SUMMARY
Homeostatic synaptic downscaling reduces neuronal excitability by modulating the number of postsynaptic receptors. Histone modifications and the subsequent chromatin remodeling play critical roles in activity-dependent gene expression. Histone modification codes are recognized by chromatin readers that affect gene expression by altering chromatin structure. We show that L3mbtl1 (lethal 3 malignant brain tumor-like 1), a polycomb chromatin reader, is downregulated by neuronal activity and is essential for synaptic response and downscaling. Genome-scale mapping of L3mbtl1 occupancies identified Ctnnb1 as a key gene downstream of L3mbtl1. Importantly, the occupancy of L3mbtl1 on the Ctnnb1 gene was regulated by neuronal activity. L3mbtl1 knockout neurons exhibited reduced Ctnnb1 expression. Partial knockdown of Ctnnb1 in wild-type neurons reduced excitatory synaptic transmission and abolished homeostatic downscaling, and transfecting Ctnnb1 in L3mbtl1 knockout neurons enhanced synaptic transmission and restored homeostatic downscaling. These results highlight a role for L3mbtl1 in regulating homeostasis of synaptic efficacy.
Graphical Abstract
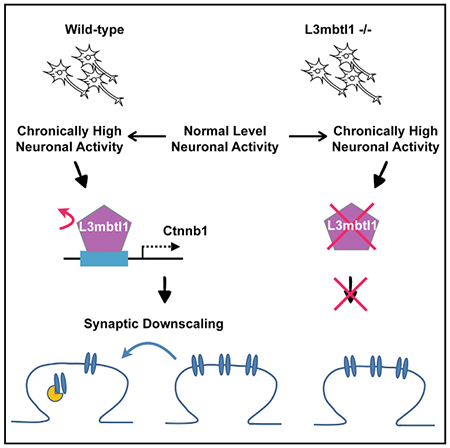
In Brief
Synaptic homeostasis is crucial for maintaining proper neuronal excitability and excitatory/inhibitory balance in the brain. Mao et al. report that an activity-dependent chromatin reader protein is required for homeostatic control of synaptic strength through the regulation of downstream target gene Ctnnb1.
INTRODUCTION
Runaway excitation of neuronal circuits induces epileptic seizures, leading to other detrimental consequences, including neuronal cell death. Neurons possess homeostatic mechanisms that compensate for activity perturbations and maintain the excitatory and inhibitory balance (E/I balance). Synaptic scaling is a specific form of homeostatic plasticity that maintains proper neuronal excitability via bidirectional regulation of synaptic strength of a neuron. Both excitatory and inhibitory synaptic strength are regulated by scaling mechanisms through changes in the abundance (scale) of postsynaptic receptors. For example, elevated neuronal activity induces calcium (Ca2+) influx followed by subsequent activation of the Ca2+/calmodulin-dependent signaling cascade and downregulation of the postsynaptic AMPA-type glutamate receptor (AMPAR)-mediated response at excitatory synapses (Turrigiano, 2011). Furthermore, elevated neuronal activity induces compensatory activation of inhibitory synaptic transmission through the recruitment of GABAA receptors (GABAARs) to inhibitory postsynaptic sites and the increased release of GABA from inhibitory terminals (Turrigiano, 2011).
A number of molecules that bridge the induction and expression steps of homeostatic synaptic downscaling have been identified. The majority of these molecules are synaptic scaffolds, kinases, and phosphatases that directly regulate synaptic function (Table S1). More than 10 genes, including Arc, Homer1a, and polo-like kinase 2 (Plk2), display altered expression levels upon an increase in neuronal activity and are critical for the expression of homeostatic synaptic downscaling (Table S1). Importantly, the inhibitors of transcription and translation dysregulate homeostatic synaptic plasticity (Ibata et al., 2008; Schanzenbacher et al., 2016). These results clearly illustrate the critical role of activity-dependent transcriptional and translational machineries in homeostatic synaptic scaling. However, the roles of epigenetic factors, particularly chromatin regulators, in this plasticity are relatively unknown.
