Abstract
FlaG homolog has been found in several bacteria including spirochetes; however, its function is poorly characterized. In this report, we investigated the role of TDE1473, a putative FlaG, in the spirochete Treponema denticola, a keystone pathogen of periodontitis. TDE1473 resides in a large gene operon that is controlled by a σ70-like promoter and encodes a putative FlaG protein of 123 amino acids. TDE1473 can be detected in the periplasmic flagella (PFs) of T. denticola, suggesting that it is a flagella-associated protein. Consistently, in vitro studies demonstrate that the recombinant TDE1473 interacts with the PFs in a dose-dependent manner and that such an interaction requires FlaA, a flagellar filament sheath protein. Deletion of TDE1473 leads to long and less motile mutant cells. Cryo-electron tomography analysis reveal that the wild-type cells have 2-3 PFs with nearly homogenous lengths (ranging from 3 to 6 μm), whereas the mutant cells have less intact PFs with disparate lengths (ranging from 0.1 to 9 μm). The phenotype of T. denticola TDE1473 mutant reported here is different from its counterparts in other bacteria, which provides insight into further understanding the role of FlaG in the regulation of bacterial cell morphogenesis and flagellation.
Keywords: Spirochete, Treponema denticola, Motility, Flagella, FlaG
INTRODUCTION
The bacterial flagellum is a sophisticated nanomachine that is composed of at least 25 structural proteins [for recent reviews, see (Erhardt et al., 2010, Mukherjee & Kearns, 2014)]. The functions of these proteins in the flagellar structure and assembly have been well studied in two model organisms, Escherichia coli and Salmonella enterica. These flagellar proteins constitute three mechanical parts: the basal body-motor complex, the flagellar hook and the filament. The basal body-motor complex is embedded within the cell envelope and works as a reversible rotary motor. The hook and filament extend outwards to the cell exterior and function as a universal joint and a propeller, respectively. The basal body is very complex and consists of several functional units: the flagellar export apparatus, the MS/C ring (rotor), the rod (driveshaft), the L/P rings (bushings), and the stator (torque generator). The motor is driven by an inward-directed electrochemical gradient of protons or sodium. The torque generated by the motor is mechanically transmitted to the filament via the rod-hook complex, leading to the rotation of the flagellar filament, which causes bacterial locomotion.
The oral bacterium Treponema denticola is an obligatory anaerobic and motile pathogen, which is implicated in both periodontal and endodontic infections (Dashper et al., 2011, Ellen & Galimanas, 2005). T. denticola belongs to the phylum of spirochetes (Charon et al., 2012). It has 2~3 periplasmic flagella (PFs) that arise from each end of the cell and extend toward the center where they overlap (Izard et al., 2009, Kurniyati et al., 2017). Structurally, the PFs of T. denticola are similar to the flagella of the model organisms, as each consists of a basal body, a hook, and a filament (Zhao et al., 2014). However, the flagellar filament of T. denticola is more complicated than its counterparts in other bacteria (Li et al., 2000b, Charon et al., 2012). It is comprised of at least one sheath protein (FlaA) and three core proteins (FlaB1, FlaB2, and FlaB3) (Ruby et al., 1997, Seshadri et al., 2004, Kurniyati et al., 2017). In contrast, the filaments of E. coli and S. enterica consist of a single polymeric protein known as the flagellin (FliC) (Samatey et al., 2001). The FlaA protein (37.0 kDa) is likely exported to the periplasmic space via the Sec secretion pathway, as its N-terminus contains a typical peptidase I cleavage site. The FlaB proteins are exported to the periplasmic space through the flagellar type III secretion apparatus (Norris et al., 1988, Norris, 1993). The FlaB proteins are ~31.0-32.0 kDa in sizes and are modified with a novel glycan, which is essential for the flagellation and motility of T. denticola (Kurniyati et al., 2017). The FlaB proteins form a core that is sheathed by FlaA. The FlaB proteins are immunologically cross-reactive (both within a given species and also between species) but have no sequence similarity and antigenic cross-reactivity to FlaA (Ruby et al., 1997, Li et al., 2000b, Li et al., 2000a, Li et al., 2008). The PFs are essential for the cell shape and motility of T. denticola (Li et al., 1996). Flagella-deficient mutants are non-motile and less helical at the regions where the PFs interact with the cell cylinder (Li et al., 1996, Charon et al., 1992, Ruby et al., 1997, Miller et al., 2016). Motility also contributes to the pathogenicity of T. denticola (e.g., non-flagellated mutants are unable to penetrate the intercellular junctions of oral epithelial cells) (Lux et al., 2001).
Some motile bacteria possess unique flagellar proteins that are absent in the model organisms (Mukherjee & Kearns, 2014, Minamino & Imada, 2015). FlaG is one of those proteins. FlaG is a small flagella-associated protein that has been studied in Aeromonas caviae, Campylobacter jejuni, Pseudomonas aeruginosa and P. fluorescens (Kalmokoff et al., 2006, Capdevila et al., 2004, Rabaan et al., 2001). In these bacteria, deletion of flaG often leads to bacterial cells with long flagellar filaments, but this usually has no impact on cell motility. In A. caviae, deletion of flaG also affects its adherence to HEp-2 cells. In this report, a FlaG homolog (TDE1473) was studied in T. denticola by using an approach involving biochemistry, genetics and cryo-electron tomography (cryo-ET). We found that the phenotype of the T. denticola flaG mutant is different from its counterparts in other bacteria. Its underlying mechanism was further explored.
RESULTS
Transcriptional analyses of a flagellar gene cluster.
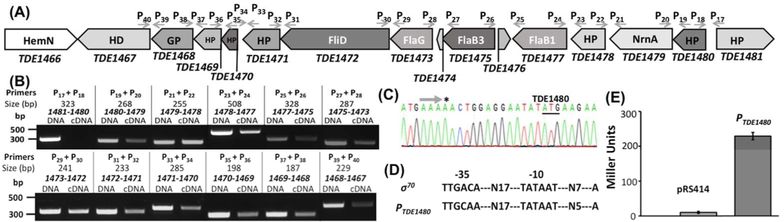
In studying T. denticola flagellar filament core proteins, we found that the flaB1 (TDE1477) and flaB3 (TDE1475) genes reside in a large gene cluster that consists of 14 open reading frames (orfs) (Figure 1A). Except for TDE1476, a small orf of only 186 bp, the rest of the genes within this cluster are transcribed from the same orientation, suggesting that these genes are co-transcribed. To test this speculation, a previously described RT-PCR (Ge et al., 1997) was conducted with twelve pairs of primers that span from TDE1467 to TDE1481 (Figure 1A). Of note, as TDE1474 (90 bp) and TDE1476 (186 bp) are too small, we did not include specific primers for these two orfs. As expected, no product was detected with the primers of P17/P18 between TDE1480 and TDE1481 as they are divergently transcribed. The rest of the primer pairs yielded positive products with the same sizes as the positive controls in which T. denticola chromosomal DNA was used as a PCR template (Figure 1B), indicating that these genes are co-transcribed and form a polycistronic operon. There is a 545 bp intergenic region upstream of TDE1480. To understand the transcriptional regulation of this operon, we mapped the transcription start site by using RLM-RACE analysis. The analysis mapped the transcription start site 14 bp upstream of the start codon of TDE1480 (Figure 1C). We searched this region for promoter-like sequences using BPROM, a promoter prediction server (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) and found a highly conserved σ70-like promoter (Figure 1D) with a −10 region showing 100% identity to the consensus sequence (TATAAT) of the E. coli σ70 promoter and a less conserved −35 region (TTGCAA) (Paget & Helmann, 2003). The identified promoter was designated as PTDE1480. A transcriptional reporter assay using lacZ showed that PTDE1480 was able to initiate the reporter gene expression in E. coli: the average β-galactosidase activity (243 ± 11 Miller units) of pRSTDE1480 is 20-fold greater than that of pRS414 (10 ± 3 Miller units), a promoterless control vector (Figure 1E). Collectively, these results indicate that these 14 genes form a large polycistronic operon that is regulated by a σ70 -like promoter.
