For their closely related species, results were less clear cut. For example, B. fragilis and B. xylanisolvens were more correlated in MZ than in DZ twins (Figure 3B; Table S3C) and would likely be heritable in a larger cohort.
We next examined genetic contributions to gut microbial functions. Among the 5,118 KOs present in 50% or more of the samples, 3,479 KOs had a higher ICC in MZ than in DZ twins (68%), and 443 KOs showed heritability, i.e., p < 0.1 between ACE and CE models (A = 0.3 ~0.6, Table S4A). We further identified 38 modules and 31 pathways whose abundances were heritable (Figure S3; Tables S4B and S4C). These included biosynthesis of branched-chain amino acids as well as proline, tyrosine, histidine and lysine, biosynthesis of the B vitamins biotin and riboflavin, as well as transporters in fructose and mannose metabolism (Figures 4 and S3; Tables S4A–S4C). Thus, a number of important functions of the gut microbiome are clearly heritable.
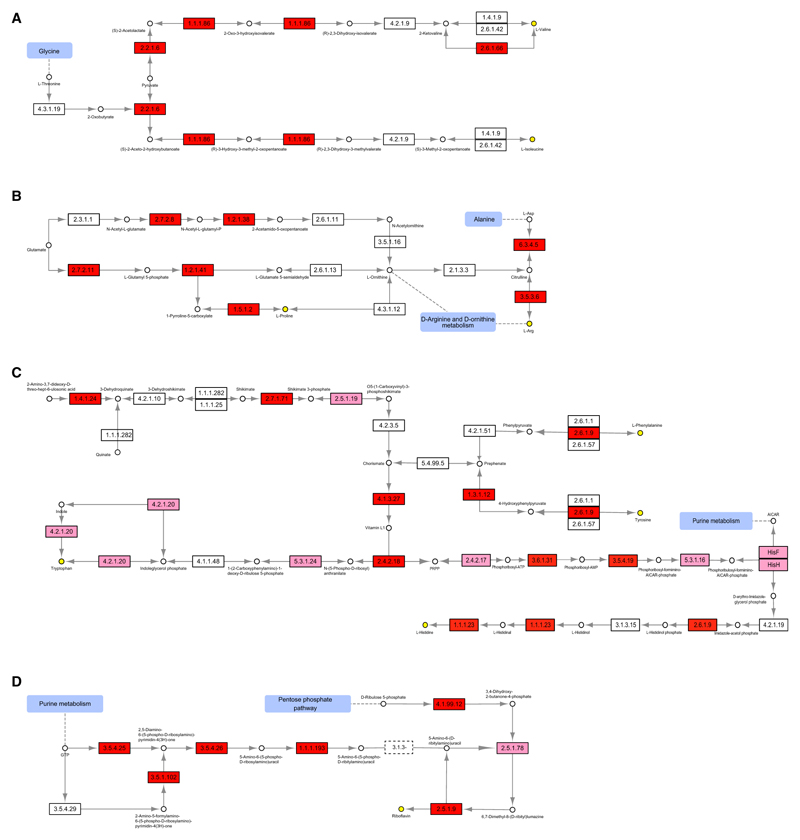
Figure 4. Heritability of Select Functions.
(A–D) Heritability of KOs in the biosynthesis of branched chain amino acids (A), arginine and proline metabolism (B), phenylalanine, tyrosine, tryptophan, and histidine biosynthesis (C), and riboflavin metabolism (D). Pink, filtered KOs and ICC MZ > ICC DZ; red, ICC MZ > ICC DZ and p < 0.1 between ACE and CE models. EC1.4.1.9, EC2.6.1.42, EC4.2.1.10, EC1.1.1.282, EC1.1.1.25, EC5.4.99.5, EC4.2.1.51, EC2.6.1.1, EC2.6.1.57, and EC4.1.1.48 are bidirectional enzymes. EC4.2.1.9, EC4.3.1.19, EC4.2.1.19, EC3.1.3.15, and EC3.5.4.25 are mapped by two or more KOs for multiple metabolic reactions. The primary pathways for L-proline (EC2.7.2.11, EC1.2.1.41, and EC1.5.1.2) and L-arginine (EC6.3.4.5 and EC3.5.3.6) biosynthesis are significantly heritable (B). More detailed data are available in Table S4A for individual KOs.
Heritability of Disease-Associated Microbes
After demonstrating host genetic impact on gut microbial taxa and functions, we explored links with the gut microbiota implicated in diseases. We chose to investigate type 2 diabetes (T2D), for which extensive genetic, epigenetic, metabolomic, and gut microbial studies were available (Karlsson et al., 2013; Mahajan et al., 2014; Manning et al., 2012; Menni et al., 2013; Morris et al., 2012; Qin et al., 2012; Scott et al., 2012; Yuan et al., 2014). Metagenome-wide association study (MWAS) on a Chinese cohort of T2D had identified 47 metagenomic linkage groups (MLGs, >100 genes) (Qin et al., 2012). All the 47 MLGs could be found in our TwinsUK cohort, although four MLGs were seen in less than 50% of the samples, and seven MLGs more abundant in the Chinese T2D patients were more abundant in the non-diabetic TwinsUK samples (Figure 5; Table S5A, ten diabetic versus 212 non-diabetic, irrelevant for heritability analysis). Thus, the Chinese T2D-associated gut microbiome was largely corroborated by this UK cohort.