Epigenetic mechanisms generally refer to changes to gene expression that persist throughout life or across generations which are not dependent on changes to DNA sequence. Epigenetic modifications, including DNA methylation, histone modifications and chromatin conformational changes, provide a crucial regulatory layer for gene expression. Methyl-CpG-binding protein 2 (Mecp2) acts as a DNA reader by binding to methylated DNA (CpG dinucleotides) and regulates homeostatic synaptic plasticity (Table S1). In addition, DNA methylation status and histone methyltransferase regulate homeostatic synaptic scaling (Table S1). However, knowledge of the functions of chromatin regulators in homeostatic synaptic plasticity is still very limited. Over 200 chromatin regulators have been identified in mammals and are sub-classified by function (e.g., readers, erasers, or writers of histone modifications). However, to the best of our knowledge, there are no reports describing a systematic analysis of chromatin regulators for potential roles in homeostatic synaptic plasticity.
Chromatin readers influence chromatin conformation by binding to specific histone modifications, thereby regulating gene expression. One of the chromatin reader genes, L3mbtl1, is a member of the MBT (malignant brain tumor) protein family and classified as a Polycomb group (PcG) protein. L3mbtl1 binds to chromatin through three tandem MBT repeats, and, in doing so, it facilitates higher-order chromatin organization via binding to methylated lysine residues in histone tails (Li et al., 2007; Min et al., 2007; Sims and Rice, 2008; Trojer et al., 2007). Specifically, L3mbtl1 binds to mono- and dimethylated histone tails, but not to trimethylated and unmethylated histone tails. Thus, L3mbtl1 protein contributes to the complex organization of chromatin as a chromatin reader (Adams-Cioaba and Min, 2009) and acts as an effector for posttranslational histone modifications (Bonasio et al., 2010). The L3mbtl1-containing polycomb repressive complex functions as a repressor in heterologous cell lines (Boccuni et al., 2003; Kalakonda et al., 2008; Trojer et al., 2007) and human pluripotent stem cells (Perna et al., 2015). The deletion of chromosome 20q12, the locus of human L3mbtl1, is associated with polycythemia vera, myelodysplastic syndrome, and acute myeloid leukemia (Gurvich et al., 2010; Perna et al., 2010). Of note, L3mbtl1 is expressed at high levels in mouse brain (Lein et al., 2007; Qin et al., 2010), and L3mbtl1 knockout (KO) mice display abnormal anxiety and depression phenotypes (Shen et al., 2015). These results suggest that L3mbtl1 may serve yet unknown functions in postmitotic neurons.
β-catenin protein, encoded by Ctnnb1, is known as a core molecule of the WNT signaling pathway and has pivotal roles in neuronal development and synaptic function by acting as a transcriptional factor in neuronal nuclei and as a component of synaptic scaffolding at excitatory synaptic sites (Takeichi, 2007). β-catenin regulates the expression of bidirectional homeostatic synaptic plasticity via interactions with cadherin, a homophilic trans-synaptic adhesion molecule important for synaptic structure and function (Okuda et al., 2007). However, the epigenetic regulatory mechanisms underlying β-catenin expression are still largely unknown.
In this study, we demonstrate that L3mbtl1 mRNA is primarily expressed in neurons, and is decreased when neuronal activity is elevated. KO of L3mbtl1 causes a reduced excitatory synaptic transmission in primary neurons and a lack of homeostatic downscaling at excitatory synapses both in hippocampal primary neurons and organotypic slice cultures. Importantly, L3mbtl1 protein binds to the transcription start sites of Ctnnb1 and Gabra2, whose products mediate homeostatic synaptic scaling and inhibitory synaptic transmission, respectively. These results provide insight into the roles of a chromatin reader molecule in synaptic function and neuronal homeostasis.