Figure 1. TDE1473 is located at a large gene cluster that is regulated by a σ70-like promoter.
(A) A diagram showing the genes adjacent to TDE1473. Arrows represent the relative positions and orientations of RT-PCR primers that span the intergenic regions between individual orfs. (B) RT-PCR analysis. For each pair of primers, chromosomal DNA was used as a positive control. The numbers below the primers are predicted sizes of RT-PCR and PCR products. (C) 5ˋ-RLM-RACE analysis. The arrow shows the sequencing direction and an asterisk (*) indicates the transcriptional start site. (D) Sequence comparison between the E. coli ſ70 promoter and the promoter sequence upstream of TDE1480 (designated as PTDE1480). (E) Transcriptional analysis of PTDE1480 using lacZ as a reporter in E. coli. For this assay, PTDE1480 was fused to the promoterless lacZ gene in the pRS414 plasmid. The promoterless pRS414 was used as a negative control. β-galactosidase activity was measured and expressed as the average Miller units of triplicate samples from two independent experiments, as previously described (Bian et al., 2013). All the primers used here are listed in Table 1.
Identification of TDE1473 as a putative FlaG.
Among these 14 orfs, four are flagellar proteins: TDE1477 (FlaB1), TDE1475 (FlaB3), TDE1473 (FlaG), and TDE1472 (hook associated protein HAP2, also known as FliD) (Seshadri et al., 2004). TDE1467 contains a conserved HD-GYP domain that is often evident in phosphoesterases responsible for bacterial second messenger c-di-GMP turnover (Schirmer & Jenal, 2009, Jenal et al., 2017). TDE1468 is a putative glycoprotease. TDE1479 belongs to the family of nanoRNase (NrnA) that hydrolyzes c-di-AMP. The rest of the seven orfs encode hypothetical proteins (Figure 1A). TDE1473 is between FliD (TDE1472) and FlaB3 (TDE1745) and consists of 123 amino acids with a predicted mass of 13.8 kDa. BLAST search showed that TDE1473 (hereafter named FlaG) belong to the family of FlaG proteins (pfam03646; COG1334) - it has 30% sequence identity to A. caviae FlaG and 42% to P. aeruginosa FlaG. Sequence alignment indicated that the C-termini of FlaG homologs are considerably conserved (Figure 2).
Figure 2. Sequence alignment of FlaG homologs.
The numbers show the positions of amino acids in the FlaG proteins from T. denticola (TDE1473, NP_972079.1), P. aeruginosa (FlaG, AAC093900) and A. caviae (FlaG, AAF19181). Only one part of the aligned sequences is presented. The alignments were conducted using the program Clustal Omega.
Isolation of flaG isogenic mutants.
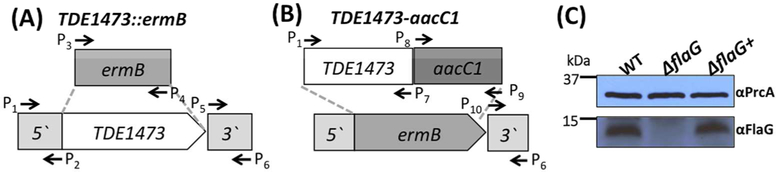
To investigate the role of FlaG, its cognate gene was deleted and in-frame replaced by the ermB cassette (Figure 3A). Erythromycin-resistant colonies that appeared on the plates were first screened by PCR for the presence of ermB (data not shown). One positive clone (ΔflaG) was further characterized by PCR and immunoblotting with a specific antibody against FlaG (αFlaG). The results showed that flaG was deleted as expected and its cognate gene product was abolished in the mutant (Figure 3C). To rule out the possibility of polar effect on its downstream gene expression, the ΔflaG mutant was rescued by replacing the ermB cassette with the full-length flaG gene, as illustrated in Figure 3B. Immunoblotting analysis showed that the expression of flaG was restored in a rescued strain (ΔflaG+) (Figure 3C).
Figure 3. Isolations of ΔflaG and its complemented strain ΔflaG+.
Diagrams showing construction of TDE1473::ermB (A) and TDE1473-aacC1 vectors (B). TDE1473::ermB was designed to in-frame replace TDE1473 with the ermB cassette; TDE1473-aacC1 was constructed to replace the ermB cassette in ΔflaG with TDE1473-aacC1. These constructs were constructed by two-step PCR. Arrows represent the relative positions and orientations of these primers, which are listed in Table 1. ermB is an erythromycin-resistance cassette; aacC1 is a gentamycin-resistance cassette. (C) Immunoblotting analysis of the wild-type, ΔflaG, and ΔflaG+ strains by using a specific antibody against FlaG (αFlaG).
FlaG affects cell motility and length.
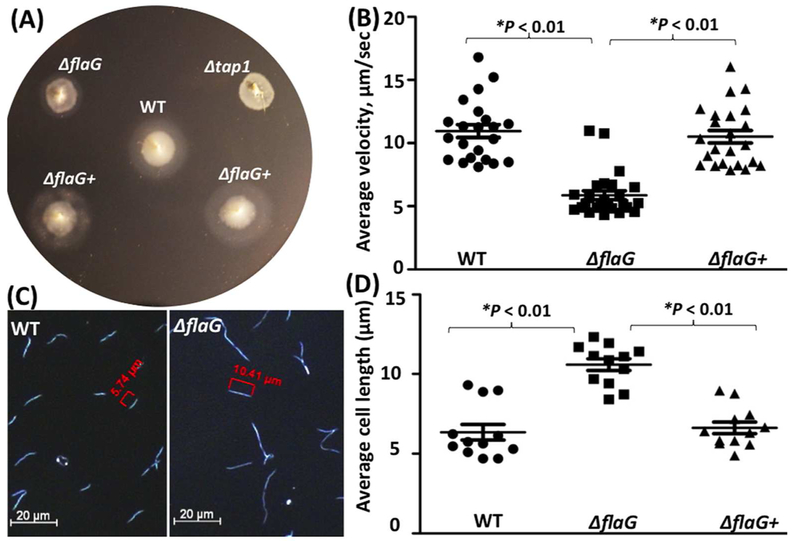
Under a dark-field microscope, in contrast to the wild-type and ΔflaG+ strains (Video 1, 3), the ΔflaG mutant swam poorly (Video 2), indicative of crippled motility. Along with the microscopic observation, the swimming plate assay showed that ΔflaG exhibited a poor swimming phenotype (Figure 4A); the average diameter of swimming rings (8.8 ± 0.19 mm, n = 5 plates) formed by the mutant is significantly (P < 0.01) smaller than that of the wild-type (11.1 ± 0.33 mm, n = 5) and ΔflaG+ (11.4 ± 0.43 mm, n = 5) strains. This was further confirmed by bacterial motion tracking analysis, where bacterial cells were tracked for at least 1 min in 1% methylcellulose. Consistent with the swimming plate assay, the tracking analysis showed that the mutant swam slower than the wild-type and ΔflaG+ strains (Figure 4B) - the average cell velocity of ΔflaG (5.9 ± 0.35 μm/s, n=22 cells) is nearly 2-fold less than that of the wild-type (10.9 ± 0.51 μm/s, n=25) and ΔflaG+ (10.5 ± 0.49 μm/s, n=23) strains. In addition to motility, under dark-field microscope, we also found that the ΔflaG mutant cells are much longer that the wild-type and complemented cells (Figure 4C, D), e.g., the average cell length of the ΔflaG mutant (10.6 ± 0.37 μm, n = 12 cells) were approximately two-fold longer (P < 0.01) than those of the wild-type (6.3 ± 0.49 μm, n = 12 cells) and the complemented cells (6.6± 0.36 μm, n = 12 cells). Of note, we also measured the growth rates and found that there was no significant difference between the wild type and the mutant (data not shown). Collectively, these results indicate that FlaG is involved in the regulation of cell morphogenesis and motility of T. denticola.
