Abstract
Lysyl oxidase like-2 (LOXL2) belongs to the lysyl oxidase (LOX) family, which comprises Cu2+- and lysine tyrosylquinone (LTQ)-dependent amine oxidases. LOXL2 is proposed to function similarly to LOX in the extracellular matrix (ECM) by promoting crosslinking of collagen and elastin. LOXL2 has also been proposed to regulate extracellular and intracellular cell signaling pathways. Dysregulation of LOXL2 has been linked to many diseases, including cancer, pro-oncogenic angiogenesis, fibrosis and heart diseases. In this review, we will give an overview of the current understandings and hypotheses regarding the molecular functions of LOXL2.
Keywords: lysyl oxidase family, lysyl oxidase like-2, lysine tyrosylquinone (LTQ), cell signaling, extracellular matrix, tumor metastasis/invasion
1. Introduction
The lysyl oxidase (LOX) family comprises five genes: lox (LOX), loxl1 (lysyl oxidase like-1, LOXL1), loxl2 (lysyl oxidase like-2, LOXL2), loxl3 (lysyl oxidase like-3, LOXL3), and loxl4 (lysyl oxidase like-4, LOXL4) (1–6). LOX family members possess a conserved carboxy-terminal (C-terminal) amine oxidase catalytic domain, which includes a His-X-His-X-His copper binding motif and a lysine tyrosylquinone (LTQ) cofactor (2,3,6–9). LOX is traditionally known to catalyze the oxidative deamination of the ε-amino group of lysines and hydroxylysines in collagen and elastin to promote crosslinking of these molecules (Scheme 1), which is essential for the tensile strength of ECM (9,10). In addition to this traditional role, LOX is proposed to have functions in transcriptional regulation, modulation of cell signaling pathways and cell adhesion (11).
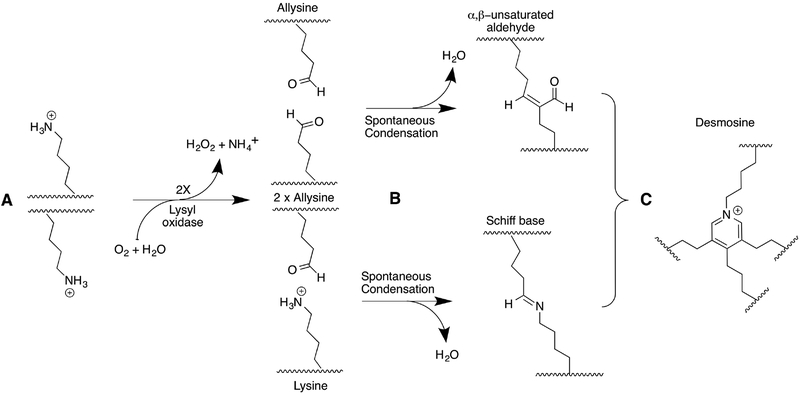
Scheme 1: LOX-initiated crosslink formation in tropocollagen and tropoelastin.
A) Lysyl oxidase catalyzes the oxidative deamination of lysine and hydroxylysine residues in tropocollagen and tropoelastin. For simplicity, only lysine residues are shown in this scheme. B) The product allysine residues spontaneously react with other allysine or lysine residues via aldol condensation or Schiff base formation. C) The bifunctional condensation products can further crosslink to form tri-, tetra-, or even pentafunctional crosslinks. Depicted is desmosine, a common tetrafunctional crosslink found in elastin. Adapted from (87).
Although all LOX family members have >50% sequence identity in the C-terminal catalytic domain (degrees of similarity and homology are summarized in Table 1), the family can be divided into two subgroups based on the primary structure of their N-termini. LOX and LOXL1 contain a highly basic propeptide (pI > 8) at their N-termini, while LOXL2, LOXL3 and LOXL4 each possess four scavenger receptor cysteine-rich (SRCR) domains at their N-termini (Fig. 1) (1,2,6–8,12). The propeptide of LOX undergoes proteolytic cleavage between Gly167 and Asp169 by bone morphogenetic protein-1 (BMP-1) in the ECM (Fig. 1) (13,14). LOXL1 has also been shown to undergo processing by BMP-1, since treatment of 56-kDa LOXL1 isolated from bovine aorta with BMP-1 produced several smaller LOXL1s (52-, 42-, and 37-kDa) (15). Additionally, the endogenous LOXL1 (~60-kDa) secreted from rat lung fibroblast (RFL-6) cells was shown to undergo processing in the culture medium to a mature form (~31-kDa) that was similar in size to secreted endogenous LOX (16). However, in this latter study, the proteolytic site was not examined. In any case, proteolytic cleavage has been shown to activate the catalytic domain of LOX and LOXL1 in the ECM (15–17). The BMP-1 cleavage sites are not conserved between LOX and LOXL1 (15,16), nor are these sites conserved in LOXL2–4.
Table 1.
Identity and homology of LOX catalytic domain in LOX family proteins
| LOX | LOXL1 | LOXL2 | LOXL3 | LOXL4 | |
|---|---|---|---|---|---|
| LOX | 77 (88) | 49 (68) | 52 (67) | 52 (67) | |
| LOXL1 | 77a (88)b | 48 (66) | 54 (66) | 51 (64) | |
| LOXL2 | 49 (68) | 48 (66) | 71 (88) | 71 (86) | |
| LOXL3 | 52 (67) | 54 (66) | 71 (88) | 72 (84) | |
| LOXL4 | 52 (67) | 51 (64) | 71 (86) | 77 (84) |
% of identity
% of homology
Figure 1: Cartoon diagram of the LOX family of proteins.
Based on their N-termini, the LOX family of proteins is divided into two subgroups. LOX and LOXL1 constitute one subfamily possessing a highly basic propeptide sequence that is proteolytically removed by bone morphogenetic protein-1 (BMP-1). The exact site of BMP-1 cleavage in LOXL1 is not defined. Additionally, LOXL1 has a proline-rich domain within its propeptide. The other subfamily comprises LOXL2–4, each of which contains 4 scavenger receptor cysteine-rich (SRCR) domains instead of a propeptide. All LOX family members possess a C-terminal amine oxidase catalytic domain containing a putative copper-binding site (His-X-His-X-His), as well as an LTQ cofactor formed from conserved Lys and Tyr residues by post-translational modification. Each member also has an N-terminal secretion signal. LOX and LOXL2–4 contain predicted N-linked glycosylation sites (i.e. Asn-X-Ser/Thr). From (50).
In contrast to LOX and LOXL1, LOXL2–4 each contain four Group A SRCR domains; each 90–115 amino acid SRCR domain contains six cysteine residues (18). The precise biological function of SRCR domains is still undetermined; however, they are generally thought to mediate homotypical or heterotypical protein-protein interactions in the extracellular matrix, and a substantial fraction of proteins containing SRCR domains act as extracellular pattern recognition receptors (reviewed in (18)).
2. Biochemical properties of LOXL2
There are currently no crystal or NMR structures of LOXL2 nor any members of the LOX family of proteins. In general, the biochemical characterization of LOXL2 has been severely hampered due to the lack of expression and purification methods for producing/isolating recombinant proteins suitable for conducting such studies. This is mostly due to the high content of Cys in LOXL2 (i.e. 10 Cys in the catalytic domain and a cumulative 24 Cys in the SRCR domains). Kim et al. produced a series of C-terminally His-tagged recombinant LOXL2s (rLOXL2s) with differing numbers of SRCR domains in Escherichia coli (E. coli) (19). These rLOXL2s were isolated as inclusion bodies and required denaturation with 6 M urea, refolding and activation with Cu2+. The activities of the resulting rLOXL2s were tested using collagen (types I, III, IV) and elastin as substrates. The SRCR domains did not have any effect on modulating the catalytic activity of LOXL2. Additionally, the activity of the rLOXL2s was not inhibited by 1 mM β-aminopropionitrile (BAPN), a potent in vitro inhibitor of LOX (Ki = 6 μM at 37°C) (20), even at 1000-fold concentration. The authors proposed that the active site of LOXL2 is structurally different from that of LOX.