RESULTS
L3mbtl1 Expression Is Downregulated by Neuronal Activity
To date, 46 genes have been reported as regulators of homeostatic up- and downscaling in hippocampal and cortical neurons (Table S1). Because the expression of many of these genes changes during induction and expression of the scaling (columns E and G in Table S1), we hypothesized that chromatin regulator molecules, whose expression is altered by neuronal activity, contribute to homeostatic synaptic scaling. To test this hypothesis, we performed unbiased transcriptome analysis by RNA sequencing (RNA-seq) of cultured primary neurons after induced homeostatic downscaling.
Hippocampal primary cultures were prepared from C57BL/6 mice and homeostatic synaptic scaling was induced by applying picrotoxin (PTX), a non-competitive GABAAR blocker, for 15 hr at days in vitro 14 (DIV 14). Total RNA isolated from control and PTX challenged samples was subjected to an unbiased transcriptome (RNA-seq) screen. RNA-seq was performed in quadruplicate for each treatment and the results were highly clustered into two treatment groups (Figure S1A). Using stringent criteria (Supplemental Experimental Procedures), we identified 1150 and 689 of up- and downregulated genes, respectively (Figure 1A; Table S2). Multiple genes previously identified as neuronal activity dependent, including Arc and Bdnf, showed a significant change from baseline in our transcriptome profiling, confirming the validity of our system (blue symbols in Figure 1A; Table S1). Importantly, 12 out of 246 chromatin regulatory genes showed changes in mRNA expression (Figure 1B; Table S2D). In particular, L3mbtl1 is the most highly downregulated molecule among chromatin regulatory genes (Figure 1B, blue symbol), with its mRNA expression decreased to 30% of the control value (0.33-fold change; Padj = 1.0 × 10−6). Quantitative PCR (qPCR) further confirmed the activity-dependent downregulation of L3mbtl1 expression (Figure S1B). In contrast, expression of L3mbtl1 mRNA was insensitive to tetrodotoxin (TTX), a voltage-gated sodium-channel blocker. These results suggest that the expression of L3mbtl1 is regulated only when neuronal activity is elevated.
Figure 1. Neuronal Activity Elevation Reduced L3mbtl1 Expression.
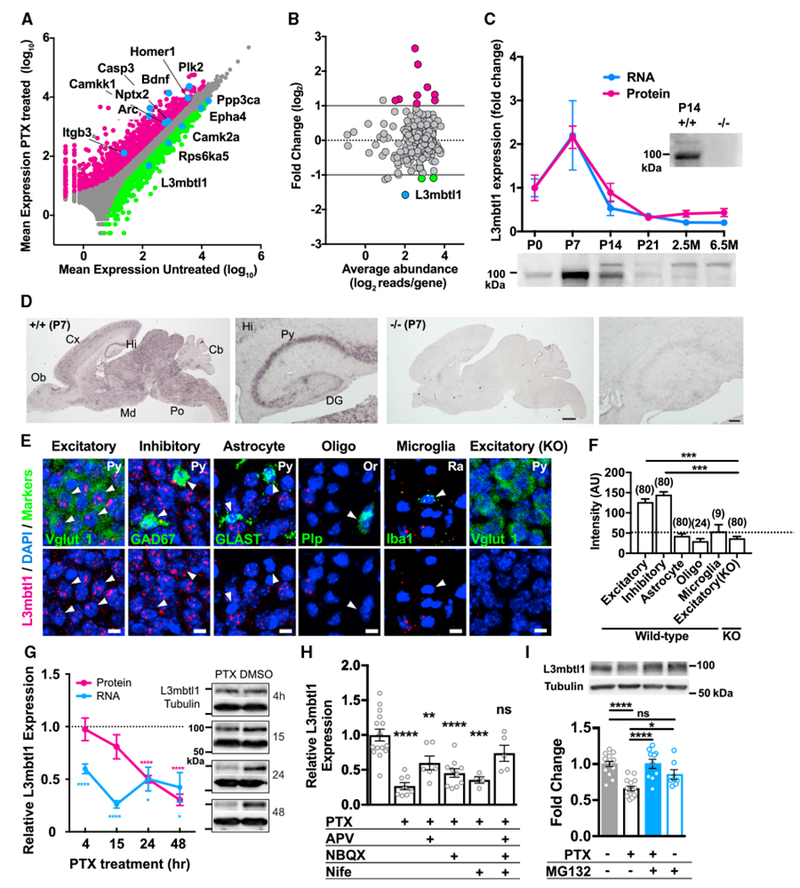
(A) Scatterplot of genes that were up regulated (magenta symbols) and downregulated (green) after 15 hr of PTX (100 μM) treatment. Gray symbols indicate genes that were not differentially expressed. See also Tables S1 and S2 and Figure S1A.