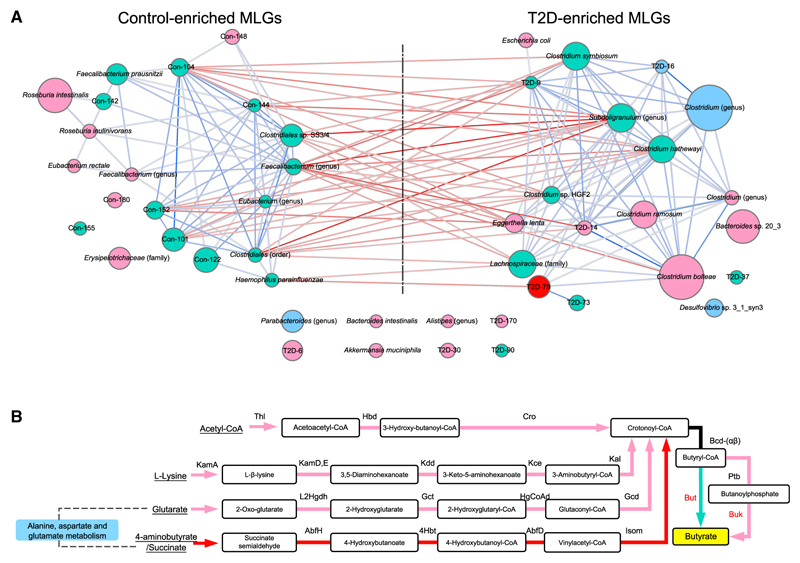
Figure 5. Heritability of T2D MLGs and Butyrate Biosynthesis Pathways.
(A) MLGs (>100 genes) from Qin et al. (2012) were profiled in the TwinsUK cohort. Blue nodes, present in less than 50% of the samples; green nodes, ICC MZ < DZ; pink nodes, ICC MZ > DZ; red nodes, ICC MZ > DZ and p < 0.1 between ACE and CE models (Table S5A). Light to dark-blue edges, Spearman’s correlation >0.4; light to dark-red edges, Spearman’s correlation < −0.4.
(B) Pathways for butyrate biosynthesis were drawn according to Vital et al. (2014). Green arrows, ICC MZ < DZ; pink arrows, ICC MZ > DZ; red arrows, ICC MZ > DZ and p < 0.1 between ACE and CE models (Table S5B). The black line from crotonoyl-CoA to butyryl-CoA was not analyzed, because it was shared by all four pathways leading to Butyryl-CoA.
Control-enriched MLGs such as Eubacterium rectale, an unclassified Faecalibacterium sp., Roseburia intestinalis, and R. inulinivorans, and T2D-enriched MLGs such as Clostridium ramosum, C. boltae, Eggerthella lenta, and Bacteroides sp. 20_3 displayed a higher ICC in MZ compared to DZ twins (49% of the 43 MLGs); T2D-79 showed significant heritability (p < 0.1 between ACE and CE models, Figure 5; Table S5A). In contrast, Clostridium sp. SS3/4, Faecalibacterium prausnitzii, Haemophilus parainfluenzae, etc. were similarly correlated in these DZ and MZ twins. We conclude that many of the T2D-associated gut microbiome components may be partly influenced by host genetics.
Functionally, 873 of the reported 1,345 T2D KO markers had a higher correlation in MZ than in DZ, of which 143 KO markers showed a significant heritability, e.g., the module for sulfur reduction enriched in T2D (Qin et al., 2012) (Figure S3; Table S3A). Enzymes for the biosynthesis of branched-chain amino acids including valine and isoleucine were also heritable (EC 2.2.1.6, EC 1.1.1.86, and EC 2.6.1.66, Figure 4A). High concentrations of circulating branched-chain amino acids were reported to be associated with insulin resistance and future diabetes risk and may partially be determined by genetic factors (Ridaura et al., 2013; Kettunen et al., 2013; Wang et al., 2011). Butyrate production, a major function depleted in individuals with T2D and a number of other conditions, also showed heritability in the 4-aminobutyrate pathway, i.e., production of butyryl-CoA from 4-aminobutyrate (Figure 5B; Table S5B). Other butyryl-CoA production pathways originating from acetyl-CoA, lysine, and glutarate had higher concordance in MZ than in DZ twins (Figure 5B; Table S5B). Bacterial bile salt metabolizing genes had been found to decrease in ulcerative colitis and T2D (Labbé et al., 2014). The bile salt hydrolase (BSH, EC 3.5.1.24), which functions as the first enzyme for deconjugation of bile acids, was significantly heritable in our cohort (Table S4A). These data indicate that the abundance of some of T2D-associated gut microbial functions might have been influenced by host genetics.