Figure 4. Deletion of flaG alters the cell length and motility.
(A) Swimming plate assay of the wild-type, ΔflaG, and ΔflaG+ strains. The assay was carried out on 0.35% agarose containing the TYGVS medium diluted 1:10 with PBS. The plates were incubated anaerobically at 37°C for 5 days. Δtap1, a non-motile mutant, was used as a control to determine initial inoculum sizes. (B) Cell-tracking analysis of the wild-type, ΔflaG, and ΔflaG+ strains. The cells were tracked in the presence of 1% methylcellulose as previously described. The results were the average of at least 20 cells and are expressed as the mean of μm/s ± standard errors of mean (SEM). (C) Dark-field microscopic images of the wild-type and ΔflaG strains. The cells were visualized under dark-field illumination at 20× magnification using a Zeiss Axiostar plus microscope. (D) Average cell lengths of the wild-type, ΔflaG, and ΔflaG+ strains. The results were the average of at least 12 cells and expressed as the mean of μm ± SEM. The data were analyzed by one-way ANOVA followed by Tukey’s multiple comparison at P < 0.01.
Deletion of flaG disrupts the flagellar hemostasis at the cell poles.
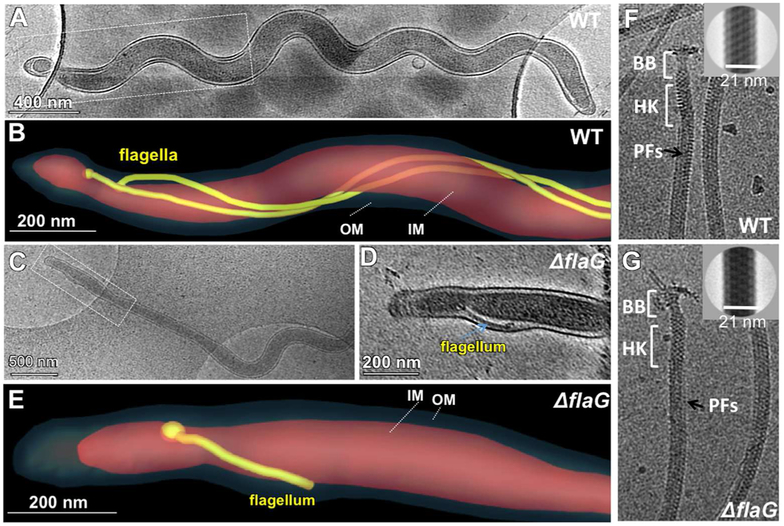
To elucidate by which mechanism FlaG affects the cell motility, cryo-ET analysis of the whole cell was carried out to directly visualize the PFs inside the wild-type and mutant cells. The wild-type cells typically have 2 to 3 long helical PFs with similar lengths (ranging from 3 to 6 μm, Table 2) arose from the cell poles, wrapped around the cell cylinders, and extended towards the central region of the cells (Figure 5A, B, and Video 4). In contrast, the mutant cells had fewer intact PFs that have disparate lengths, ranging from 0.1 to 9 μm (Table 2, Figure 5C, D, E, and video 5), e.g., the shortest is only 0.1 μm and the longest reaches 9 μm. In the rescued strain, both flagellar length and number restored to the level found in the wild type. High-resolution cryo-ET analysis further revealed that deletion of flaG had no notable impact on the number (Table 2) and structure of hook-basal bodies (HBBs) and the diameter of the PFs (Figure 5F, G). Collectively, these results indicate that deletion of flaG altered the length of flagellar filaments but had no impact on the HBBs.
Table 2.
Cryo-ET reconstructions of intact cells of WT, ΔflaG, and ΔflaG+ strains show the filament length and the numbers of the flagellar filament and motor at each cell pole
| Strains | Flagellar length | Motor number* | Filament number** |
|---|---|---|---|
| ΔflaG (cell 1) | ~0.3 μm | 1 | 1 |
| >3.5 μm | |||
| ~0.4 μm | 2 | 2 | |
| ΔflaG (cell 2) | ~0.3 μm | 2 | 2 |
| ~0.1 μm | |||
| >8.0 μm | 2 | 2 | |
| ~0.1 μm | |||
| ΔflaG (cell 3) | ~0.4 μm | 2 | 2 |
| ~0.1 μm | |||
| >9.0 μm | 2 | 2 | |
| ~0.1 μm | |||
| WT (cell 1) | >3.0 μm | 2 | 2 |
| >4.0 μm | 3 | 3 | |
| WT (cell 2) | >6.0 μm | 2 | 2 |
| >5.0 μm | 2 | 2 | |
| WT (cell 3) | >3.0 μm | 2 | 2 |
| >4.0 μm | 2 | 2 | |
| ΔflaG+ (cell 1) | >4.0 μm | 2 | 2 |
| >3.0 μm | 2 | 2 |
Number of flagellar motors visualized at the cell poles;
Number of flagellar filaments visualized at the cell poles.
Figure 5. Cryo-ET analysis of the wild-type and ΔflaG cells and isolated PFs.
(A) A cryo-EM image shows a wild-type cell. (B) Surface rendering of the cell pole (boxed region) shows the long PFs (colored in yellow) between the inner membrane (IM, pink) and the outer membrane (OM, dark gray). (C) A cryo-EM image shows a ΔflaG mutant cell. (D) A slice (boxed region) from the cryo-ET reconstruction of one cell pole. (E) Surface rendering of the cell pole shows the short flagellum. The flagellum is about 0.3 μm in length. Cryo-EM images of purified PFs from the wild type (F) and ΔflaG (G). The diameter of the filaments with sheath is about 21 nm (right upper corner). PFs: periplasmic flagella; BB: basal body; HK: Hook.
FlaG impairs the level of flagellins.
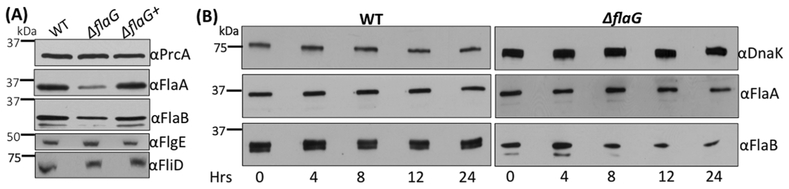
The flagellar filaments of T. denticola consist of one sheath protein FlaA and three core proteins FlaBs (i.e., FlaB1, FlaB2, and FlaB3). We reasoned that the short filaments in the ΔflaG mutant might be a result of reduction of flagellin proteins. To test this hypothesis, we measured the levels of these flagellins in the mutant by using immunoblotting analysis. In supporting this hypothesis, we found that the level of FlaA and FlaBs in the mutant was decreased substantially (Figure 6A). Of note, the molecular weights of FlaB1 (31.3 kDa) and FlaB2 (31.6 kDa) are too close to be separated on SDS-PAGE. In contrast, the levels of the hook FlgE and the filament cap FliD proteins remained unchanged. To investigate by which mechanism FlaG impairs the levels of FlaA and FlaB proteins, a protein turnover assay was carried out to monitor their stability. After arresting protein synthesis with addition of spectinomycin, while the levels of FlaA and FlaBs remained unchanged in the wild type during a course of 24 hours, the amounts of FlaA and the FlaBs in the mutant decreased approximately 30% and 60%, respectively, at 24 hours (Figure 6B). These results indicate that FlaG controls the level of FlaA and FlaB most likely through promoting their stability in T. denticola.