(B) MA plot (log ratio-mean average plot) of chromatin regulators that were up regulated (magenta symbols) and downregulated (green) after PTX treatment (fold change > 2, Padj < 0.01). See also Table S2 and Figure S1B.
(C) Developmental profile of L3mbtl1 mRNA (blue circles) and protein (magenta circles) levels in mouse hippocampus from postnatal 0–21 days (P0–21) to adulthood (2.5 and 6.5 months). mRNA and protein expression were normalized to the control gene, Gapdh, and the immunoblotting signal of L3mbtl1 at P0, respectively. N = 3–4 mice. Bottom: immunoblotting images of L3mbtl1 at different postnatal days. The pattern of L3mbtl1 signal changed during development. The upper bands, presumably due to posttranslational modification, were observed from P14 hippocampus. Note that these two bands are not detected in the P14 L3mbtl1 KO sample(−/−,P14).
(D) Chromogenic in situ hybridization against P7 brain sections prepared from wild-type (left) and L3mbtl1 KO (right) mouse brains, including highly magnified images of the hippocampus. Cb, cerebellum; Cx, neocortex; Hi, hippocampus; Md, midbrain; Ob, olfactory bulb; Po, pons; DG, dentate gyrus; Py, pyramidal cell layer. Scale bars, 1 mm (high magnification); 100 μm (low magnification).
(E) Fluorescence in situ hybridization (FISH) images in the CA3 regions against L3mbtl1 and cell-type-specific markers, Vglut1 (excitatory neurons [excitatory]), GAD67 (inhibitory neurons [inhibitory]), GLAST (astrocyte), Plp (oligodendrocyte [Oligo]), or Iba1 (microglia). Or, stratum oriens; Py, pyramidal cell layer; Ra, stratum radiatum. Scale bars, 10 μm. Arrows indicate cells expressing respective cell markers.
(F) Summary bargraph of cell-type-specific expression showing the mean peak intensity of L3mbtl1 signals in distinct cell types in wild-type (+/+) and L3mbtl1 KO (−/−) mice. The number in the parentheses above each column indicates the number of cells analyzed.
(G) Time course of L3mbtl1 expression (mRNA, blue circles; protein, magenta circles) after application of PTX. mRNA and protein expression, relative to Gapdh and tubulin, respectively, were normalized to untreated samples. One sample t test was performed to compare mean value to 1. N = 4–6.
(H) Pharmacological characterization of activity-dependent downregulation of L3mbtl1. Primary cultures were incubated with PTX alone or with D-APV (APV, 50 μM), NBQX(5 μM), and nifedipine (Nife, 10 μM) for 15 hr. Expression of L3mbtl1 was normalized to that of Gapdh. One-way ANOVA with Sidak’s multiple comparisons test compares each condition with control: N = 4–15. See also Figure S2.
(I) The proteasome inhibitor, MG-132, blocked activity-dependent degradation of L3mbtl1. Nuclear lysates were prepared from primary cultures 24 hr after PTX and/or MG-132 (10 μM). Top: immunoblot images of L3mbtl1 and tubulin. Bottom: quantification of immunoblotting. Expression of L3mbtl1 was normalized to that of tubulin. One-way ANOVA with Sidak’s multiple comparisons test. N= 7–16 independent culture batches.
Data shown are means ± SEM. ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05; ns, not significant.