Besides the T2D-associated gut microbiome, Prevotella copri reported to be elevated in new-onset rheumatoid arthritis (NORA) patients (Scher et al., 2013; Zhang et al., 2015) also appeared heritable (A = 0.419, 90% CI = 0.031 ~0.564, Table S3D), consistent with heritability of the Prevotella genus (Figure 3A; Table S3B). Peptostreptococcus stomatis found to be more abundant in patients with colorectal cancer (Feng et al., 2015; Yu et al., 2015; Zeller et al., 2014) also showed evidence of heritability (A = 0.496, 90% CI = 0.210 ~0.639, p = 0.017 between ACE and CE, Table S3D), consistent with previous estimates for the Peptostreptococcaceae family (Davenport et al., 2015; Goodrich et al., 2016; O’Connor et al., 2014; Org et al., 2015). Functionally, 31 of the 51 previously reported gut microbial modules (GMMs) differentially enriched in individuals with a low or high gut microbial gene richness were more concordant (higher ICC) in MZ compared to DZ twins; six of the GMMs showed clear heritability (p < 0.1 between ACE and CE models), including high richness-associated modules such as cysteine biosynthesis/homocysteine degradation and low richness-associated modules such as N-acetylglucosamine degradation (Table S5C). These results demonstrate heritability in the abundance of taxa and functions of the gut microbiome, which have previously been associated with complex diseases.
Concordance in Microbial SNPs
To explore the twins’ gut microbiome at higher resolution, we looked for SNPs in the microbiome (Figure 6), which could offer further information on the extent to which the gut microbiome remained concordant between adult twins.
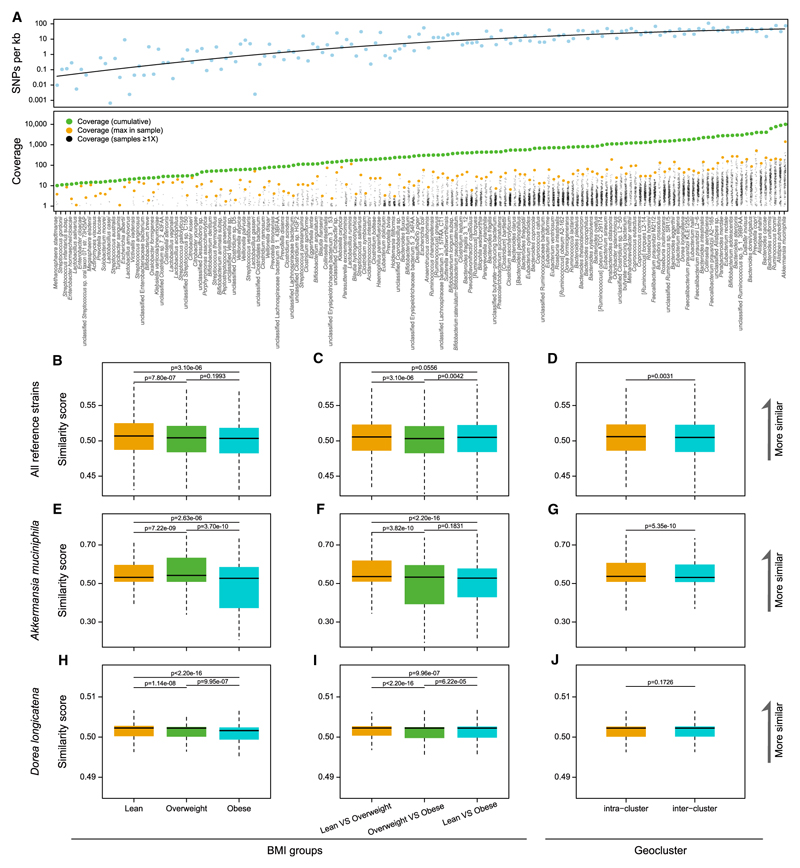
Figure 6. Gut Microbiome SNPs Detected in the TwinsUK Cohort.
(A) SNP density in the 152 reference bacterial genomes with a cumulative coverage of at least 10x in the 250 samples. The bacterial genomes were ordered according to the cumulative coverage (green circles, Table S6). The coverage in each sample (black circles) was also plotted, with the maximum coverage among samples highlighted in beige.
(B, E, and H) SNP similarity score within lean, overweight, and obese groups, calculated from all 152 reference genomes (B), for A. muciniphila (E) or D. longicatena only (H). p values from Wilcoxon rank-sum tests. 130 subjects in the lean group have a normal BMI (18.50–24.99) except for four underweight subjects (<18.5).
(C, F, and I) SNP similarity score between the different BMI groups, calculated from all 152 reference genomes (C), for A. muciniphila (F) or D. longicatena only (I). p values from Wilcoxon rank-sum tests.
(D, G, and J) SNP similarity score within and between geographic regions (Figure 2) calculated from all 152 reference genomes (D), for A. muciniphila (G) or D. longicatena only (J). p values from Wilcoxon rank-sum tests.
After removing redundancy within species (STAR Methods), 152 bacterial or archaeal genomes were present in the cohort with a cumulative sequencing depth of at least 10x (the criterion used in a previous study [Schloissnig et al., 2013]; Table S6). Between one to dozens of SNPs/kilobase were identified for each genome, increasing with the cumulative sequencing depth until around 500x (Schloissnig et al., 2013) (Figure 6A). A high cumulative SNP density was found for bacteria such as Akkermansia muciniphila, Ruminococcus bromii, Bacteroides uniformis, and Roseburia hominis. These results demonstrate a high degree of sequence variations in the gut microbiome that could only be captured by metagenomic shotgun sequencing and would be useful for future analysis of microbial strains.