Vadasz et al. were the first to express full-length rLOXL2 in Chinese hamster ovary (CHO) cells (21). The rLOXL2 was secreted into the culture medium and was detected by immunoblotting as two isoforms (~100 kDa and ~65 kDa). The rLOXL2 was not isolated from the medium for further biochemical analysis. Instead, oxidation of collagen by the rLOXL2 was checked using crude conditioned medium, and it was confirmed that the crude medium containing rLOXL2 oxidized substantially more collagen than crude medium from LOXL2-negative cells or from LOXL2-expressing cells incubated with 0.1 mM D-penicillamine, a Cu2+- chelator. However, oxidation of collagen by rLOXL2 was not inhibited by the addition of 390 μM BAPN.
Hollosi et al. expressed full-length rLOXL2 in immortalized nontransformed mammary epithelial (MCF-10A) and non-metastatic breast cancer (MCF-7) cells (22). rLOXL2 was detected in the culture media as a mixture of ~100-kDa and ~60-kDa proteins. The concentration of ~60-kDa rLOXL2 increased in the media in a time-dependent fashion, and a cocktail of protease inhibitors was shown to inhibit the processing. These results indicated that the ~60-kDa form was the proteolytic product of the ~100-kDa rLOXL2 in the culture media. In this study, the media fraction of the stable cells was directly used for the activity assay. The activity of rLOXL2-containing culture medium towards 1,5-diaminopentane was 1.5- to 3-fold higher than those without rLOXL2.
The kinetic parameters (Km ≈ 1 mM and kcat ≈ 0.02 s−1) of LOXL2 in the oxidation of 1,5-diaminopentane and spermine in solution (pH 8.0) were first determined for a commercially available C-terminally His-tagged rLOXL2 (a mixture of ~87-kDa and ~60-kDa forms) produced in murine myeloma cells (23). BAPN was found to inhibit the oxidation of 1,5-diaminopentane (IC50 = 5.0 ± 1.4 μM) and spermine (IC50 = 3.8 ± 0.2 μM) by this rLOXL2. Additionally, a murine monoclonal antibody (AB0023) specifically targeting the fourth SRCR domain of LOXL2 was shown to inhibit the amine oxidase activity of the commercially available rLOXL2 in vitro (IC50 = 62 ± 6 nM for 1,5-diaminopentane; 55 ± 11 nM for spermine; and 61 ± 4 nM for collagen I). It was proposed that AB0023 is an allosteric regulator of the LOX catalytic domain of LOXL2. In a subsequent study, AB0023 was shown to effectively impede tumor growth in primary and metastatic xenograft models of breast and ovarian cancer. Additionally, treatment with AB0023 inhibited or even reversed lung and liver fibrosis in mouse models of these diseases. Collectively, these data implicate that secreted LOXL2 has potential as a therapeutic target in some cancers and fibrotic diseases (24).
Our group produced C-terminally StrepII-tagged rLOXL2s lacking the first three or all four SRCR domains (i.e. Δ1–3SRCR- and Δ1–4SRCR-LOXL2s) in the culture medium of Drosophila Schneider 2 cells (25). This was the first time that highly pure (> 95%) and soluble rLOXL2s had been isolated. The presence of the LTQ cofactor, its precursor residues (Lys653 and Tyr689), and N-linked glycans at Asn455 and Asn644 were determined by mass spectrometry (25,26). The N-glycosylation site at Asn644 in the LOX catalytic domain of LOXL2 is unique to the SRCR domain-containing LOXLs (LOXL2, LOXL3 and LOXL4); it is not conserved in LOX or LOXL1. However, the N-glycosylation site at Asn455 in the fourth SRCR domain is unique to LOXL2. Disruption of N-glycosylation at either site by site-directed mutagenesis (N455Q, N644Q mutants of rLOXL2) or treatment with tunicamycin (an inhibitor of UDP-GlcNAc—dolichylphosphate GlcNAc-1-phosphate transferase) (27,28) revealed that the N-linked glycans at Asn455 and Asn644 are independently essential for protein folding and secretion of rLOXL2 from S2 cells.
The two truncated rLOXL2s competently catalyze the amine oxidation in vitro (25). For tropoelastin oxidation, the Km values were determined to be 0.59 ± 0.13 μM (for Δ1–3SRCRLOXL2) and 0.62 ± 0.17 μM (for Δ1–4SRCR-LOXL2), and kcat values were determined to be 2.04 ± 0.17 min−1 and 0.69 ± 0.07 min−1, respectively, at pH 8.0. These values are comparable to the reported values for native LOX from bovine aorta (29,30), suggesting that the LOX family of proteins comprises intrinsically “slow” enzymes. In addition, the data suggest that the 4th SRCR domain plays a positive allosteric role in modulating the activity of the LOX catalytic domain of LOXL2.
3. The role of Cu2+ in the amine oxidase activity of LOXL2
Members of the LOX family of proteins possess three highly conserved His (His626-XHis628-X-His630 in LOXL2) in the LOX catalytic domain that are proposed to be the binding site for Cu2+ (12). These His are postulated to form an octahedral coordination complex with Cu2+ based on an X-band EPR spectrum of Cu2+-containing LOX isolated from bovine aorta (bLOX) (31). The role of Cu2+ in the catalysis of LOX remains controversial; whether Cu2+ plays a redox role is debated (31,32). Gacheru et al proposed that Cu2+ plays an essential role in the catalytic cycle of bLOX (31), as originally proposed for copper amine oxidases that contain 2,4,5-trihydroxyphenylalanine quinone (TPQ), another tyrosine-derived quinone cofactor. They removed Cu2+ from bLOX using a copper-chelator (2,2’-bipyridyl) in the presence of 6 M urea, and found that the activity of bLOX toward elastin, n-hexylamine, and p-hydroxybenzylamine was completely abrogated. Reconstitution of the apo-form of bLOX with 1 mole Cu2+ per mole enzyme restored 96% of the original activity toward elastin, and also restored the ability to oxidize p-hydroxybenzylamine. In contrast, Tang et al removed copper from bLOX by a similar chelation process, and reported that the apo-bLOX retained 65% phenylhydrazine-titratable LTQ cofactor and catalyzed multiple turnovers of benzylamine at 50–60% of the rate of WT-bLOX. The activity of apo-bLOX was completely abolished by BAPN. Consequently, the authors concluded that Cu2+ is not required for the oxidation of some small amine substrates or hydrazone (azo) formation with phenylhydrazine. Instead, they proposed that Cu2+ plays a structural role to stabilize the LTQ cofactor or the enzyme. It should be noted that the authors reported that a UV-vis spectroscopic change accompanied the removal of Cu2+, and suggested that the LTQ cofactor underwent some modification. The exact nature and consequence of this modification has yet to be determined.
In order to clarify whether the catalytic activity of LOXL2 is required for the Snail1-mediated repression of E-cadherin (discussed in greater detail later in this review) (33), a double mutant of rLOXL2 in which two of the three conserved His residues were mutated to Gln (i.e. H626Q/H628Q) was expressed in human embryonic kidney 293T (HEK293T) cells (34). The activity of the H626Q/H628Q mutant was assessed using rLOXL2 immunoprecipitated from cell extracts. The double mutant exhibited no observable catalytic activity in the oxidation of benzylamine, as expected.
4. Biogenesis of LTQ in LOXL2
Phenylhydrazine-derivatized LTQ was originally identified in bLOX by mass spectrometry and resonance Raman spectroscopy (9). Sequence alignment of bLOX with human LOXL2 indicates that Lys653 and Tyr689 are the predicted precursors for the LTQ crosslink in LOXL2. The presence of LTQ in LOXL2 was determined by electrospray mass spectrometry after trypsin digestion of the phenylhydrazine-derivatized form of Δ1–3SRCR-LOXL2 (25). The result confirmed that Lys653 and Tyr689 are the precursors for the LTQ cofactor in LOXL2.