L3mbtl1 Is an MBT Gene Highly Expressed in Neurons during Early Development
We further addressed the expression profiles of L3mbtl1 in the hippocampus. Expression of both L3mbtl1 mRNA and protein reached peak levels during postnatal days 7 to 14 after which expression decreased and remained relatively low thereafter (Figure 1C). Chromogenic in situ hybridization indicated wide expression of L3mbtl1 mRNA in the brain with highest intensity in the hippocampus (Figure 1D). The hippocampus exhibited strong signals in the pyramidal cell layer but less so in the dentate gyrus. The hybridizing signals were completely abolished in L3mbtl1 KO mice (Figure 1D, right). Double-label fluorescent in situ hybridization in the CA3 region confirmed that L3mbtl1 mRNA is expressed in both excitatory and inhibitory neurons, but the signal intensities of L3mbtl1 in non-neuronal cells (astrocytes, oligodendrocytes, and microglia) were equivalent to the background level in L3mbtl1 KO (Figures 1E and 1F). These data demonstrate that L3mbtl1 is predominantly expressed in neurons of postnatal brains.
Characterization of Neuronal Activity-Dependent Downregulation of L3mbtl1
To further characterize the regulation of L3mbtl1 expression by neuronal activity, total RNA was extracted from PTX-treated and control hippocampal neuronal cultures at different time points, followed by qPCR. Consistent with the results of RNA-seq, L3mbtl1 mRNA levels started to decrease 4 hr after application of PTX and reached the lowest level (less than 30% of control level) after 15 hr (Figure 1G). Expression of L3mbtl1 protein exhibited a steady reduction and reached 30% of the control group at 48 hr of PTX treatment.
Next, we performed pharmacological experiments (Figure 1H) and observed that blockage of AMPAR, NMDA receptor (NMDAR), or L-type Ca2+ channels alone did not fully rescue PTX-induced downregulation of L3mbtl1 mRNA, possibly due to Ca2+ influx through remaining Ca2+-permeable receptors and/or channels (Figure S2). In contrast, simultaneous inhibition of AMPAR, NMDAR, and L-type Ca2+ channels completely abolished the neuronal activity-dependent decrease of L3mbtl1 expression. These results suggest that neuronal L3mbtl1 downregulation requires the activation of downstream signaling cascades through these receptors and channels.
In cell lines, DNA double-stranded breaks (DSBs) induce rapid dissociation of L3mbtl1 protein from chromatin, followed by degradation via the ubiquitin-mediated proteasomal pathway (Acs et al., 2011). We asked whether induction of homeostatic downscaling induces the ubiquitination and degradation of L3mbtl1. To address this possibility, we cultured hippocampal neurons in the presence of the proteasomal inhibitor MG-132 and induced homeostatic downscaling with PTX. Nuclei were harvested 24 hr after PTX treatment with or without MG-132, followed by measurement of L3mbtl1 expression. Consistent with Figure 1G, 24 hr of PTX treatment significantly reduced the expression of L3mbtl1 (Figure 1I). However, MG-132 blocked the down regulation of L3mbtl1 protein by PTX, suggesting a role for ubiquitin-mediated proteasomal signaling in L3mbtl1 downregulation. MG-132 itself did not cause a significant reduction of L3mbtl1 protein.
Loss of Homeostatic DownScaling, Reduced Excitatory Synaptic Strength, and Surface GluA1 Expression in Primary L3mbtl1 KO Neurons
If the decreases of L3mbtl1 transcripts and protein are part of an essential switch to induce homeostatic synaptic downscaling, then loss of L3mbtl1 should prevent homeostatic synaptic scaling upon activity elevation. To test this hypothesis, we prepared hippocampal primary cultures from wild-type and L3mbtl1 KO mice and compared amplitudes and frequencies of AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) from neurons in which downscaling had been induced by PTX treatment for 48 hr (Figure 2). We found that L3mbtl1 KO cultured neurons failed to induce homeostatic scaling-down by PTX treatment, whereas wild-type neurons were capable of inducing bidirectional up- and downscaling. Homeostatic scaling up by TTX was intact in L3mbtl1 KO cultures, suggesting that the effects of L3mbtl1 on homeostatic synaptic scaling are unidirectional. Interestingly, we observed that, under basal conditions, L3mbtl1 KO neurons showed significantly smaller mEPSC amplitudes than that of wild-type, suggesting that a lack of L3mbtl1 reduces the number and/or conductance of AMPA receptors per synapse (Figures 2B and 2C). This observation raises the possibility that the lack of homeostatic downscaling results from the floor effect of reduced basal AMPAR-mediated response. The frequencies of mEPSCs were not changed by L3mbtl1 genetic deletion or drug treatments, suggesting that the number of active excitatory synapses and/or presynaptic release probability remain unaffected (Figure 2C). Taken together, we conclude that L3mbtl1 regulates basal synaptic transmission and homeostatic synaptic downscaling at excitatory synapses in primary neurons.