Figure 6. The flagellin proteins are decreased in the ΔflaG mutant.
(A) Immunoblotting analysis of the wild-type, ΔflaG, and ΔflaG+ strains. Equivalent amounts of whole-cell lysates were analyzed by SDS-PAGE and then probed against different antibodies as labeled. PrcA was used as a loading control as previously described (Kurniyati et al., 2017). (B) Protein turnover assay. Spectinomycin was added to the bacterial cultures to arrest protein translation. Samples were withdrawn at the indicated time points and subjected to immunoblotting with antibodies against DnaK, FlaA, or the FlaBs. DnaK was used as a sample loading control.
FlaG is a minor flagella-associated protein.
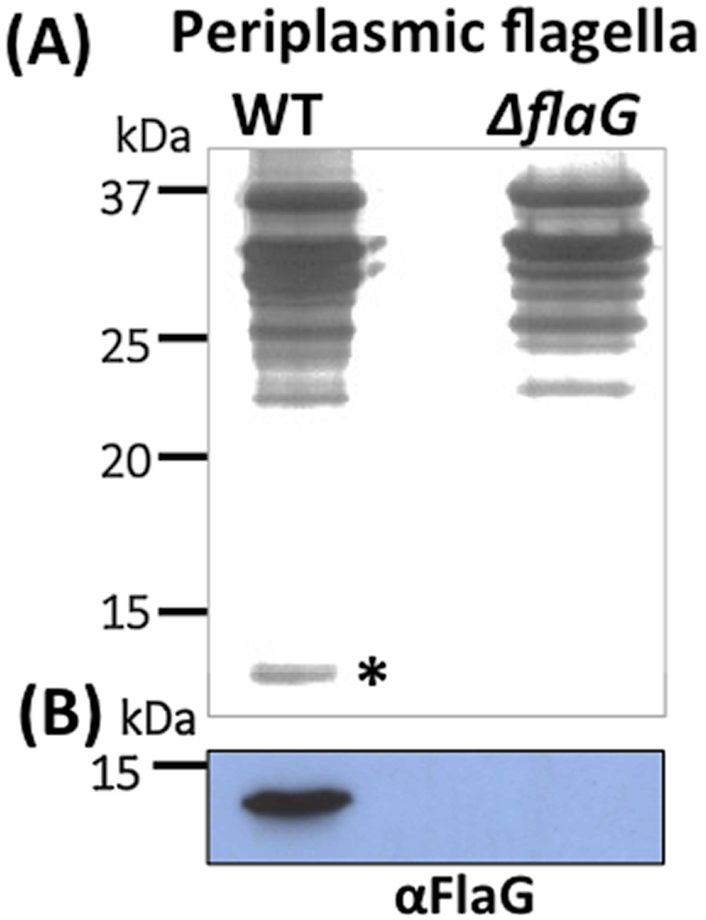
Deletion of FlaG affects the stability of flagellin proteins, suggesting that FlaG is a flagella-associated protein. To test this speculation, we isolated the PFs from the wild-type and ΔflaG strains and compared their protein profiles by using SDS-PAGE. On Coomassie blue-stained gels, there was no notable difference between the wild type and ΔflaG with respect to their protein compositions (data not shown). In the silver-stained gel, which is more sensitive than Coomassie staining, a very faint band with a similar mass to FlaG (13.8 kDa) was observed in the PFs isolated from the wild type but not in the one from ΔflaG (Figure 7A). This band was recognized by αFlaG (Figure 7B). It is worth mentioning that we also carried out an immunoelectron microscopy (IEM) study, attempting to determine the location of FlaG on the PFs; however, there was no notable difference between the wild type and the mutant with respect to gold particle distribution patterns (data not shown). One reasonable explanation is that the amount of FlaG is too low to be detected by IEM. It is also possible that FlaG in its native state or inside the PFs where is not accessible to the antibody. Collectively, these results indicate that FlaG is part of the PFs but its level is considerably lower than that of the flagellin proteins.
Figure 7. FlaG was detected in the isolated PFs.
(A) SDS-PAGE analysis of the PFs isolated from the wild-type and ΔflaG strains, followed by silver staining. Asterisk points to FlaG. (B) Immunoblotting analysis of the PFs isolated from the wild-type and ΔflaG strains with a specific antibody against FlaG (αFlaG).
The binding of FlaG to the PFs depends on the sheath protein FlaA.
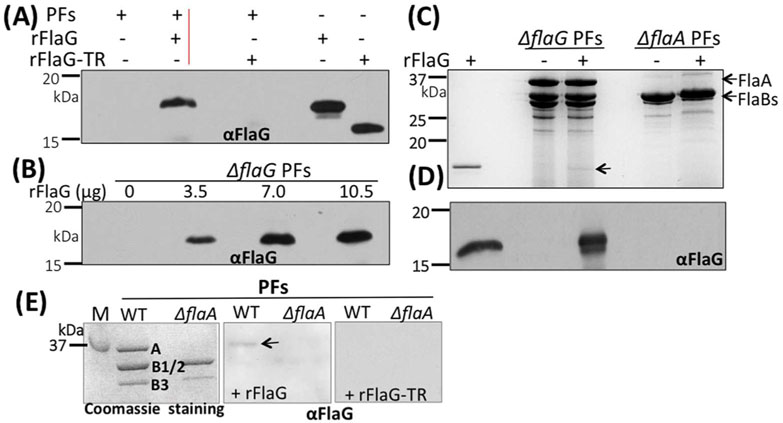
FlaG was detected in the isolated PFs (Figure 7) and the deletion of flaG impairs the level of FlaA and FlaBs (Figure 6), suggesting that there might be an interplay between FlaG and the flagellin proteins. To test this hypothesis, the PFs isolated from ΔflaG were incubated with different concentrations of recombinant FlaG protein (rFlaG). After the incubation and washing with PBS, the PFs were collected and subjected to SDS-PAGE, followed by immunoblotting. The results showed that rFlaG bound to the PFs in a dose-dependent manner (Figure 8A, B). The C-terminus of FlaG contains a conserved region (Figure 2). Deletion of this region (15 aa from the C-terminus of FlaG) totally abolished the binding of rFlaG to the PFs (right lane, Figure 8A), indicating that the interaction between FlaG and the PFs is specific and perhaps mediated by this conserved region. In addition, we also found that rFlaG failed to bind to the PFs isolated from ΔflaA, a flaA deletion mutant, and that there was no FlaG detected in the PFs of ΔflaA by the immunoblots with αFlaG (Figure 8C, D), indicating that FlaG might interact with the sheath protein FlaA. To substantiate this proposition, we first attempted pull down assays by affinity chromatography using recombinant FlaA and FlaG proteins. Unfortunately, recombinant FlaA proteins (i.e., His-tagged and GST-tagged FlaA) are insoluble. As an alternative approach, far-western blotting analysis was carried out. For this experiment, the PFs isolated from the wild type and ΔflaA were separated on SDS-PAGE, transferred to PVDF membranes, co-incubated with either rFlaG or rFlaG-TR, and then probed against αFlaG. If FlaG binds to FlaA, we expect that the complex of FlaA/rFlaG would be recognized by αFlaG. Consistent with this expectation, in the presence of rFlaG, FlaA was detected by αFlaG in the wild type PFs, but not in the ΔflaA PFs (middle image, Figure 8E). Of note, in the presence of rFlaG-TR, there was no FlaA signal detected by αFlaG (right image, Figure 8E). Taken together, these results indicate that FlaG is a part of the flagellar filaments, where it most likely interacts with the sheath protein FlaA.