To explore the possible link between gut microbiome SNPs and host phenotype, we divided the subjects into different BMI groups (lean, BMI <5; overweight, ≤25, BMI <30; obese, BMI ≥30), a significant influencing factor on the gut microbiome (Figure 6; Table S2A). Interestingly, the lean group shared a greater proportion of gut microbiome SNPs as analyzed by SNP similarity score between individuals (Schloissnig et al., 2013) than the overweight or the obese groups (p = 7.80e-07 and p = 3.10e-06, Wilcoxon rank-sum test, Figure 6B). For the normal weight-related bacterium, A. muciniphila (Chevalier et al., 2015; Everard et al., 2013; Lukovac et al., 2014) showed decreased SNP similarity in the obese group compared to the overweight group (Figures 6E and 6F). The overweight group had the largest number of total SNPs in A. muciniphila, with on average more than 300 SNPs per sample (Figure S4A). Overall, the SNPs were distributed evenly across the A. muciniphila genome except for the repeat regions (Figure S4B). Besides BMI, samples from the same geographic region showed greater sharing of SNPs than samples from different regions (Figures 6D and 6G), possibly reflecting strain-level differences. For the bacterium D. longicatena (Figure 2; Table S3C), in contrast, a different trend was observed for BMI, while no difference could be seen for geographic regions (Figures 6H–6J).
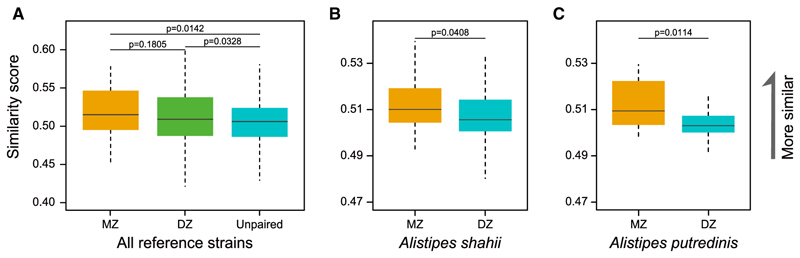
We next compared twin siblings and found that the similarity score for gut microbial SNPs was significantly higher between twins than between unpaired samples (p = 0.0142 with MZ, p = 0.0328 with DZ, one-tailed Wilcoxon rank-sum test, Figure 7A) and slightly higher between MZ than between DZ pairs both before and after downsizing to the same sequencing amount, and after correcting for age-started living apart or years lived apart (p = 0.1085, 0.3144, 0.2134, 0.1689, one-tailed Wilcoxon rank-sum test, Figures S5A and S5B). For some microbes like the bile-resistant bacteria Alistipes shahii and A. putredinis, the difference was significant between MZ and DZ twins (Figures 7B and 7C). Thus, although we found variation, overall genetically identical twins have very similar SNPs in their gut microbiome even as middle-aged or senior adults, suggesting host genetic contributions to the fine structures of the gut microbiome.
Figure 7. Greater Sharing of Gut Microbiome SNPs between Twins.
(A) SNP similarity score between twins compared to unrelated samples, calculated from all 152 reference genomes. p = 0.0142 between MZ and unpaired samples, p = 0.1805 between MZ and DZ, p = 0.0328 between DZ and unpaired, one-tailed Wilcoxon rank-sum test.
(B and C) SNP similarity score between MZ twins compared to DZ twins for A. shahii (B) and A. putredinis (C). p = 0.0408 and 0.01144, respectively, one-tailed Wilcoxon rank-sum test.
A recent analysis on SNPs in the Human Microbiome Project (HMP) samples suggested drifting of gut microbial strains in healthy adults (Li et al., 2016). We explored this over a much longer timescale. While age did not show a significant correlation with SNP similarity score between twins (consistent with results from gene abundances [Table S2B]), age started living apart positively correlated with SNP similarity score, and years lived apart negatively correlated with SNP similarity score (p < 0.05 for Spearman’s and Kendall’s but not for Pearson’s cc, Table S7). This is also true when we only look at twins that started living apart between 16 and 24 years old, the range that most of the samples fall into (Tables S1A and S7). Thus, no longer sharing a household, perhaps in a critical time period, appeared to contribute more to the divergence of gut microbial strains between twins than chronological age.
Discussion
We report an updated gut microbial reference gene catalog containing 11.4 million genes from 1,517 fecal samples and 511 sequenced gut-related bacteria or archaeal genomes, a comprehensive resource for metagenomic, metatranscriptomic, and metaproteomic studies on the human or mouse-associated microbiome around the world (Li et al., 2014; Qin et al., 2010; Thaiss et al., 2014; Wang and Jia, 2016).