Figure 2. L3mbtl1 Is Required for Basal Excitatory Synaptic Transmission and Homeostatic Downscaling in Hippocampal Primary Neurons.
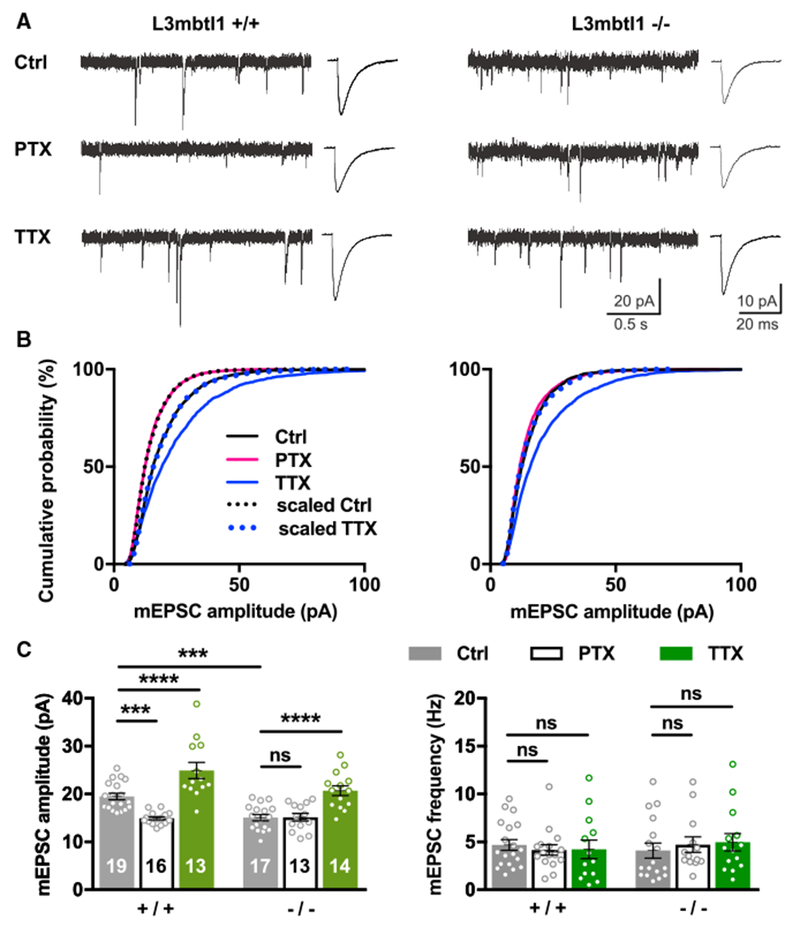
(A–C) Hippocampal primary cultures were prepared from wild-type (L3mbtl1 +/+)or L3mbtl1 KO (L3mbtl1 −/−) mice. They were treated with PTX (100 μM), TTX (1 μM), or DMSO (0.1%) as a control for 48 hr.
(A) Representative mEPSC traces (left) and average mEPSC event traces (right).