In both Δ1–3SRCR-LOXL2 and Δ1–4SRCR-LOXL2, ~20% of the LTQ could be titrated with phenylhydrazine (Moon et al., unpublished data). These results implicate that the 4th SRCR domain does not play a significant role in the biogenesis of the LTQ cofactor in LOXL2. The modest level of LTQ biogenesis is not unprecedented for tyrosine-derived quinone cofactors. Titration with phenylhydrazine revealed only ~50% formation of the LTQ cofactor in native LOX isolated from bovine calf aorta (32). Also, in recombinant CAOs from yeast and human cells only ~20% TPQ (another tyrosine-derived quinone cofactor) formation was detected (35,36). Moreover, neither recombinant D. melanogaster lysyl oxidase-like 1 (rDmLOXL-1) produced in S2 cells nor rLOXL2 produced in E. coli contains LTQ upon initial isolation. These recombinant proteins must undergo denaturation, chelation, and reactivation with Cu2+ to produce ~50% and ~20% LTQ, respectively (37,38). However, this activation system did not increase the amount of phenylhydrazine-titratable TPQ cofactor in a human CAO (36), and has likewise been ineffective in increasing the phenylhydrazine-titratable LTQ cofactor in our rLOXL2s.
The LTQ cofactor is proposed to undergo autocatalytic biogenesis, requiring only Cu2+ and O2, analogous to the mechanism proposed for the TPQ cofactor biogenesis in CAOs (39,40). In this mechanism, the 1,4-addition of the e-amino group of the conserved Lys to a dopaquinone (DPQ) derived from the conserved Tyr produces the reduced form of the LTQ cofactor (aminocatechol). The subsequent O2 oxidation of the catechol yields the LTQ cofactor (Fig. 2) (9,41). In attempt to trap the DPQ intermediate, a Lys residue was incorporated into the active site of a bacterial CAO by site-directed mutagenesis (42). X-ray crystallography revealed that the mature mutant (D298K) contained an LTQ-like quinone (Fig. 3), strongly supporting the common intermediacy of DPQ in LTQ and TPQ biogenesis. The electron density of the LTQ-like quinone indicated that the quinone is stabilized as the para-iminoquinone tautomer via a hydrogen bond interaction with the conserved Tyr in the active site, resulting in a ~50 nm blue-shift in λmax (Scheme 2). The λmax at ~502 nm observed in our rLOXL2 and in purified bLOX (9) suggest that such a hydrogen bond interaction is absent in the active site of rLOXL2 and bLOX.
Figure 2: Structure of the LTQ cofactor and proposed pathway of LTQ cofactor biogenesis.
A) Structure of LTQ, the tyrosine-derived cofactor of LOXL2. B) The proposed mechanism for the biogenesis of the LTQ cofactor. Steps: (i) autocatalytic oxidation of the precursor peptidyl tyrosine residue by O2 and Cu2+; (ii) O2 oxidation of dihydroxyphenylalanine (DOPA) to dopaquinone (DPQ); (iii) 1,4-addition of the ε-amino group of the side chain of a peptidyl lysine residue (Lys653 in human LOXL2) to DPQ yields the reduced form of LTQ, where neutral amino side chain of Lys653 is a nucleophile; (iv) Oxidation of reduced form of LTQ to LTQ by O2. Adapted from (9,41).
Figure 3. X-ray crystal structure of LTQ-like quinone in D298K mutant of a bacterial CAO.
Active site structures of mature D298K-AGAO and the putative DPQ intermediate detected during X-ray snapshot analysis of TPQ biogenesis in WT-AGAO. (A) DPQ intermediate (PDB: 1IVV)(88), (B) D298K (PDB: 2YX9). Cu2+ is shown as an orange sphere, water molecules are shown as light-blue spheres, hydrogen bonding interactions are represented by blue lines, and ligand interactions are represented by purple lines. Val282 and Asn381 (white) form the edges of a wedge-shaped pocket. Hydrogen bonding distances are denoted in angstroms. From (42).
Scheme 2: LTQ-LTI tautomerism.
The LTQ-like quinone (λmax=504 nm) formed in D298KAGAO is in equilibrium with its iminoquinone tautomer (LTI, λmax=454 nm). The LTI form is thermodynamically favored as it is stabilized by the hydrogen bonding interaction with the conserved Tyr284 in the active site. When the hydrogen bonding interaction was disrupted via site-directed mutagenesis (i.e. D298K/Y284F-AGAO), only the LTQ-like form was detected. From (42).
To examine whether LTQ cofactor formation is a prerequisite for maturation of LOXL2 (i.e. proper folding, N-glycosylation and secretion), MCF-7 cells stably expressing K653R- and K653S mutants of rLOXL2 were selected (43). K653R- and K653S-LOXL2 are expected to lack the LTQ cofactor. Similar to wild-type (WT) LOXL2, K653R-LOXL2 (~100-kDa) was secreted, whereas K653S-LOXL2 (~75-kDa) was detected exclusively in the soluble cell lysate. These data demonstrate that when the positive charge is conserved at residue 653 (i.e. K653R), the LOXL2 maturation pathway is not sufficiently sensitive to distinguish between the precursor form of LOXL2 (containing unmodified Lys and Tyr) and mature LOXL2 (containing the LTQ cofactor). This may account for the sub-stoichiometric amount of LTQ cofactor detected in recombinant and native LOX(L)s, as described earlier. Interestingly, in the absence of the positive charge (i.e. K653S), N-glycosylation was completely inhibited and the K653S-LOXL2 was only detected in the soluble cell lysate. The elimination of the positive charge in the active site seems to induce some conformational change that impedes N-glycosylation and ultimately prevents secretion of LOXL2.
5. LOXL2 in Disease
Dysregulation of the LOX-family of proteins is strongly associated with heritable connective tissue and fibrotic disorders (44–46), cardiovascular diseases (47,48), and cancers. In cancers, the LOX-family of proteins is thought to play multiple roles in processes important to cancer progression, namely cell growth, adhesion, motility, and invasion (49,50).
The correlation between LOXL2 expression and tumor progression is dependent upon tissue type. LOXL2 expression is decreased in ovarian tumors (51,52). However, increased LOXL2 expression is associated with poor prognosis in patients with colon and esophageal tumors (53), as well as oral squamous cell carcinomas, laryngeal squamous cell carcinomas (24,54), and head and neck squamous cell carcinomas (55). Additionally, increased LOXL2 expression has been found to promote gastric cancer (56) and breast cancer metastasis (57). Some highly invasive human breast cancer cell lines are reported to have elevated levels of LOXL2 mRNA (33,58).
Recently, LOXL2 has been considered as a promising therapeutic target for invasive/metastatic breast cancers (56,59). LOXL2 is almost absent in poorly invasive/non-metastatic MCF-7 breast cancer cells, but is highly expressed in invasive/metastatic MDA-MB-231, MDA-MB-435, and 4T1 breast cancer cells (57,60). Inhibition of the production or activity of LOXL2 by shRNA, small molecule inhibitors or AB0023 reverted the phenotype of these cells and reduced their invasiveness in vitro and in vivo (animal models) (24,33,56). In addition, when MCF-7 cells stably expressing LOXL2 were transplanted into the fourth mammary fat pad of nude mice, they produced tumors with many fibrotic foci that were capable of invading surrounding blood vessels, nerves and muscle tissue (57). These results strongly suggest roles for LOXL2 in oncogenic cell signaling pathways.
6. Secreted LOXL2 in Breast Cancer Metastasis/Invasion
Because the oncogenic functions of LOXL2 have been most studied in the context of breast cancers, the remainder of this review will focus on recent discoveries in this field, with references to other pathological contexts, where appropriate.