Furthermore, we demonstrate a widespread concordance in the composition, SNPs, and functional capacity of the gut microbiome between twins and specifically an increased concordance among MZ twins over DZ twins, consistent with host genetic influence. To the best of our knowledge, this is the only large cohort of twins surveyed by metagenomic shotgun sequencing to date, providing high-resolution information for taxa and potential functions. Compared to a previous 16S rRNA gene amplicon sequencing on twins, however, the sample size is relatively small and no replication cohort is available (Goodrich et al., 2014). Early work on cross-bred mice (another important and more controlled source of genetic associations) has identified a number of quantitative trait loci (QTL) associated with bacteria taxa in the gut (Benson et al., 2010; O’Connor et al., 2014; Org et al., 2015). It is likely that several microbes reported as non-heritable here will attain significance with a larger cohort with greater power. For example, a previous study in mice identified a chromosomal region associated with Roseburia spp. abundance, which overlapped with liver and adipose eQTLs (expression QTLs) of the Irak4 gene in mice fed a high-fat, high-sucrose (HF/HS) diet (Org et al., 2015). The upper limit of 90% CI for three Roseburia species ranged between 0.2 and 0.5 in our cohort (Table S3D), and a larger cohort is expected to raise the lower limit of 90% CI further above zero.
As in other studies of the gut microbiome, diet is a confounding factor. Aspects of dietary preference are heritable (Teucher et al., 2007), and it remains to be explored how much these are influenced by heritable components of the gut microbiome and vice versa.
Immune traits are also highly heritable and human genetic associations have been found for all major immune cell types (Roederer et al., 2015). Although the gut microbiota is known to interact with host metabolism and immune function, only a handful of human genes, most notably Fut2, Nod2, and the major histocompatibility complex (MHC) locus, have been implicated in differences in the gut microbiota (Frank et al., 2011; Goodrich et al., 2014; Goto et al., 2014; Khachatryan et al., 2008; Lukovac et al., 2014; Maslowski et al., 2009; McKnite et al., 2012; Pickard et al., 2014; Rausch et al., 2011; Rehman et al., 2011; Scher et al., 2013; Trompette et al., 2014; Wacklin et al., 2011). Interestingly, P. copri was reported to be more abundant in patients lacking the major susceptibility allele, HLA-DRB1 (Scher et al., 2013), suggesting that its host genetic association may involve non-MHC loci or MHC alleles analyzed by higher-resolution methods than is commonly employed (Zhou et al., 2016). Although metagenome-wide association studies (MWAS) currently explore associations between the relative abundance of microbial genes with diseases (Le Chatelier et al., 2013; Cotillard et al., 2013; Feng et al., 2015; Karlsson et al., 2013; Qin et al., 2012, 2014; Zhang et al., 2015), high-resolution analyses of the SNPs and structural variations both in microbial genomes and in the human genome are expected to fully realize the power of this method. Further studies on the interaction between host genes and the gut microbiome could help explain the missing heritability in many complex diseases, although this would involve much larger sample sizes.
With the current cohort, a portion of the disease-associated gut microbiome appear to be explained by environmental factors and thus in theory may be readily amenable to microbiome-based therapeutics. Individual-specific features of the gut microbiome are determined not only by genes, but also by factors such as geographic location (or geographically associated environmental and lifestyle effects) and the time and duration of sharing a home environment, which deserve further study in larger and younger cohorts. All of these are important to consider in studies on human diseases and could be useful in microbiome-based forensics and disease treatments (Franzosa et al., 2015; Lax et al., 2014; Ridaura et al., 2013). Our results suggest that heritable components of the gut microbiome are worth investigating for a number of complex diseases, and controlling for microbiome factors might facilitate future explorations in disease genetics. Similarly, investigations of the gut microbiota need to account for genetic influences that are an important component of the differences between individuals and could influence response to therapies such as probiotics or non-autologous fecal transplants.
Star★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical Commercial Assays | ||
| BOX,TruSeq PE CLUSTER KIT v3-cBot-HS, BOX 1 of 2 | Illumina(Hiseq 2000) | PE-401-3001-1 |
| BOX,TruSeq PE CLUSTER KIT v3-cBot-HS, BOX2 of 2 | Illumina(Hiseq 2000) | PE-401-3001-2 |
| FC, PE HiSeq Flow Cell v3 - Grafted | Illumina(Hiseq 2000) | PE-401-3001-FC |
| TruSeq SBS KIT-HS v3 (200 CYCLES) BOX 1 of 2 | Illumina(Hiseq 2000) | FC-401-3001-1 |
| TruSeq SBS KIT-HS v3 (200 CYCLES) BOX 2 of 2 | Illumina(Hiseq 2000) | FC-401-3001-2 |
| Deposited Data | ||
| Shotgun-sequenced reads with human sequences removed | This paper | EBI: ERP010708 |
| Reference catalog of 11.4 million genes and other related data | This paper | http://dx.doi.org/10.5524/100253 |
| Reference catalog of 9.9 million genes | Li et al., 2014 | http://gigadb.org/dataset/100064 |
| Software and Algorithms | ||
| Blast | NCBI | https://blast.ncbi.nlm.nih.gov |
| Soap2 | Li et al., 2009 | http://soap.genomics.org.cn/ |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| BWA | Li et al., 2009 | http://bio-bwa.sourceforge.net/ |
| mOTU | Sunagawa et al., 2013 | http://www.bork.embl.de/software/mOTU/ |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to the Lead Contact Huijue Jia (jiahuijue@genomics.cn), who holds responsibility for fulfillment of these requests.