(B) Cumulative distribution of mEPSC amplitudes obtained from L3mbtl1 wild-type and KO primary cultures under three different conditions. Scaled distributions were plotted by multiplying the distribution by a scaling factor and removing values that fell below the detection threshold. Kolmogorov-Smirnov (KS) tests for wild-type distributions: ctrl versus PTX, p = 1.6 × 10−31; ctrl versus TTX, p = 5.9 × 10−18; scaled ctrl versus PTX, not significant; scaled TTX versus ctrl, not significant. KS tests for KO distributions: ctrl versus PTX, not significant; ctrl versus TTX, p = 1.1 × 10−24; scaled TTX versus ctrl, not significant.
(C) Quantification of mean mEPSC amplitudes (left) and frequencies (right).
****p < 0.0001, ***p < 0.001; ns, not significant; two way ANOVA with Sidak’s multiple comparisons test. Numbers in each bar represent the number of neurons tested (N = 5–8 mice were tested). Data shown are means ± SEM.
See also Figures S3 and S4.
Removal of AMPARs at postsynaptic sites is an established mechanism for downscaling (Pozo and Goda, 2010). We therefore examined the level of surface GluA1, one of the major AMPAR subunits in hippocampal primary neurons, prepared from wild-type and L3mbtl1 KO mice (Figure S3). Importantly, the PTX-induced downregulation of both surface GluA1 and Vglut1-colocalized synaptic GluA1 were absent in L3mbtl1 KO neurons. Total GluA1 protein level was unchanged in L3mbtl1 KO culture compared with that in wild-type. These results suggest that L3mbtl1 controls downscaling and basal mEPSC amplitude through regulating synaptic AMPAR levels without affecting total expression of AMPARs.
The strength of inhibitory synaptic transmission is also regulated bidirectionally dependent on neuronal activity (Turrigiano, 2011). To address whether L3mbtl1 is involved in inhibitory synaptic scaling, we induced homeostatic scaling by applying bicuculline, a competitive GABAAR antagonist, and TTX in wild-type and KO neurons, and compared amplitudes and frequencies of GABAAR-mediated miniature inhibitory postsynaptic currents (mIPSCs) (Figure S4). We found that L3mbtl1 KO neurons showed reduced basal mIPSC amplitudes compared with those of wild-type, but both exhibited intact homeostatic up- and downscaling. Therefore, in contrast to its effects on excitatory synapses, the role of L3mbtl1 on inhibitory synaptic function is small.
Loss of Homeostatic Downscaling in Organotypic L3mbtl1 KO Neurons
The results described above were obtained in dissociated primary cultures, raising the question as to whether the function of L3mbtl1 may be different in a more physiological experimental system. Hippocampal CA3 pyramidal neurons in organotypic slice cultures have been shown to self-restore synaptic connectivity and spontaneous network activity to a level resembling the in vivo situation (Takahashi et al., 2010). Furthermore, it has been reported that CA3 synapses are capable of inducing homeostatic scaling (Lee et al., 2013; Mitra et al., 2011). Therefore, we recorded mEPSCs from hippocampal CA3 pyramidal neurons in organotypic slice cultures. We found that PTX treatment induced scaling down in wild-type, but not L3mbtl1 KO, neurons (Figure 3). In contrast to the results from primary cultures, basal synaptic transmission was not significantly different between wild-type and L3mbtl1 KO neurons in slice culture (Figures 2C and 3C). This discrepancy could be due to the differences in experimental systems; for example, dissociated primary cultures have been shown to exhibit higher network activity compared to organotypic hippocampal neurons that display neuronal activity similar to in vivo conditions (Cingolani and Goda, 2008; Takahashi et al., 2010). While loss of L3mbtl1 in primary cultures impairs basal excitatory synaptic transmission, our results suggest that L3mbtl1 specifically regulates homeostatic downscaling, but not basal excitatory synaptic strength, in hippocampal CA3 neurons.
Figure 3. L3mbtl1 Is Required for Homeostatic Downscaling in Hippocampal CA3 Synapses.