An understanding of the functions of secreted LOXL2 in promoting breast cancer invasion and metastasis has begun to emerge in the last fifteen years. Akiri et al. were the first to report that tumors derived from MCF-7 breast cancer cells ectopically expressing rLOXL2 exhibited increased invasiveness in vivo (i.e. in nude mice)(57). Consistent with this, Hollosi et al. later reported that ectopic expression of rLOXL2 in MCF-7 cells and MCF-10A normal breast epithelial cells induces epithelial-to-mesenchymal transition (EMT) in these cells (22). EMT is a cellular process in which epithelial cells lose their characteristic cell-cell adhesions and cell polarity, and acquire invasive and migratory traits generally associated with the mesenchymal phenotype (Fig. 4) (61–63). EMT is an important process in the progression of localized tumors, as it enables immobile epithelial cells within the tumor to transform into cells that are capable of invading adjacent and distant tissues. EMT can be induced by growth factors and ECM components, and the progression of EMT is coordinated by complex networks of signaling proteins and transcription factors (reviewed in (64)). Consequently, most of the research performed since Hollosi et al.’s discovery of the positive correlation between LOXL2 expression and the onset of EMT has sought to define the signaling pathways in which LOXL2 participates to contribute to the induction of EMT (22).
Figure 4: Epithelial-to-Mesenchymal transition (EMT).
EMT is a process in which epithelial cells are transformed into mesenchymal cells. Changes in the tumor microenvironment (e.g. growth factors, cytokines, and extracellular matrix components) have been shown to induce EMT. During EMT, cells downregulate expression of cell-cell adhesion and cell polarity proteins, while upregulating expression of proteins that confer invasive and migratory traits. Adapted from (61).
However, elucidating a discrete role for secreted LOXL2 in breast cancer invasion has been complicated by the fact that LOX protein expression is also elevated in some of the invasive cell lines commonly used to study the functions of LOXL2 (e.g. MDA-MB-231, MDAMB-435 and 4T1 cells) (24,65,66). Stiffening of the ECM (via LOX-induced crosslinking of collagen and elastin) has been linked to alteration of cellular mechanotransduction through activation of integrin signaling pathway, leading to EMT and cell proliferation and invasion of breast and colorectal cancers (59,67,68). LOXL2 is generally proposed to function similarly to LOX in the ECM to promote stiffening and alteration of the ECM by its LOX amine oxidase activity. To support this, LOXL2 has been shown to colocalize with stroma-activated fibroblasts and fibrillar collagen in biopsies of human colon and larynx squamous cell carcinoma tissues (24). LOXL2 is also proposed to be responsible for the aberrant deposition level of dense collagen around hepatocytes in Wilson’s disease, and primary biliary cirrhosis (21). It can be challenging to differentiate the contributions made to EMT and tumor progression by LOX and LOXL2, since they are often present in the same pathological microenvironment.
In order to compare the potencies of secreted LOXL2 and secreted LOX in breast cancer metastasis/invasion, tumors generated from MDA-MB-435 breast cancer cells (expressing both LOX and LOXL2) were transplanted in nude mice (24). The tumors were then treated with AB0023 (an antibody specific for the 4th SRCR domain of LOXL2; AB0023 was described earlier in this review) or M64 (a LOX-specific antibody against a peptide in the LOX catalytic domain). Treatment with AB0023 was effective in reducing tumor volume, the extent of crosslinked collagen, and microvessel density. AB0023 also inhibited the activation of fibroblasts. In contrast, M64 was virtually ineffective in all these respects. In vitro, the addition of conditioned medium from MDA-MB-231 cells (expressing endogenous LOX and LOXL2) was shown to induce EMT-like morphological change in MCF-7 cells. Treatment with AB0023 was sufficient to revert the phenotype change. Addition of conditioned medium from HEK cells stably expressing rWT-LOXL2 to MCF-7 cells induced a similar EMT-like morphological change, while the addition of conditioned medium containing a catalytically incompetent rLOXL2 mutant (Y689F) induced no such effect. Based on these results, LOXL2 was proposed to play more significant roles than LOX in ECM remodeling and formation of the tumor microenvironment.
LOXL2 expression has also been linked to upregulation of tissue inhibitor of metalloproteinase-1 (TIMP-1) and MMP-9, thereby promoting ECM degradation to enable subsequent metastatic dissemination of MDA-MB-231 and 4T1 cells (69). Additionally, LOXL2 secreted from invasive breast cancer cells has been shown to activate stroma-derived fibroblasts, possibly through activation of FAK/Src (59,65). In both studies, reactive oxygen species (ROS) produced by LOXL2 during the amine oxidation of the ECM substrates were proposed to activate ErbB2/Erk2 and FAK/Src signaling pathways, the mechanism originally proposed for LOX (Fig. 5A) (11,66,67,70). In MCF-10A cells, rLOXL2 was also shown to promote cell invasion via activation of ErbB2 (HER2), a cell surface receptor for human epidermal growth factor (59).
Figure 5: Proposed roles for LOXL2 in breast cancer metastasis and invasion.
A) Proposed mechanism whereby secreted LOXL2 induces ECM stiffening to activate oncogenic signaling pathways. B) Proposed mechanism whereby intracellular LOXL2 regulates Snail1 or H3K4me3 to repress E-cadherin expression, eventually leading to EMT.
Because the ability of α-smooth muscle actin (α-SMA)-positive fibroblasts (i.e. activated fibroblasts) to promote tumor progression has been demonstrated in breast, prostate, pancreatic, and skin cancer mouse models (65,71–76), Barker et al. investigated whether LOXL2 secreted from breast cancer cells could activate cancer-associated fibroblasts (CAFs) (65). The authors determined that tumors derived from orthotopically implanted 4T1 mouse breast cancer cells (expressing high levels of secreted LOXL2) displayed substantially greater α-SMA expression than tumors derived from cells in which the expression of LOXL2 was suppressed by shRNA. A positive correlation between secreted LOXL2 concentration and several traits of activated fibroblasts was also demonstrated, and addition of a LOXL2-specific antibody to the growth medium decreased the degree of activation. The authors hypothesized that secreted LOXL2 was activating the fibroblasts by engaging integrins and components of focal adhesion complexes. This hypothesis was supported by data showing that α-SMA and activated forms of FAK and Akt were upregulated when cells were cultured in the presence of secreted LOXL2. Consistent with this hypothesis, an antibody against β3 integrin abrogated this effect. Finally, the authors demonstrated that activated fibroblasts expressed ~1.8-fold greater LOXL2 mRNA, suggesting that activation of fibroblasts by tumor-derived LOXL2 could initiate a feed-forward loop to further stimulate tumor progression.
These reports have substantially advanced knowledge of the functions of LOXL2 in breast cancer progression; however, there are some critical issues that need to be addressed in defining the functions of LOXL2 in the ECM. To date, there has been no biochemical study to evaluate the proposed function of LOXL2 in oxidizing collagen and elastin to initiate their crosslinking, leading to stiffening of the ECM. In addition, the activation of ErbB2/Erk2 and FAK/Src pathways by ROS could be attributed to both LOX and LOXL2. As a consequence, further studies are necessary to evaluate the proposed functions of LOXL2 in the ECM of the tumor microenvironment and disentangle them from the roles of LOX.
7. Intracellular Functions of LOXL2 in Breast Cancer Metastasis/Invasion
Intracellular functions of LOXL2 in breast cancer metastasis/invasion have also been proposed (Fig. 5B), owing to the detection of perinuclear expression patterns of LOXL2 in basal-like (invasive/metastatic) breast and laryngeal squamous cell carcinomas (54,77,78). LOXL2 has been proposed to induce EMT by downregulating E-cadherin, either by stabilizing Snail1 transcription factor or deaminating trimethylated Lys4 of histone H3 [H3K4me3] (33,79). Snail1 is one of the four essential EMT-activating transcription factors and its stability is regulated by its phosphorylation status (80,81). Peinado et al reported that LOXL2 interacts with the N-terminal SNAG domain of Snail1, and that this interaction is essential for LOXL2-induced stabilization of Snail1 at the protein level (33). Among the conserved Lys residues in the Snail family of proteins, four of them (i.e. Lys9, Lys16, Lys98 and Lys137) are within the N-terminus (i.e. not in the C-terminal zinc-finger domain). The authors correlated the interaction between LOXL2 and a series of Lys-to-Arg single and double mutants of Snail1 with the degree of E-cadherin suppression. They discovered that Lys98 and Lys137 are essential for Snail1 stabilization, E-cadherin repression, and induction of EMT. Thus, they proposed that LOXL2 oxidizes Lys98 and/or Lys137 to induce some undefined conformational change that protects Snail1 from GSK3b-catalyzed phosphorylation and subsequent proteasomal degradation (Fig. 6).