Experimental Model and Subject Details
The study included 35 MZ and 92 DZ female twin pairs with a mean age of 61 years (range 36-80 years of age), who were unselected for any phenotypes and are representative of the participants within the TwinsUK database. None of the women was pregnant or lactating at the time of sampling. Most of them were postmenopausal. Fecal samples were collected by the participates and refrigerated at home for no more than 2 days, before they were taken to King’s College London for storage at −80°C. The samples were transported on dry ice to Cornell University for DNA extraction using the PowerSoil kit (MoBio), as in (Goodrich et al., 2014). The DNA samples were then transported on dry ice to BGI-Shenzhen. 1 MZ and 3 DZ samples failed to yield an Illumina library. The study was approved by the institutional review boards at King’s College London and BGI-Shenzhen. All the remaining feces and DNA samples have been returned to Prof. Timothy D. Spector at King College London (tim.spector@kcl.ac.uk).
Method Details
Metagenomic sequencing and assembly
Paired-end metagenomic sequencing was performed on the Illumina HiSeq2000 platform with a read length of 100 bp (insert size 350 bp). Low quality or human reads (according to alignment to hg19) were removed, and the high-quality sequencing reads were then de novo assembled into contigs using SOAPdenovo v1.06 in the MOCAT pipeline (Kultima et al., 2012; Li et al., 2014). On average, 7.24 Gb of high-quality non-human sequences were obtained per sample (Table S1B).
Gene catalog construction
GeneMark v2.7d was used to predict genes from the assembled contigs. Redundant genes were removed using CD-HIT of the MOCAT pipeline (95% identity, 90% overlap) (Kultima et al., 2012), resulting in a non-redundant gene catalog of 5,901,478 non-redundant genes. This gene catalog was further integrated into an existing gut microbial reference catalog of 9,879,896 genes (IGC) using CD-HIT (Kultima et al., 2012; Li et al., 2014), resulting in a final catalog of 11,446,577 genes (Figure S1; Tables S1B and S1C). Relative abundances of the genes were computed by aligning high-quality sequencing reads to the reference gene catalog as previously described (Qin et al., 2012).
Taxonomic annotation and abundance calculation
Genes from the existing reference gene catalog inherited their original taxonomic annotation (Li et al., 2014). Taxonomic assignment of the newly included genes was performed using the same in-house pipeline, through BLASTN alignment to 3,449 bacterial or archaeal genomes or draft genomes from the National Center for Biotechnology Information (NCBI) and the European Molecular Biology Laboratory (EMBL) (Li et al., 2014; Mende et al., 2013). For each gene, only the top 10% highest-scoring alignments covering ≥ 80% of gene length and identity ≥ 65% were retained. Each gene was assigned the taxonomy of the alignment(s) with 50% or higher consensus above the similarity threshold for taxonomic rank (> 65% for phylum, > 85% for genus and > 95% for species). Our phylogenetic annotation pipeline ensures unique assignment to phylum, genus and species for each gene, and minimizes ambiguity (Arumugam et al., 2011; Li et al., 2014; Qin et al., 2012). The relative abundance of a taxon was calculated from the relative abundance of its genes. Taxa containing less than 10 genes were removed.
mOTU and MLG profiling
High-quality reads in each sample were aligned to the 79268 sequences of mOTU reference with default parameters (Mende et al., 2013; Sunagawa et al., 2013), and 597 species-level mOTUs were identified.
Similarly, abundance of the 47 MLGs from the Chinese T2D study (Qin et al., 2012) in each TwinsUK sample was determined by aligning high-quality reads to genes in the MLGs. The co-occurrence network was constructed according to Spearman’s correlation between the MLGs in the TwinsUK cohort, and visualized by Cytoscape 3.0.2.
Functional annotation according to KEGG
Genes from IGC inherited their original KO annotation (Li et al., 2014). KO assignment of the newly included genes was performed using the same procedure. Putative amino acid sequences were translated from the gene catalogs and aligned against the proteins/domains in the KEGG databases (release 59.0, with animal and plant genes removed) using BLASTP (v2.2.24, default parameter except that −e1e − 5a6 − b50 − FFm8). Each protein was assigned to a KO by the highest scoring annotated hit(s) containing at least one high-scoring segment pair (HSP) scoring over 60 bits. KO pathways of different heritability were highlighted in iPath (Yamada et al., 2011).
Quantification and Statistical Analysis
PERMANOVA on the influence of phenotypes
We applied permutational multivariate analysis of variance (PERMANOVA) on the gene abundance profile of the samples to assess impact from each of the factors listed (Anderson, 2001; Feng et al., 2015) (Table S2A). We used Bray-Curtis distance and 9999 permutations in R (3.10, vegan package (Feng et al., 2015; Zapala and Schork, 2006)). Similar results were obtained for Jensen-Shannon distance.
Correlation between numerical phenotypes and microbial distance between twins
Bray-Curtis distance between the gut microbial gene abundance profile of paired twins was assessed for correlation with the difference between twins in the phenotypes listed (Table S2B). Both linear and non-linear correlations were considered using Pearson’s, Spearman’s and Kendall’s correlation coefficients.