Figure 8. FlaG binds to the PFs in a FlaA-dependent manner.
(A)& (B) rFlaG binds to the filaments in a dose-dependent manner. For these experiments, different amounts of rFlaG as labeled were co-incubated with the PFs (18 μg) isolated from ΔflaG for 16 hrs, washed three times with PBS, and then subjected to SDS-PAGE, followed by immunoblotting with ⟨FlaG. (C) rFlaG fails to bind to the PFs isolated from the flaA mutant (ΔflaA). Similar to the above experiments, here rFlaG was co-incubated with the PFs isolated from ΔflaG and ΔflaA strains, and then subjected to SDS-PAGE, followed by Coomassie blue staining (top image) or immunoblotting with αFlaG (lower image). Arrow points to rFlaG detected. (D) Far-western blotting analysis. The PFs isolated from the wild type and ΔflaA strains were separated by SDS-PAGE (left image), transferred to PVDF membranes, incubated with rFlaG (middle image) or rFlaG-TR (C-terminus 15 aa were truncated, right image), washed 4 times with T-PBS, and finally probed with αFlaG. The blots were developed using a horseradish peroxidase secondary antibody with an enhanced ECL, as previously described (Kurniyati et al., 2017). Arrow points to FlaA detected. M: marker; A: FlaA; B1/2: FlaB1, FlaB2; B3: FlaB3.
DISCUSSION
As a newly identified flagellar protein that is absent in E. coli and S. enterica, the role of FlaG remains largely unknown. The results shown here provide some insights into understanding the role of FlaG in flagellation and motility as well as a potential mechanism involved. FlaG homologs have been found in numerous flagellated bacteria, including both Gram-negative and Gram-positive, but so far have been studied only in A. caviae, C. jejuni, P. aeruginosa and P. fluorescens (Kalmokoff et al., 2006, Capdevila et al., 2004, Rabaan et al., 2001). In these bacteria, inactivation of flaG resulted in bacterial cells with long flagellar filaments but maintained normal cell motility. As such, it has been speculated that FlaG functions as a negative regulator that controls the length of flagellar filaments. In this report, we found that deletion of flaG disrupts the flagellar homeostasis at the cell poles of T. denticola, e.g., deletion of flaG leads to mutant cells with the PFs ranging from 0.1 to 9 μm in length, whereas the wild type cells with the PFs ranging from 3 to 6 μm in length (Figure 5, Table 2). In addition, the flaG mutant is less motile than the wild type (Figure 4). The observed phenotype is different to what has been reported for FlaG homologs in other bacteria (albeit they have only 30 to 42% sequence identity). This difference between FlaG homologs in different bacteria suggests that they might have variable functions among different bacterial species.
Several lines of evidence shown in this report indicate that FlaG is a minor flagellar filament protein or filament-associated protein. First, the flaG gene is located between the fliD (TDE1472) and flaB3 (TDE1745) genes and is co-transcribed with other flagellin genes (Figure 1). Second, FlaG is detected in the isolated PFs (Figure 7) and interacts with the filaments in a FlaA-dependent manner (Figure 8). Third, deletion of flaG impairs the level and stability of FlaA and FlaB proteins (Figure 6). Finally, deletion of flaG alters the flagellar length and number of intact PFs (Figure 5, Table 2). Based on these results, we propose that, as a flagellar filament protein or filament-associated protein, FlaG most likely maintains the stability of flagellin proteins (i.e., FlaA and FlaBs) and/or facilitates the assembly of flagellar filaments in the periplasmic space of T. denticola and that loss of FlaG would result in flagellin protein turnover, which in turn would impair the filament length and number, as well as the cell motility of T. denticola. We are currently attempting to solve the crystal structure of FlaG. If successful, it will help us elucidate the role of FlaG at a molecular level.
Compared to other bacteria, one of the unique aspects about spirochetes is their distinct, long and spiral morphology with polar PFs. The sizes of the spirochetes, the numbers of PFs attached at each end, and whether the PFs overlap at the middle of the cells vary from species to species (Charon et al., 2012). For instance, the Lyme disease spirochete Borrelia burgdorferi has 7-11 PFs that form a ribbon wrapping around the cell cylinders and its average cell size can reach up to 23 μm in length (Jutras et al., 2016). In contrast, Leptospira species have singular short PF at each end of the cells (Wunder et al., 2016). In this report, we found that the average size of T. denticola cells is approximately 6.3 μm in length, which is much shorter than that of B. burgdorferi. In addition, whole cell cryo-ET analysis revealed that T. denticola cells generally have 2-3 long PFs that arise from the cell poles and extend along the cell cylinders. For the first time, we measured the length of PFs in situ by using cryo-ET and found that the PFs at the wild-type cell poles are nearly homogenous in terms of the length. Similar to other bacteria, spirochetes divide by binary fission. Given their unique cell morphology and arrangement of PFs, spirochetes must finely tune their flagellar biosynthesis and assembly to synchronize with cell division and elongation; however, the underlying mechanism remains unknown. Here we found that deletion of flaG not only disrupts the flagellar homeostasis at the cell poles but also alters the cell length. Thus, it is conceivable to speculate that FlaG may function as a molecular ruler to orchestrate the processes of cell growth and flagellar elongation during cell division, e.g., acting as a chaperon to shuffle equal amounts of the flagellin proteins to two newly formed cell poles. Deletion of flaG disrupts this equilibrium and leads to biased allocation of flagellin proteins at the cell poles, resulting in abnormal PFs with disparate lengths, as evident in the flaG mutant. Interestingly, BLAST analysis revealed that FlaG homologs are found only in some species from the genus of Treponema and Spirochaeta, suggesting that spirochetes may have evolved different mechanism(s) to orchestrate the processes of cell growth and flagellar elongation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Treponema denticola strain ATCC 35405 (wild type) was used in this study. Cells were grown in tryptone-yeast extract-gelatin-volatile fatty acids-serum (TYGVS) medium at 37°C in an anaerobic chamber in presence of 85% nitrogen, 10% carbon dioxide and 5% hydrogen (Kurniyati et al., 2013, Kurniyati et al., 2017). T. denticola isogenic mutants were grown with appropriate antibiotic(s) for selective pressure as needed: erythromycin (50 μg/ml) and/or gentamicin (20 μg/ml). Escherichia coli DH5α strain was used for DNA cloning and BL21-CodonPlus (DE3)-RIL (Agilent, Santa Clara, CA) for preparing recombinant proteins. The E. coli strains were cultivated in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Isolation of periplasmic flagella (PFs).