Figure 6: Proposed mechanism of LOXL2 oxidation and stabilization of Snail1.
Intracellular LOXL2 is proposed to deaminate Lys98 and Lys137 in the SNAG domain of Snail1. Following the modification by LOXL2, an undetermined conformational change in Snail1 inhibits phosphorylation by GSK3β and ultimately suppresses the degradation of Snail1.
An alternative intracellular role for LOXL2 was proposed by Herranz et al., who reported that ectopic expression of LOXL2 in MCF-7 cells results with the downregulation of trimethylated H3K4 [H3K4me3] in the E-cadherin promoter (79). H3K4 trimethylation is generally associated with actively transcribed genes (reviewed in (82)). The authors reported that the LOXL2-catalyzed downregulation was specific for H3K4me3; dimethylated and monomethylated H3K4, as well as Lys9(me3) and Lys27(me3), apparently did not serve as substrates for intracellular LOXL2. Furthermore, downregulation of H3K4me3 was not detected in MCF-7 cells expressing a catalytically incompetent LOXL2 mutant (the mutation was not specified), indicating that the phenomenon is dependent upon LOX amine oxidase activity. To analyze the molecular mechanism by which H3K4me3 is downregulated, the authors isolated Flag-tagged WT and catalytically inactive mutant rLOXL2 from Sf9 insect cells. These rLOXL2s were then incubated with various H3 peptides. Based on the results of attenuated total reflection-Fourier transform infrared spectroscopy (ATR-FTIR) and mass spectrometry analyses, the authors proposed a novel mechanism whereby LOXL2 downregulates H3K4me3 via two steps: formation of an alcohol via deamination (cleavage of the terminal trimethylamine), and subsequent oxidation of the product alcohol to an aldehyde (Fig. 7A). This mechanism is distinct from that of the Jumonji C (JMJC)-containing family of demethylases, which employ an Fe2+- and α-ketoglutarate-dependent dioxygenase reaction to catalyze demethylation of H3K4me3 (reviewed in (83,84)) (Fig. 7B). Moreover, it should be noted that there is no precedent for LOX family members or the related CAOs catalyzing deamination of a trimethylamino group or oxidation of alcohols.
Figure 7: Proposed mechanisms for regulating methylated H3K4.
A) A novel mechanism proposed by Herranz et al. whereby LOXL2 deaminates H3K4(me3) via two steps: formation of an alcohol via deamination, and subsequent oxidation of alcohol to aldehyde. From (79). B) A scheme of the mechanism by which a Jumonji C (JMJC)-containing lysine demethylase, which is an Fe2+- and α-ketoglutarate-dependent dioxygenase, catalyzes demethylation of H3K4me3. Adapted from (84). C) A proposed mechanism for lysine side chain oxidation by LOXL2.
Intracellular LOXL2 has also been proposed to regulate cell polarity in basal breast cancer cells by disrupting tight junctions (such as claudin1, occludin, and ZO-1) and cell polarity complexes (such as Lgl2 and Par3) (77). Tight junction proteins play critical roles in maintaining the cell polarity. It was proposed that LOXL2 interferes with the expression and organization of tight junction and cell polarity complexes by transcriptionally downregulating cell polarity (LLGL2) and tight junction (CLDN1) genes independently of Snail1. Additionally, silencing LOXL2 with shRNA in MDA-MB-231 cells resulted in upregulation of these genes, accompanied by reversion of phenotype (i.e. mesenchymal-to-epithelial transition). However, no substantial changes in E-cadherin expression were associated with expression of LOXL2, nor was the catalytic domain of LOXL2 required for repression of the LLGL2 or CLDN1 gene promoters. Therefore, repression of these genes represents a new mechanism by which LOXL2 can regulate the phenotype of breast cancer cells.
An added twist which complicates the unambiguous attribution of specific functions to either intracellular LOXL2 or extracellular LOXL2 is the possibility that the perinuclear LOXL2 detected in basal-like breast cancer cells could originate from overexpressed secreted LOXL2. In a recent study of LOXL2 in keratinocyte differentiation, keratinocytes were shown to internalize secreted LOXL2 (85). The authors observed that expression of LOXL2 and differentiation are antagonistic processes in HaCaT skin keratinocytes: LOXL2 promoter activity was downregulated when the cells were cultured in conditions that stimulate differentiation, while the expression of involucrin (a differentiation marker) was suppressed when the cells were forced to overexpress rLOXL2 or were cultured in the presence of exogenous rLOXL2. To determine the relationship between the LOXL2 enzyme activity and suppression of cell differentiation, two mutants lacking either the critical tyrosine precursor for the LTQ cofactor (Y689F-LOXL2) or else the entire catalytic domain were used as controls. The ectopic overexpression of these mutant rLOXL2s in HaCaT cells also inhibited involucrin expression, even under culture conditions that would otherwise stimulate differentiation. Additionally, BAPN was unable to abolish the LOXL2-induced inhibition of HaCaT cell differentiation. These data collectively suggest that the catalytic activity of LOXL2 is not required for its function in suppression of differentiation markers. However, addition of AB0023 (which targets the fourth SRCR domain of LOXL2) to the culture medium inhibited the suppression of involucrin by any of these rLOXL2s. Based on these data, the authors surmised that the 4th SRCR domain of extracellular LOXL2 is responsible for repressing the expression of involucrin.
The authors hypothesized that the 4th SRCR domain might bind to a cell membrane-anchored receptor that transduces inhibitory signals, and predicted that such a receptor might also internalize LOXL2. Their hypothesis was supported by the discovery that HaCaT cells could internalize epitope-tagged Y689F-LOXL2 at 37 °C, and that the internalization was inhibited by AB0023 and excess untagged rLOXL2. These results indicated that internalization of LOXL2 is mediated by a specific high-affinity LOXL2 receptor. It is not unreasonable to predict that this receptor may also be present on the surface of breast cancer cells. Moreover, the discovery that secreted LOXL2 could be re-internalized would not be unprecedented in the LOX family. When fluorescently labeled purified bLOX was added to the culture medium of smooth muscle cells, time-dependent accumulation of mature bLOX in the nuclei was observed over the course of 4 hours (86). Internalization was not prevented when BAPN-inhibited bLOX was used instead of catalytically active bLOX, indicating that catalytic activity is unimportant for the phenomenon, similar to the findings for LOXL2 in HaCaT cells.
Since LOXL2 may be re-internalized, it has been difficult to unambiguously distinguish the functions of secreted LOXL2 from intracellular LOXL2. To accurately understand the molecular roles of LOXL2 in breast cancer, new approaches are urgently needed to dissect the functions of extracellular and intracellular LOXL2s. Toward this end, the post-translational modifications of LOXL2 were recently characterized and used to direct the subcellular localization of the enzyme. Following the discovery that N-linked glycosylation at Asn455 and Asn644 are essential for secretion of truncated rLOXL2s from Drosophila S2 cells (25), similar studies were undertaken in human breast cancer cells (43). Site-directed mutagenesis was employed to produce full-length rLOXL2s with mutations in the three predicted N-linked glycosylation sites: N288Q-LOXL2, N455Q-LOXL2, and N644Q-LOXL2. These mutants were stably overexpressed in MCF-7 cells, and the rLOXL2s were characterized. N288Q-LOXL2 was secreted into the culture medium as an ~100-kDa protein, identical in molecular mass to endogenous LOXL2 from MDA-MB-231 cells and WT rLOXL2 secreted from MCF-7 cells. This suggests that Asn288 is most likely not N-glycosylated. In contrast, N455Q- and N644QLOXL2 were not secreted, and were instead only found in the soluble cell lysate as ~75-kDa proteins, as determined by SDS-PAGE and protein immunoblotting. Treatment of MDA-MB-231 cells with tunicamycin, an N-linked glycosylation inhibit6or, also prevented secretion of endogenous LOXL2. These data confirm that the N-linked glycans at Asn455 and Asn644 are essential for LOXL2 secretion from these luminal and basal type breast cancer cells.