Heritability analysis
Heritability of traits including genera, MLGs, KOs, KO pathways or modules was estimated as previously described (Goodrich et al., 2014). Traits present in less than 50% of the individuals were not analyzed because they mostly could not fulfil the requirement for normal distribution even after Box-Cox transformation. Abundances of the remaining traits were scaled to make sure the minimum nonzero is 1 and then subjected to one-parameter Box-Cox transformation for normal distribution, and multiple linear regression to eliminate influence from the number of sequencing reads per sample, age, and age started living apart by using the powerTransform command from the R package ‘car’ and offset of 1. The traits were further filtered according to their ICCs using two criteria: 1) r(MZ) > r(DZ); 2) r(MZ) > 0 and p < 0.01, i.e., r(MZ) is significantly greater than 0. These steps were expected to satisfy the model-assumed conditions and increase the proportion of heritable traits in the pool. All ICC calculations were generated with the ‘icc’ command from the R package ‘irr’.
Heritability is calculated by using the R package ‘OpenMx’. The twin-based ACE model and its submodels were used to estimate heritability of each trait, which were weighted to correct for the unequal number of MZ and DZ pairs. The equation of the objective function is:
The p value of the A component was regarded as the difference between the ACE and the CE models, i.e., inclusion of the genetic component better explains the data than using the common and unique environmental components only. Although heritability studies traditionally only report heritability and confidence intervals, here we controlled for multiple testing on the p values of the ICC-filtered traits (977 total) according to Storey’s FDR method (Storey, 2002) as a stringent measure. For a p value of 0.1 between ACE and CE, the q-value is 0.05 (Tables S4, S5, and S6).
Phylogenetic tree for gut microbial genera
The phylogenetic relationship according to the NCBI database (http://www.ncbi.nlm.nih.gov/Taxonomy/Selector/taxse.cgi) for the 120 genera detected in at least 50% of the samples was uploaded to the Interactive Tree of Life server (http://itol.embl.de) to plot a phylogenetic tree.
SNP identification and similarity score
Sequencing reads from the 250 TwinsUK samples were aligned to 983 reference genomes or draft genomes of gut bacteria or archaea (previously identified as human gut microbes from all 3449 bacteria or archaea genomes or draft genomes available from NCBI (Li et al., 2014)) using SOAP2 with identity ≥ 90% (Li et al., 2009). To eliminate redundancy, the genomes were clustered according to their MUMi distances (Deloger et al., 2009) into 343 species-level clusters (MUMi > 0.5). Representative genomes from each cluster were identified according to three criteria: i) The genome recruited the highest number of reads in its cluster; ii) The genome had a cumulative depth of at least 10-fold from all samples; iii) At least 40% of the genome was covered by reads from a single sample (Schloissnig et al., 2013). A total of 152 genomes fulfilled these criteria and were used as references for SNP calling.
An in-house script was used to call SNPs using SAMtools (Li, 2011). Single nucleotide variants were considered as SNPs if they had a frequency of at least 1% and were supported by at least 4 reads. Shared allele similarity score were calculated as previously described (Schloissnig et al., 2013).
Data and Software Availability
The accession number for metagenomic shotgun sequencing data for all 250 samples after removal of human sequences reported in this paper is European Bioinformatic Institute (EBI): ERP010708. Other relevant data have been deposited to the GigaScience Database (GigaDB) (http://dx.doi.org/10.5524/100253).
Supplementary Material
Supplemental Information includes five figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.10.004.
Highlights.
Metagenomic data from the UK confirm saturation of the gut microbiome gene content
Sharing of household and geographic region influences the similarity of gut microbiome
Much of gut microbial composition and functions is heritable
Microbial SNPs are often shared between twins and slowly diverge after living apart
Acknowledgments
The TwinsUK resource is funded by the Wellcome Trust 105022/Z/14/Z and from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. T.D.S. is an NIHR Senior Investigator. The sample collection and DNA extraction were supported by NIH grants RO1 DK093595 and DP2 OD007444 to Ruth E. Ley and her team at Cornell University. Collections in the UK were handled by Ms. Gabriela Surdulescu and the TwinsUK lab and administration teams. At BGI, the metagenomic study was supported by the Shenzhen Municipal Government of China (JSGG20140702161403250, DRC-SZ[2015]162, CXB201108250098A, JSGG20160229172752028, and JCYJ20160229172757249) and the Fund for Science and Technology Development (FDCT) from Macao (grant 077/2014/A2). We gratefully acknowledge colleagues at BGI-Shenzhen for DNA quality control, library construction, sequencing, and helpful discussions. We also thank Drs. Ruth E. Ley, Andrew G. Clark, and Julia K. Goodrich from Cornell University for DNA preparation and commenting on the manuscript.
Footnotes
Author Contributions
T.D.S., J.T.B., and K.J.W. oversaw the sample collection and provided phenotypic information according to the TwinsUK database. T.D.S. directed the project at KCL, and H.J., J.L., and Q.F. directed the project at BGI-Shenzhen. H. Xie, R.G., H.Z., Z.L., B.Q., and X.C. performed the bioinformatic analyses and prepared figures and texts for the manuscript. B.C. and H. Xia also helped checking the phenotypes. H.J., T.D.S., H.Z., J.T.B., and M.A.J. wrote the manuscript. All authors contributed to the revision of the manuscript.