The PFs were isolated as previously described (Kurniyati et al., 2017). Briefly, 500 ml of the late-logarithmic-phase T. denticola culture (~108 cells/ml) was centrifuged at 8,000×g for 20 min at 4°C. The cells were washed four times with phosphate-buffered saline (PBS, pH 7.4) and once with T1 buffer (0.15 M Tris-HCl, pH 6.8). The final cell pellets were resuspended in 30 ml of T1 buffer and then mixed with 3 ml of 10% Triton X-100. The mixture was incubated for 1 hour at room temperature and then 3 ml of mutanolysin (200 μg/ml) and 300 μl of T2 buffer (0.1 M Tris-HCl buffer, pH 6.8) were added and mixed slowly. The mixtures were incubated for 2 hours at room temperature and then at 4°C overnight. Followed the incubation, 600 μl of 0.1 M MgSO4 and 600 μl of T2 buffer were added and incubated for 5 min at room temperature. The resultant samples were centrifuged at 17,000×g for 15 min at 4°C. The supernatant fractions were collected and mixed with 2 ml of 20% PEG 8000 (Alfa Aesar, London, UK) in 1.0 M NaCl. The mixtures were mixed thoroughly, incubated for 30 min on ice and then centrifuged at 27,000×g for 30 min at 4°C. The resultant pellets were suspended in an alkaline solution (0.1 M KCl, 0.5 M sucrose, 0.1% Triton X-100, 50 mM sodium bicarbonate, pH 11) and incubated for 1 hour on ice, followed by centrifugation at 80,000×g for 45 min at 4°C. The obtained flagellar filament pellets were washed once in 20 mM Tris-HCl buffer (pH 8), suspended in water, and stored at 4°C for further analyses.
Electrophoresis and immunoblotting analyses.
Sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis (SDS-PAGE), native PAGE and immunoblotting were carried out as previously described (Kurniyati et al., 2017). Briefly, equal amounts of T. denticola whole cell lysates (~10-50 μg) were separated on SDS-PAGE, transferred to PVDF membranes and then probed with different antibodies, including antibodies against T. denticola FlaG (raised in this study), FlgE (Miller et al., 2016), PrcA (Godovikova et al., 2010), T. pallidum FlaBs (Norris, 1993), and Borrelia burgdorferi FliD (Li et al., unpublished data). The immunoblots were developed using a horseradish peroxidase secondary antibody with an enhanced chemiluminescence (ECL).
Preparing recombinant FlaG protein and its antiserum.
The DNA fragment that encodes the full-length FlaG (from aa 1 to 123, named rFlaG) and its truncate version (from aa 9 to 108, designated as rFlaG-TR) were PCR amplified with primers P13/P14 and P15/P16 (see details in Table 1) using Pfx DNA polymerase (Life Technologies, Grand Island, NY). The resultant PCR products were cloned into either pET200/D-TOPO or pET100/D-TOPO expression vectors (Life Technologies). The resulting plasmids were then transformed into BL21-CodonPlus (DE3)-RIL (Agilent). The expression of recombinant proteins was induced with 1 mM isopropyl β-D-1-thiogalactosidase (IPTG). The recombinant proteins were first purified using Ni-NTA agarose under native conditions according to the manufacturer’s protocol (Qiagen, Valencia, CA). The resultant crude proteins were further purified using size-exclusion chromatography (SEC) with a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences, Marlborough, MA). The concentrations of purified proteins were determined using a NanoDrop 2000 (Thermo Scientific). To raise antibodies against FlaG, ~5 mg of purified rFlaG was used to immunize two rats (2.5 mg for each animal) following a standard immunization procedure.
Table 1.
Oligonucleotide primers used in this study
| Primers | Sequences (5’-3’) | Note a |
|---|---|---|
| P1 | ATGATTATTAATCACAATAT | 5ˋ portion for TDE1473 inactivation; [F] |
| P2 | GAATATTTTATATTTTTGTTCATAAAAAACCTCCTTGGCTTTA | 5ˋ portion for TDE1473 inactivation; [R] |
| P3 | ATGAACAAAAATATAAAATATTCTC | Erythromycin B cassette (ermB); [F] |
| P4 | TTATTTCCTCCCGTTAAATAATAG | Erythromycin B cassette (ermB); [R] |
| P5 | TATTTAACGGGAGGAAATAATCGGAAATTTCCCTTTTTTAG | 3ˋ portion for TDE1473 inactivation; [F] |
| P6 | CAAAGGAATCGTAATTGTCTG | 3ˋ portion for TDE1473 inactivation; [R] |
| P7 | CATCGTTGCTGCTGCGTAACATCTATATTGATTCATCGAATAA | 5ˋ portion for TDE1473 mutant |
| P8 | ATGTTACGCAGCAGCAACG | Gentamicin cassette (aacC1); [F] |
| P9 | TTAGGTGGCGGTACTTGGGTC | Gentamicin cassette (aacC1); [R] |
| P10 | GACCCAAGTACCGCCACCTAATCGGAAATTTCCCTTTTTTAG | 3ˋ portion forTDE1473 mutant complementation; [F] |
| P11 | ATTATTGTTTATTGCAAGAGC | 3ˋ flanking region of TDE1473, ΔTDE1473 PCR analysis; [R] |
| P12 | ATGAGTATAGAAATAAACGGC | TDE1473, ΔTDE1473 PCR analysis; [F] |
| P13 | CACCAGTATAGAAATAAACGGCATAG | TDE1473 recombinant protein; [F] |
| P14 | CTATATTGATTCATCGAATAA | TDE1473 recombinant protein; [R] |
| P15 | CACCGGGCACCAAGCAGCATTACAAC | TDE1473 protein-truncate; [F] |
| P16 | CTACTGCAGTCTTTGTATTGCCTC | TDE1473 protein-truncate; [F] |
| P17 | GGTACCCATGAGCTATCATTC | TDE1481; co-RT-PCR [R] |
| P18 | CATCCGCCTCCTCAATCTTTTG | TDE1480; co-RT-PCR [R] |
| P19 | GGAAGCCCCATATGGGGTG | TDE1480; co-RT-PCR [F] |
| P20 | CATCGGGCTCTTTATGACCTG | TDE1479; co-RT-PCR [R] |
| P21 | GCAAGATAGCCGCATCATTTG | TDE1479; co-RT-PCR [F] |
| P22 | CTCCATTGTACGTTTGATATC | TDE1478; co-RT-PCR [R] |
| P23 | CTGAAAGGGAGCTTTCCCGTC | TDE1478; co-RT-PCR [F] |
| P24 | GAAGCCTGAACAGCGAGTTC | TDE1477; co-RT-PCR [R] |
| P25 | ATAGCCAGCTTGTAATGTCTC | TDE1477; co-RT-PCR [F] |
| P26 | GTGATTAATAATCATATGAC | TDE1475; co-RT-PCR [R] |
| P27 | CACAAGCCAATGCTCAGCCTC | TDE1475; co-RT-PCR [F] |
| P28 | GCTGCTTGGTGCCCTATGCCG | TDE1473; co-RT-PCR [R] |
| P29 | GACTGCAGGCAAGAATACTTG | TDE1473; co-RT-PCR [F] |
| P30 | CTGCCTCCAGGAATCATCCTG | TDE1472; co-RT-PCR [R] |
| P31 | GAAATACGGAATAATGGACGG | TDE1472; co-RT-PCR [F] |
| P32 | CTAAGACTTGGGTTATAGTGC | TDE1471; co-RT-PCR [R] |
| P33 | GATATTGCAGAATATGAATTG | TDE1471; co-RT-PCR [F] |
| P34 | GTATATATTGCTGTTCCGAC | TDE1470; co-RT-PCR [R] |
| P35 | CTGCGTCCGCCGTTTAATCG | TDE1470; co-RT-PCR [F] |
| P36 | CTTCAATATCCAAACCTTGGG | TDE1469; co-RT-PCR [R] |
| P37 | CGGCATAATGGTAAATGATG | TDE1469; co-RT-PCR [F] |
| P38 | GATCAGTATGGCAGAATGTC | TDE1468; co-RT-PCR [R] |
| P39 | CCTTGATAGAAGCAGACAAAG | TDE1468; co-RT-PCR [F] |
| P40 | CGGATTGTCCTGATCGTCTTG | TDE1467; co-RT-PCR [R] |
| P41 | AAACATCCGCCTCCTCAATC | TDE1473 5ˋRACE outer primer |
| P42 | GCTTTAATCCCGCTCTTCGG | TDE1473 5ˋRACE inner primer |
| P43 | GAATTCTTTTTATTCCTAGGTGAAAC | TDE1480 promoter; [F] |
| P44 | GGATCCATATTCCTCCAGTTACCCCA | TDE1480 promoter; [R] |
Underlined sequences are engineered restriction cut sites for DNA cloning; [F] forward; [R] reverse.