Interestingly, 75-kDa LOXL2 was also was detected the cytosol of MDA-MB-231 cells and MCF-7 cells expressing WT-rLOXL2, even when these cells were not treated with tunicamycin. This might be explained by the aforementioned possibility that secreted LOXL2 could be re-internalized (85). Alternatively, it may indicate that overexpression of LOXL2 saturates the glycosylation pathway, preventing a fraction of the LOXL2 from being glycosylated and forcing it to remain inside the cell. Based on experiments in which the cell nuclei were fractionated from the other intracellular components, each of the 75-kDa LOXL2s (both endogenous and recombinant) has an apparent affinity for the nucleus.
Treating the lysates of MCF-7 cells stably expressing N455Q- or N644Q-LOXL2s with peptide-N-glycosidase F to remove any remaining N-linked glycans had no effect on the molecular mass of the rLOXL2s, suggesting that they completely lack N-glycans. Because WT-LOXL2 is predicted to be ~84-kDa after the signal peptide is removed, the molecular mass of the 75-kDa intracellular LOXL2 suggests that the protein is proteolytically processed. This must occur at the N-terminus, since the C-termini of N455Q- and N644Q-LOXL2 bear a short epitope tag for immunodetection. The specific site of the processing was not determined, due to the presence of a blocking group that prevented N-terminal sequencing.
Upon selecting MCF-7 cells stably transfected with each of the rLOXL2s (i.e. WT, N288Q, N455Q, N644Q, and the catalytically incompetent K653R and K653S mutants), the authors discovered that cells expressing high levels of catalytically active ~75-kDa intracellular LOXL2 (i.e. N455Q- and N644Q-LOXL2s) underwent spontaneous morphological changes indicative of EMT. The morphological changes were accompanied by transcriptional upregulation of mesenchymal markers (e.g. vimentin and fibronectin) and downregulation of epithelial markers (E-cadherin, occludin, claudin-1, and estrogen receptor alpha [ERα]), as would be expected for cells experiencing EMT. Because transcription of the genes encoding E-cadherin, occludin, claudin-1, ERα and fibronectin is known to be influenced by Snail1, the authors also performed qRT-PCR and protein immunoblotting to determine the transcript and protein levels of Snail1 in each cell line. Importantly, Snail1 protein (but not Snail1 mRNA) was upregulated in MCF-7 cells expressing the catalytically active nuclear associated rLOXL2s; however, Snail1 protein was not upregulated in the other cell lines. This finding is consistent with the hypothesis that catalytically active intracellular LOXL2 drives EMT by stabilizing Snail1 against degradation (43).
Finally, a Matrigel in vitro invasion assay demonstrated that MCF-7 cells expressing N455Q or N644Q LOXL2 were highly invasive (even more than MDA-MB-231 cells) in vitro, whereas the other cell lines were not significantly more invasive than MCF-7 cells transfected with an empty vector. The single exception was the cell line expressing WT LOXL2, which will be discussed further below. The increased invasiveness of cells expressing catalytically active nuclear associated LOXL2 was attributed to the increased expression of fibronectin and MT1-MMP, a transmembrane matrix metalloproteinase (MMP) whose gene also happens to be controlled by Snail1. Because the invasion assay was conducted in the absence of fibroblasts, it is likely that MT1-MMP participated directly in ECM degradation.
As mentioned above, MCF-7 cells expressing WT LOXL2 were more invasive than the negative control in vitro. This was accompanied by a low penetrance of morphological transformation and transcriptional changes indicative of EMT; only a very minor population of MCF-7 cells expressing WT-LOXL2 exhibited mesenchymal morphology. This minor population of cells may have contributed to the slightly increased invasiveness of MCF-7 cells expressing WT-LOXL2, compared to the negative controls. It is possible that consequences of overexpressing secreted LOXL2 may require a long time-frame (i.e. a high concentration of LOXL2) and a tumor microenvironment where complex interactions between LOXL2 and proteins secreted by fibroblasts are necessary to induce EMT and promote invasion in vivo (43).
8. Paradox
Two issues concerning LOXL2 are paradoxical. The first issue is whether the amine oxidase activity of LOXL2 is essential to induce EMT. As originally proposed by Peinado et al. (33), we have demonstrated that the LOX amine oxidase activity of LOXL2 plays an essential role in inducing EMT and promoting invasion of MCF-7 breast cancer cells via posttranslational stabilization of Snail1 transcription factor (43). Additionally, Herranz et al. has reported that LOXL2 suppresses EMT via deamination of H3K4me3 in a LOX amine oxidase-dependent fashion (79). In contrast, Cuevas et al. have recently reported that the catalytic activity of LOXL2 is not essential for interaction with Snail1 and induction of EMT in Madin-Darby canine kidney epithelial (MDCK) cells (34). This study employed two catalytically incompetent forms of LOXL2: H626Q/H628Q (lacking two His of the putative copper binding site) and ΔLOXL2 (lacking 2/3 of the catalytic domain including the copper binding site, an LTQ precursor residue at Lys653, and the N-glycosylation site at Asn644). Both forms of LOXL2 were able to induce EMT to a similar extent as WT-LOXL2. WT-LOXL2 and H626Q/H628Q LOXL2 were detected both in the culture medium and cell lysate, but ΔLOXL2 was only detected in the cell lysate. Additionally, it is intriguing that WT-LOXL2 was able to induce EMT in MDCK cells, in contrast to reports by us and others that WT-LOXL2 is not effective in inducing EMT in MCF-7 cells (34,43) It is not clear whether these discrepancies are due to the use of different cell types; further study is necessary.
The second issue concerns the inhibitory effect of BAPN on LOXL2. As we have described in this review, conflicting evidences have been reported. A plurality of reports state that BAPN is ineffective in inhibiting LOXL2 in the conditioned media of MCF-7 (22), COS-7 and CHO cells (21). However, in solution, our group has found that BAPN inhibits Δ1–3SRCRLOXL2 (Ki = 1.60 ± 0.30 μM, IC50 =1.31 ± 0.04 μM, unpublished results) in a competitive fashion, as reported for LOX (Ki = 6 μM)(20). Additionally, BAPN was shown to inhibit the oxidation of 1,5-diaminopentane (IC50 = 5.0 ± 1.4 μM), spermine (IC50 = 3.8 ± 0.2 μM), and fibrillar collagen I (IC50 not reported) by rLOXL2 isolated from murine myeloma cells (23). Further study is necessary to understand this discrepancy; however, it is possible that BAPN may be metabolized or unstable in cell culture, thus preventing it from being an effective inhibitor of LOXL2.
9. Summary
The majority of studies of LOXL2 have so far focused on its in vivo roles, particularly in tumor metastasis/invasion. In addition to the proposed extracellular function of LOXL2 in ECM remodeling, recent studies have provided strong evidence for intracellular functions of LOXL2 in the induction of epithelial plasticity. Additionally, recent efforts to biochemically characterize extracellular and intracellular LOXL2s have revealed that secreted LOXL2 is N-glycosylated, and that N-glycosylation at Asn455 and Asn644 is essential for secretion. In contrast, the intracellular LOXL2 is unglycosylated and N-terminally processed, and has apparent affinity for the cell nucleus. LOXL2 is known to exhibit LOX amine oxidase activity in solution; however, its in vivo substrates and binding partners have not been fully characterized. Furthermore, the role of each SRCR domain is unclear, although the fourth SRCR domain seems to allosterically modulate the activity of the LOX catalytic domain. Importantly, the lack of crystal or NMR structures for any member of the LOX family of proteins severely hinders elucidation of the structure-function correlations of LOXL2 and LOX.