References
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Stojanović O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanović A, Hagemann S, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: An integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. ANR MicroObes consortium Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. PLoS ONE. 2015;10:e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloger M, El Karoui M, Petit M-A. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009;191:91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity (Edinb.) 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341 doi: 10.1126/science.1237439. 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:E2930–E2938. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JKK, Waters JLL, Poole ACC, Sutter JLL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JTT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic Determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345 doi: 10.1126/science.1254009. 1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species article. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4 doi: 10.1038/ncomms3151. 2151. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikäinen LP, Kangas AJ, Soininen P, Würtz P, Silander K, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2013;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultima JR, Sunagawa S, Li J, Chen W, Chen H, Mende DR, Arumugam M, Pan Q, Liu B, Qin J, et al. MOCAT: A metagenomics assembly and gene prediction toolkit. PLoS ONE. 2012;7:e47656. doi: 10.1371/journal.pone.0047656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0115175. e115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. MetaHIT consortium Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, Hutník K, Royrvik EC, Cunliffe B, Lawson DJ, et al. Wellcome Trust Case Control Consortium 2 The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. MetaHIT Consortium An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. MetaHIT Consortium. [DOI] [PubMed] [Google Scholar]
- Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, Huerta-Cepas J, Nieuwdorp M, Salojarvi J, Voigt AY, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- Liebman M, Al-Wahsh IA. Probiotics and other key determinants of dietary oxalate absorption. Adv Nutr. 2011;2:254–260. doi: 10.3945/an.111.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. 2014;5 doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MCY, Prokopenko I, et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A, Hivert M-F, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu C-T, Bielak L, Prokopenko I, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JWM, Wang X, Kachman SD, Auwerx J, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0039191. e39191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende DR, Sunagawa S, Zeller G, Bork P. Accurate and universal delineation of prokaryotic species. Nat Methods. 2013;10:881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- Menni C, Fauman E, Erte I, Perry JRB, Kastenmüller G, Shin SY, Petersen AK, Hyde C, Psatha M, Ward KJ, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Wellcome Trust Case Control Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Maes HHM. Methodology for Genetic Studies of Twins and Families (Kluwer) 1992 [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, et al. MetaHIT Consortium Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. MetaHIT Consortium. [DOI] [PubMed] [Google Scholar]
- O’Connor A, Quizon PM, Albright JE, Lin FT, Bennett BJ. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm Genome. 2014;25:583–599. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, Mehrabian M, Pan C, Knight R, Gunsalus R, et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25:1558–1569. doi: 10.1101/gr.194118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KSS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- Rausch P, Rehman A, Künzel S, Häsler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A, Sina C, Gavrilova O, Häsler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Quaye L, Spector TD, Nestle FO, Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y. The genetic architecture of the human immune system : A bioresource for autoimmunity and disease pathogenesis resource. Cell. 2015;161:1–17. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Mägi R, Strawbridge RJ, Rehnberg E, Gustafsson S, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol. 2002;64:479–498. [Google Scholar]
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10:1196–1199. doi: 10.1038/nmeth.2693. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359–1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Taras D, Simmering R, Collins MD, Lawson PA, Blaut M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52:423–428. doi: 10.1099/00207713-52-2-423. [DOI] [PubMed] [Google Scholar]
- Teucher B, Skinner J, Skidmore PML, Cassidy A, Fairweather-Tait SJ, Hooper L, Roe MA, Foxall R, Oyston SL, Cherkas LF, et al. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Res Hum Genet. 2007;10:734–748. doi: 10.1375/twin.10.5.734. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyakht AV, Kostryukova ES, Popenko AS, Belenikin MS, Pavlenko AV, Larin AK, Karpova IY, Selezneva OV, Semashko TA, Ospanova EA, et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun. 2013;4 doi: 10.1038/ncomms3469. 2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitasalo L, Niemi L, Ashorn M, Ashorn S, Braun J, Huhtala H, Collin P, Mäki M, Kaukinen K, Kurppa K, Iltanen S. Early microbial markers of celiac disease. J Clin Gastroenterol. 2014;48:620–624. doi: 10.1097/MCG.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5 doi: 10.1128/mBio.00889-14. e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, Partanen J, Aranko K, Mättoö J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jia H. Metagenome-wide association studies: Fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking longterm dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: Interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–W415. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2015-309800. Published online September 25, 2015. [DOI] [PubMed] [Google Scholar]
- Yuan W, Xia Y, Bell CG, Yet I, Ferreira T, Ward KJ, Gao F, Loomis AK, Hyde CL, Wu H, et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat Commun. 2014;5 doi: 10.1038/ncomms6719. 5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapala MA, Schork NJ. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci USA. 2006;103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Boöhm J, Brunetti F, Habermann N, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- Zhou F, Cao H, Zuo X, Zhang T, Zhang X, Liu X, Xu R, Chen G, Zhang Y, Zheng X, et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat Genet. 2016;48:740–746. doi: 10.1038/ng.3576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.