Incubation of PFs with rFlaG and far-western blot.
The PFs (18 μg) isolated from ΔflaG or ΔflaA mutants were incubated with either different amounts of rFlaG (0, 3.5, 7, and 10.5 μg) or rFlaG-TR (7 μg) for 16 hours at 4° C. After the incubation, the PFs were collected by centrifugation, washed three times with ice-cold PBS, and resuspended in 25 μl Laemmli sample loading buffer. The resultant samples were boiled for 5 min and then subjected to SDS-PAGE, followed by either silver staining or immunoblotting analyses. Far-western blots were carried out as previously described (Wu et al., 2007). Briefly, following SDS-PAGE, the proteins were transferred to PVDF membranes, incubated with rFlaG, washed 4 times with T-PBS, and finally probed against αFlaG. The blots were developed using a horseradish peroxidase secondary antibody with an enhanced ECL.
Reverse transcription-PCR (RT-PCR), quantitative RT-PCR (qRT-PCR), and RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE).
Total T. denticola RNA was isolated using TRI reagent (Sigma-Aldrich), following the manufacturer’s instructions. The resultant samples were treated with Turbo DNase I (Ambion, Austin, TX) at 37°C for 2 hours to eliminate genomic DNA contamination. Followed the treatment, PCR was carried out to ensure that there was no DNA contamination. Otherwise, the RNA samples were treated with DNase again till DNA contamination was totally eliminated. The resultant RNA samples were re-extracted using acid phenol-chloroform, precipitated in isopropanol, and washed once with 70% ethanol. The RNA pellets were dissolved in RNase-free water. cDNA was generated from 1 μg purified RNA using AMV reverse transcriptase and random primers included in the Kit (Promega, Madison, WI) and then PCR amplified with Taq DNA polymerase (Takara, Mountain View, CA). qRT-PCR was conducted using iQ SYBR Green Supermix and a MyiQ thermal cycler (Bio-Rad). The transcript of dnaK (TDE0628) was used as an internal control to normalize the qRT-PCR data (Bian et al., 2013). The results were expressed as the normalized difference of the threshold cycle (ΔΔCT) between the wild type and ΔflaG. The primers for RT-PCR and qRT-PCR are listed in Table 1. RLM-RACE was performed using the First Choice RLM-RACE kit (Ambion), according to the manufacturer’s protocol. Purified T. denticola RNA was reversed transcribed to cDNA with a 5ˋ RACE adapter, followed by PCR amplifications with primers P41/P42 as listed in Table 1. The resultant PCR products were cloned into pGEM-T Easy vector and sequenced at the Roswell Park Cancer Institute DNA Sequencing Laboratory (Buffalo, NY).
β-galactosidase activity assay.
A fragment spanning from nucleotides −125 to −1 of TDE1480, the first gene in the operon (see the details in Figure 1), was PCR amplified with primers P41/P42, generating a fragment with engineered EcoRI and BamHI cut sites at the 5ˋ and 3ˋ ends, respectively. The obtained fragment was in-frame fused to the promoterless lacZ gene in the pRS414 plasmid (a gift from R. Breaker, Yale University), creating pRSTDE1480. The resultant plasmid was transformed into E. coli DH5α. β-galactosidase activity was measured and expressed as average Miller units of triplicate samples from two independent experiments.
Construction of an isogenic mutant of TDE1473 and its complemented strain.
The vector TDE1473::ermB (Figure 3A) was constructed to replace the entire TDE1473 gene with a previously documented erythromycin B resistance cassette (ermB) (Goetting-Minesky & Fenno, 2010). This vector was constructed by two-step PCR and DNA cloning as previously described (Bian & Li, 2011). Briefly, the TDE1473 upstream flanking region and ermB were PCR amplified with primers P1/P2 and P3/P4, respectively, and then fused together with primers P1/P4, generating Fragment 1. The downstream region of TDE1473 was PCR amplified with primers P5/P6, and then fused to the Fragment 1 by PCR using primers P1/P6. The obtained DNA fragment was cloned into pGEM-T Easy vector (Promega), generating the vector of TDE1473::ermB (Figure 3A). To delete TDE1473, TDE1473::ermB was linearized by using NotI, transformed into T. denticola wild-type cells via electroporation, and then plated on semi-solid agar plates containing erythromycin (50 μg/ml), as previously described. The deletion was confirmed by PCR and immunoblotting. The resultant mutant was designated as ΔflaG.
A similar method to that described above was used to construct TDE1473-aacC1 (Figure 3B), a vector for cis-complementing ΔflaG. To construct this vector, the full-length TDE1473 gene and aacC1 cassette were PCR amplified with primers P1/P7 and P8/P9, respectively, and then fused together with primers P1/P9, generating Fragment 1. The downstream region of TDE1473 was PCR amplified with primers P10/P6, and then fused to Fragment 1 by PCR using primers P1/P6, generating TDE1473-aacC1 (Figure 3B). To complement the mutant, TDE1473-aacC1 was linearized and transformed into ΔflaG via electroporation and then plated on semi-solid agar plates containing 20 μg/ml of gentamicin. The obtained antibiotic-resistant colonies were screened and characterized by PCR and immunoblotting. The resultant complemented clones were designated as ΔflaG+.
Bacterial swimming plate assay and motion tracking analysis.
A swimming plate assay was performed as previously described (Kurniyati et al., 2017). Briefly, 3 μl of bacterial cultures (109 cells/ml) were inoculated onto 0.35% agarose containing TYGVS medium diluted 1:10 with PBS. The plates were incubated anaerobically at 37°C for 5 to 7 days. The diameters of swimming rings were measured in millimeters. Δtap1, a previously constructed non-motile mutant (Limberger et al., 1999), was included as a control to determine the initial inoculum size. The average diameters of each strain were calculated from three independent plates, and the results are represented as the mean of diameters ± standard error of the mean (SEM). The velocity of bacterial cells was measured using a computer-based bacterial tracking system as previously described (Bian et al., 2013). The data were statistically analyzed by one-way ANOVA followed by Tukey’s multiple comparison at P < 0.01.
Protein turnover assay.
A protein turnover assay was carried out as previously described (Kurniyati et al., 2017, Motaleb et al., 2004). Briefly, T. denticola strains were first grown to late-logarithmic-phase (~108 cells/ml). Subsequently, spectinomycin was added into the cultures to a final concentration of 100 μg/ml, followed by incubation at 37° C for up to 24 hours. Samples (5.0 ml) were harvested at the indicated time points and subjected to immunoblotting analysis.