The development of specific inhibitors of LOXL2 is greatly needed, since they would have strong potential to serve as effective therapies against metastatic/invasive tumors and fibrosis. Antibodies specific for the fourth SRCR domain of LOXL2 (e.g. AB0023 and its humanized variant, AB0024) appear quite promising in this respect, as they have been shown to reduce tumor progression and invasion in mice. Small molecule cell-permeable inhibitors of LOXL2, which could target both intracellular and extracellular LOXL2, are also likely to have great therapeutic value, should they be discovered.
ACKNOWLEGEMENT
This work was supported, in part, by Bridge Fund (2112114–099), Proof-of-Concept Fund (INS0072694–985) from the University of Kansas (KU) and Cancer Biology Pilot Grant (KAN0072264–908) by KU Cancer Center.
References
- 1.Kenyon K, Modi WS, Contente S, and Friedman RM (1993) J Biol Chem 268, 18435–18437 [PubMed] [Google Scholar]
- 2.Kim Y, Boyd CD, and Csiszar K (1995) J Biol Chem 270, 7176–7182 [DOI] [PubMed] [Google Scholar]
- 3.Saito H, Papaconstantinou J, Sato H, and Goldstein S (1997) J Biol Chem 272, 8157–8160 [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Dai J, Tang R, Zhao W, Zhou Z, Wang W, Ying K, Xie Y, and Mao Y (2001) Matrix Biol 20, 153–157 [DOI] [PubMed] [Google Scholar]
- 5.Maki JM, and Kivirikko KI (2001) Biochem J 355, 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jourdan-Le Saux C, Tronecker H, Bogic L, Bryant-Greenwood GD, Boyd CD, and Csiszar K (1999) J Biol Chem 274, 12939–12944 [DOI] [PubMed] [Google Scholar]
- 7.Asuncion L, Fogelgren B, Fong KS, Fong SF, Kim Y, and Csiszar K (2001) Matrix Biol 20, 487–491 [DOI] [PubMed] [Google Scholar]
- 8.Csiszar K (2001) Prog Nucleic Acid Res Mol Biol 70, 1–32 [DOI] [PubMed] [Google Scholar]
- 9.Wang SX, Mure M, Medzihradszky KF, Burlingame AL, Brown DE, Dooley DM, Smith AJ, Kagan HM, and Klinman JP (1996) Science 273, 1078–1084 [DOI] [PubMed] [Google Scholar]
- 10.Smith-Mungo LI, and Kagan HM (1998) Matrix Biol 16, 387–398 [DOI] [PubMed] [Google Scholar]
- 11.Payne SL, Fogelgren B, Hess AR, Seftor EA, Wiley EL, Fong SF, Csiszar K, Hendrix MJ, and Kirschmann DA (2005) Cancer Res 65, 11429–11436 [DOI] [PubMed] [Google Scholar]
- 12.Lopez KM, and Greenaway FT (2011) J Neural Transm 118, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 13.Seve S, Decitre M, Gleyzal C, Farjanel J, Sergeant A, Ricard-Blum S, and Sommer P (2002) Connect Tissue Res 43, 613–619 [PubMed] [Google Scholar]
- 14.Cronshaw AD, Fothergill-Gilmore LA, and Hulmes DJ (1995) Biochem J 306 (Pt 1), 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borel A, Eichenberger D, Farjanel J, Kessler E, Gleyzal C, Hulmes DJ, Sommer P, and Font B (2001) J Biol Chem 276, 48944–48949 [DOI] [PubMed] [Google Scholar]
- 16.Thomassin L, Werneck CC, Broekelmann TJ, Gleyzal C, Hornstra IK, Mecham RP, and Sommer P (2005) J Biol Chem 280, 42848–42855 [DOI] [PubMed] [Google Scholar]
- 17.Grimsby JL, Lucero HA, Trackman PC, Ravid K, and Kagan HM (2010) J Cell Biochem 111, 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez VG, Moestrup SK, Holmskov U, Mollenhauer J, and Lozano F (2011) Pharmacol Rev 63, 967–1000 [DOI] [PubMed] [Google Scholar]
- 19.Kim YM, Kim EC, and Kim Y (2011) Mol Biol Rep 38, 145–149 [DOI] [PubMed] [Google Scholar]
- 20.Tang SS, Trackman PC, and Kagan HM (1983) J Biol Chem 258, 4331–4338 [PubMed] [Google Scholar]
- 21.Vadasz Z, Kessler O, Akiri G, Gengrinovitch S, Kagan HM, Baruch Y, Izhak OB, and Neufeld G (2005) J Hepatol 43, 499–507 [DOI] [PubMed] [Google Scholar]
- 22.Hollosi P, Yakushiji JK, Fong KS, Csiszar K, and Fong SF (2009) Int J Cancer 125, 318–327 [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez HM, Vaysberg M, Mikels A, McCauley S, Velayo AC, Garcia C, and Smith V (2010) J Biol Chem 285, 20964–20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barry-Hamilton V, Spangler R, Marshall D, McCauley S, Rodriguez HM, Oyasu M, Mikels A, Vaysberg M, Ghermazien H, Wai C, Garcia CA, Velayo AC, Jorgensen B, Biermann D, Tsai D, Green J, Zaffryar-Eilot S, Holzer A, Ogg S, Thai D, Neufeld G, Van Vlasselaer P, and Smith V (2010) Nat Med 16, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 25.Xu L, Go EP, Finney J, Moon H, Lantz M, Rebecchi K, Desaire H, and Mure M (2013) Journal of Biological Chemistry 288, 5357–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebecchi KR, Go EP, Xu L, Woodin CL, Mure M, and Desaire H (2011) Anal Chem 83, 8484–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller RK, Boon DY, and Crum FC (1979) Biochemistry 18, 3946–3952 [DOI] [PubMed] [Google Scholar]
- 28.Heifetz A, Keenan RW, and Elbein AD (1979) Biochemistry 18, 2186–2192 [DOI] [PubMed] [Google Scholar]
- 29.Bedell-Hogan D, Trackman P, Abrams W, Rosenbloom J, and Kagan H (1993) The Journal of biological chemistry 268, 10345–10350 [PubMed] [Google Scholar]
- 30.Shah MA, Scaman CH, Palcic MM, and Kagan HM (1993) The Journal of biological chemistry 268, 11573–11579 [PubMed] [Google Scholar]
- 31.Gacheru SN, Trackman PC, Shah MA, O’Gara CY, Spacciapoli P, Greenaway FT, and Kagan HM (1990) J Biol Chem 265, 19022–19027 [PubMed] [Google Scholar]
- 32.Tang C, and Klinman JP (2001) J Biol Chem 276, 30575–30578 [DOI] [PubMed] [Google Scholar]
- 33.Peinado H, Del Carmen Iglesias-de la Cruz M, Olmeda D, Csiszar K, Fong KS, Vega S, Nieto MA, Cano A, and Portillo F (2005) EMBO J 24, 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuevas EP, Moreno-Bueno G, Canesin G, Santos V, Portillo F, and Cano A (2014) Biol Open 3, 129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DuBois JL, and Klinman JP (2006) Biochemistry 45, 3178–3188 [DOI] [PubMed] [Google Scholar]
- 36.Heuts DP, Gummadova JO, Pang J, Rigby SE, and Scrutton NS (2011) J Biol Chem 286, 29584–29593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bollinger JA, Brown DE, and Dooley DM (2005) Biochemistry 44, 11708–11714 [DOI] [PubMed] [Google Scholar]
- 38.Herwald SE, Greenaway FT, and Lopez KM (2010) Protein Expr Purif 74, 116–121 [DOI] [PubMed] [Google Scholar]
- 39.Cai D, and Klinman JP (1994) J Biol Chem 269, 32039–32042 [PubMed] [Google Scholar]
- 40.Matsuzaki R, Fukui T, Sato H, Ozaki Y, and Tanizawa K (1994) FEBS Lett 351, 360–364 [DOI] [PubMed] [Google Scholar]
- 41.Mure M, Wang SX, and Klinman JP (2003) J Am Chem Soc 125, 6113–6125 [DOI] [PubMed] [Google Scholar]
- 42.Moore RH, Spies MA, Culpepper MB, Murakawa T, Hirota S, Okajima T, Tanizawa K, and Mure M (2007) J Am Chem Soc 129, 11524–11534 [DOI] [PubMed] [Google Scholar]
- 43.Moon HJ, Finney J, Xu L, Moore D, Welch DR, and Mure M (2013) J Biol Chem 288, 30000–30008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byers PH, Narayanan AS, Bornstein P, and Hall JG (1976) Birth Defects Orig Artic Ser 12, 293–298 [PubMed] [Google Scholar]
- 45.Khakoo A, Thomas R, Trompeter R, Duffy P, Price R, and Pope FM (1997) Clin Genet 51, 109–114 [DOI] [PubMed] [Google Scholar]
- 46.Soares MB, de Lima RS, Rocha LL, Vasconcelos JF, Rogatto SR, dos Santos RR, Iacobas S, Goldenberg RC, Iacobas DA, Tanowitz HB, de Carvalho AC, and Spray DC (2010) J Infect Dis 202, 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zibadi S, Vazquez R, Larson DF, and Watson RR (2010) Cardiovasc Toxicol 10, 190–198 [DOI] [PubMed] [Google Scholar]
- 48.Lopez B, Gonzalez A, Hermida N, Valencia F, de Teresa E, and Diez J (2010) Am J Physiol Heart Circ Physiol 299, H1–9 [DOI] [PubMed] [Google Scholar]
- 49.Payne SL, Hendrix MJ, and Kirschmann DA (2007) J Cell Biochem 101, 1338–1354 [DOI] [PubMed] [Google Scholar]
- 50.Finney J, Moon HJ, Ronnebaum T, Lantz M, and Mure M (2014) Arch Biochem Biophys 546, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, and Morin PJ (2000) Cancer Res 60, 6281–6287 [PubMed] [Google Scholar]
- 52.Ono K, Tanaka T, Tsunoda T, Kitahara O, Kihara C, Okamoto A, Ochiai K, Takagi T, and Nakamura Y (2000) Cancer Res 60, 5007–5011 [PubMed] [Google Scholar]
- 53.Fong SF, Dietzsch E, Fong KS, Hollosi P, Asuncion L, He Q, Parker MI, and Csiszar K (2007) Genes Chromosomes Cancer 46, 644–655 [DOI] [PubMed] [Google Scholar]
- 54.Peinado H, Moreno-Bueno G, Hardisson D, Perez-Gomez E, Santos V, Mendiola M, de Diego JI, Nistal M, Quintanilla M, Portillo F, and Cano A (2008) Cancer Res 68, 4541–4550 [DOI] [PubMed] [Google Scholar]
- 55.Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, Cmelak AJ, Levy S, Slebos RJ, and Yarbrough WG (2006) Cancer Res 66, 8210–8218 [DOI] [PubMed] [Google Scholar]
- 56.Peng L, Ran YL, Hu H, Yu L, Liu Q, Zhou Z, Sun YM, Sun LC, Pan J, Sun LX, Zhao P, and Yang ZH (2009) Carcinogenesis 30, 1660–1669 [DOI] [PubMed] [Google Scholar]
- 57.Akiri G, Sabo E, Dafni H, Vadasz Z, Kartvelishvily Y, Gan N, Kessler O, Cohen T, Resnick M, Neeman M, and Neufeld G (2003) Cancer Res 63, 1657–1666 [PubMed] [Google Scholar]
- 58.Kirschmann DA, Seftor EA, Fong SF, Nieva DR, Sullivan CM, Edwards EM, Sommer P, Csiszar K, and Hendrix MJ (2002) Cancer Res 62, 4478–4483 [PubMed] [Google Scholar]
- 59.Chang J, Nicolau M, Cox TR, Wetterskog D, Martens JW, Barker HE, and Erler JT (2013) Breast Cancer Res 15, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagaraja GM, Othman M, Fox BP, Alsaber R, Pellegrino CM, Zeng Y, Khanna R, Tamburini P, Swaroop A, and Kandpal RP (2006) Oncogene 25, 2328–2338 [DOI] [PubMed] [Google Scholar]
- 61.Thiery JP, and Sleeman JP (2006) Nat Rev Mol Cell Biol 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 62.Thiery JP, Acloque H, Huang RY, and Nieto MA (2009) Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 63.Tiwari N, Gheldof A, Tatari M, and Christofori G (2012) Semin Cancer Biol 22, 194–207 [DOI] [PubMed] [Google Scholar]
- 64.Foroni C, Broggini M, Generali D, and Damia G (2012) Cancer Treat Rev 38, 689–697 [DOI] [PubMed] [Google Scholar]
- 65.Barker HE, Bird D, Lang G, and Erler JT (2013) Mol Cancer Res 11, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor MA, Amin JD, Kirschmann DA, and Schiemann WP (2011) Neoplasia 13, 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, and Weaver VM (2009) Cell 139, 891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker AM, Bird D, Lang G, Cox TR, and Erler JT (2013) Oncogene 32, 1863–1868 [DOI] [PubMed] [Google Scholar]
- 69.Barker HE, Chang J, Cox TR, Lang G, Bird D, Nicolau M, Evans HR, Gartland A, and Erler JT (2011) Cancer Res 71, 1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erler JT, and Weaver VM (2009) Clin Exp Metastasis 26, 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erez N, Truitt M, Olson P, Arron ST, and Hanahan D (2010) Cancer Cell 17, 135–147 [DOI] [PubMed] [Google Scholar]
- 72.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, and Logsdon CD (2008) Cancer Res 68, 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, and Cunha GR (1999) Cancer Res 59, 5002–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, and Weinberg RA (2005) Cell 121, 335–348 [DOI] [PubMed] [Google Scholar]
- 75.Orimo A, and Weinberg RA (2006) Cell Cycle 5, 1597–1601 [DOI] [PubMed] [Google Scholar]
- 76.Shimoda M, Mellody KT, and Orimo A (2010) Semin Cell Dev Biol 21, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, Santos V, Montes A, Morales S, Castilla MA, Rojo-Sebastian A, Martinez A, Hardisson D, Csiszar K, Portillo F, Peinado H, Palacios J, and Cano A (2011) EMBO Mol Med 3, 528–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cano A, Santamaria PG, and Moreno-Bueno G (2012) Future Oncol 8, 1095–1108 [DOI] [PubMed] [Google Scholar]
- 79.Herranz N, Dave N, Millanes-Romero A, Morey L, Diaz VM, Lorenz-Fonfria V, Gutierrez-Gallego R, Jeronimo C, Di Croce L, Garcia de Herreros A, and Peiro S (2012) Molecular Cell 46, 369–376 [DOI] [PubMed] [Google Scholar]
- 80.Peinado H, Olmeda D, and Cano A (2007) Nat Rev Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 81.De Craene B, and Berx G (2013) Nat Rev Cancer 13, 97–110 [DOI] [PubMed] [Google Scholar]
- 82.Shilatifard A (2008) Curr Opin Cell Biol 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kooistra SM, and Helin K (2012) Nat Rev Mol Cell Biol 13, 297–311 [DOI] [PubMed] [Google Scholar]
- 84.Upadhyay AK, Horton JR, Zhang X, and Cheng X (2011) Curr Opin Struct Biol 21, 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lugassy J, Zaffryar-Eilot S, Soueid S, Mordoviz A, Smith V, Kessler O, and Neufeld G (2012) Journal of Biological Chemistry 287, 3541–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nellaiappan K, Risitano A, Liu G, Nicklas G, and Kagan HM (2000) J Cell Biochem 79, 576–582 [DOI] [PubMed] [Google Scholar]
- 87.Rucker RB, Kosonen T, Clegg MS, Mitchell AE, Rucker BR, Uriu-Hare JY, and Keen CL (1998) Am J Clin Nutr 67, 996S–1002S [DOI] [PubMed] [Google Scholar]
- 88.Kim M, Okajima T, Kishishita S, Yoshimura M, Kawamori A, Tanizawa K, and Yamaguchi H (2002) Nat Struct Biol 9, 591–596 [DOI] [PubMed] [Google Scholar]