Cryo-EM sample preparation, data collection and image processing.
The frozen-hydrated specimens of T. denticola were prepared as previously described (Kurniyati et al., 2017). Briefly, T. denticola cultures were mixed with 10 nm colloidal gold solutions and then deposited on a freshly glow-discharged, holey carbon grid for about one minute. The grids were blotted with a small piece of filter paper for ~4 seconds and then rapidly plunged in liquid ethane using a gravity-driven plunger apparatus. The frozen-hydrated specimens of T. denticola were transferred to a 300 kV electron microscope (Polara, FEI Company) that is equipped with a field emission gun and a direct detection detector (Gatan K2 Summit). To generate three-dimensional (3D) reconstructions of whole bacterial cells, tomographic package SerialEM (Mastronarde, 2005) was used to acquire multiple tilt series along the cell at 4,500 × magnification. The pixel size at the specimen level is 8.2 Å. All tilt series were collected in the low-dose mode with ~10 μm defocus. A total dose of 50 e−/Å2 is distributed among 35 tilt images covering angles from - 51° to +51° at tilt steps of 3°. The tilt series were aligned and reconstructed by IMOD (Kremer et al., 1996). Multiple reconstructions from different segments of the same cell were integrated into one large reconstruction. In total, we generated eight 3D reconstructions of the entire cells from the wild type (3 cells), ΔflaG (3 cells) and the complemented strain (2 cells). Tomographic package IMOD was used to take 2D snapshots of the reconstructions and generate 3D surface rendering of the reconstructions.
Online Supplementary Videos
Supplementary Material
Tracking WT cells in 1% methylcellulose.
Tracking the flaG mutant (ΔflaG) in 1% methylcellulose.
Tracking the complemented strain (ΔflaG+) in 1% methylcellulose.
Whole cell Cryo-ET analysis of WT.
Cryo-ET analysis of the flaG mutant. Note: only one end of the cells was reconstructed and the mutant was labeled as ΔTDE1473 in the movie.
ACKNOWLEDGEMENTS
This research was supported by a grant (No. 31728002) from National Natural Science Foundation of China, Public Health Service Grants DE023080 to C. Li; AI087946 to J. Liu; and KJ2014ZD33 to Y. Min. We are grateful for the technical assistance from Dustin Morado and Juyi Wang.
REFERENCES
- Bian J & Li C, (2011) Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector. Appl Environ Microbiol 77: 4573–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Liu X, Cheng YQ & Li C, (2013) Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. J Bacteriol 195: 3897–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila S, Martinez-Granero FM, Sanchez-Contreras M, Rivilla R & Martin M, (2004) Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150: 3889–3897. [DOI] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA & Wolgemuth CW, (2012) The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66: 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Block SM, Curci K, Ruby JD, Kreiling JA & Limberger RJ, (1992) Morphology and dynamics of protruding spirochete periplasmic flagella. J Bacteriol 174: 832–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Seers CA, Tan KH & Reynolds EC, (2011) Virulence factors of the oral spirochete Treponema denticola. J Dent Res 90: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen RP & Galimanas VB, (2005) Spirochetes at the forefront of periodontal infections. Periodontol 2000 38: 13–32. [DOI] [PubMed] [Google Scholar]
- Erhardt M, Namba K & Hughes KT, (2010) Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2: a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Old IG, Saint Girons I & Charon NW, (1997) Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J Bacteriol 179: 2289–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godovikova V, Wang HT, Goetting-Minesky MP, Ning Y, Capone RF, Slater CK & Fenno JC, (2010) Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (dentilisin) complex. J Bacteriol 192: 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetting-Minesky MP & Fenno JC, (2010) A simplified erythromycin resistance cassette for Treponema denticola mutagenesis. J Microbiol Methods 83: 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M & Radolf JD (2009) Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191: 7566–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A & Lori C, (2017) Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. [DOI] [PubMed] [Google Scholar]
- Jutras BL, Scott M, Parry B, Biboy J, Gray J, Vollmer W& Jacobs-Wagner C, (2016) Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc Natl Acad Sci U S A 113: 9162–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J & Szymanski CM, (2006) Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol 188: 4312–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN & McIntosh JR, (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76. [DOI] [PubMed] [Google Scholar]
- Kurniyati K, Kelly JF, Vinogradov E, Robotham A, Tu Y, Wang J, Liu J, Logan SM & Li C, (2017) A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol Microbiol 103: 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurniyati K, Zhang W, Zhang K & Li C, (2013) A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola. Mol Microbiol 89: 842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Corum L, Morgan D, Rosey EL, Stanton TB & Charon NW, (2000a) The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J Bacteriol 182: 6698–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Motaleb A, Sal M, Goldstein SF & Charon NW, (2000b) Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol 2: 345–354. [PubMed] [Google Scholar]
- Li C, Wolgemuth CW, Marko M, Morgan DG & Charon NW, (2008) Genetic analysis of spirochete flagellin proteins and their involvement in motility, filament assembly, and flagellar morphology. J Bacteriol 190: 5607–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruby J, Charon N & Kuramitsu H, (1996) Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol 178: 3664–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberger RJ, Slivienski LL, Izard J & Samsonoff WA, (1999) Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol 181: 3743–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Miller JN, Park NH & Shi W, (2001) Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infection and immunity 69: 6276–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51. [DOI] [PubMed] [Google Scholar]
- Miller MR, Miller KA, Bian J, James ME, Zhang S, Lynch MJ, Callery PS, Hettick JM, Cockburn A, Liu J, Li C, Crane BR & Charon NW, (2016) Spirochaete flagella hook proteins self-catalyse a lysinoalanine covalent crosslink for motility. Nat Microbiol 1: 16134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T & Imada K, (2015) The bacterial flagellar motor and its structural diversity. Trends Microbiol 23: 267–274. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Sal MS & Charon NW, (2004) The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J Bacteriol 186: 3703–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S & Kearns DB, (2014) The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet 48: 319–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ, (1993) Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema Pallidum Polypeptide Research Group. Microbiol Rev 57: 750–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ, Charon NW, Cook RG, Fuentes MD & Limberger RJ, (1988) Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J Bacteriol 170: 4072–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS & Helmann JD, (2003) The sigma70 family of sigma factors. Genome Biol 4: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan AA, Gryllos I, Tomas JM & Shaw JG, (2001) Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infection and immunity 69: 4257–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF & Charon NW, (1997) Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol 179: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M & Namba K, (2001) Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410: 331–337. [DOI] [PubMed] [Google Scholar]
- Schirmer T & Jenal U, (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7: 724–735. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM & Paulsen IT, (2004) Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A 101: 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li Q & Chen XZ, (2007) Detecting protein-protein interactions by Far western blotting. Nat Protoc 2: 3278–3284. [DOI] [PubMed] [Google Scholar]
- Wunder EA, Figueira CP, Benaroudj N, Hu B, Tong BA, Trajtenberg F, Liu J, Reis MG, Charon NW, Buschiazzo A, Picardeau M & Ko AI, (2016) A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol Microbiol. 101:457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Norris SJ & Liu J, (2014) Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53: 4323–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tracking WT cells in 1% methylcellulose.
Tracking the flaG mutant (ΔflaG) in 1% methylcellulose.
Tracking the complemented strain (ΔflaG+) in 1% methylcellulose.
Whole cell Cryo-ET analysis of WT.
Cryo-ET analysis of the flaG mutant. Note: only one end of the cells was reconstructed and the mutant was labeled as ΔTDE1473 in the movie.