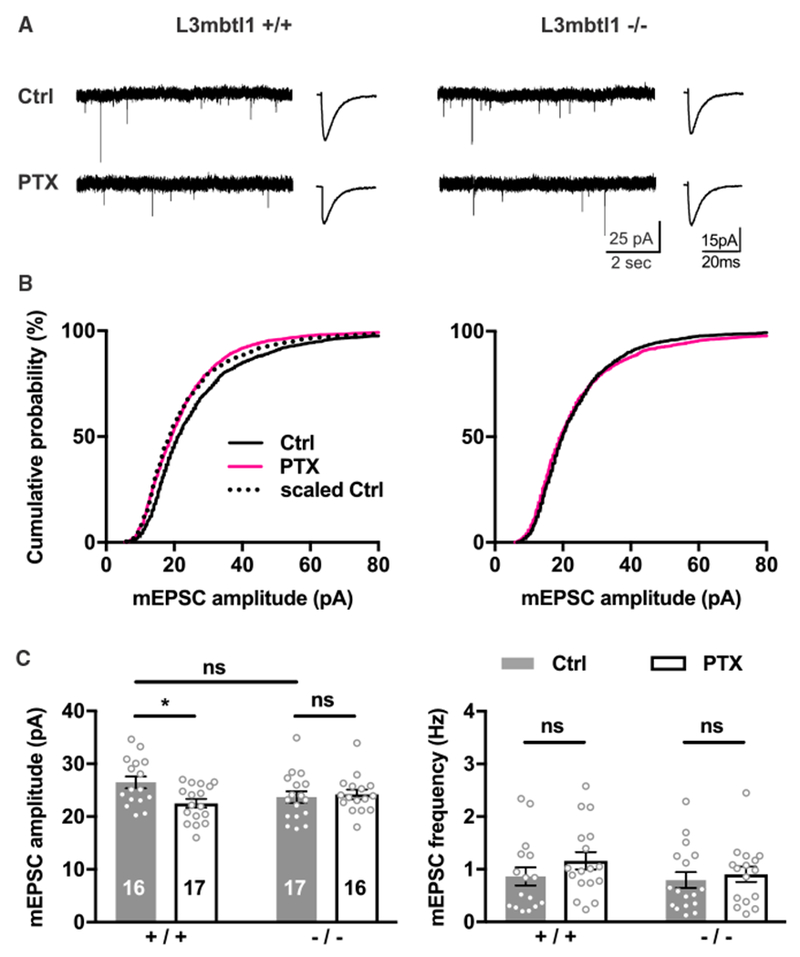
(A–C) Organotypic hippocampal slice cultures of wild-type or L3mbtl1−/− mice were treated with PTX or DMSO for 48 hr. mEPSC were recorded from CA3 pyramidal neurons.
(A) Representative mEPSC traces (left) and average mEPSC event traces (right) are shown.
(B) Cumulative distribution of mEPSC amplitudes. KS tests for distributions in wild-type group: ctrl versus PTX, p = 2.8 × 10−11; scaled ctrl versus PTX, not significant.
(C) Mean mEPSC amplitude (left) and frequency (right).
*p < 0.05, ns, not significant; two-way ANOVA test; N = 6 mice were tested. Data shown are means ± SEM.
Acute Knockdown of L3mbtl1 Causes a Lack of Homeostatic Downscaling in Hippocampal CA3 Pyramidal Neurons
It is possible that developmental effects underlie the loss of homeostatic downscaling in L3mbtl1 KO neurons. To address the role of L3mbtl1 in postnatal neurons, we acutely knocked down (KD) L3mbtl1 and measured homeostatic downscaling in CA3 pyramidal neurons. Small hairpin RNA (shRNA) directed against L3mbtl1 (Figures S5A and S5B) was transfected into organotypic hippocampal slice cultures from wild-type mice using a biolistic gene gun that allows us to test cell-autonomous function of genes of interest. Importantly, L3mbtl1 KD blocked homeostatic downscaling in CA3 pyramidal neurons without affecting basal synaptic strength (Figure 4), a finding consistent with the results obtained from L3mbtl1 KO slice cultures (Figure 3). This result suggests that L3mbtl1 plays a critical and cell-autonomous role in homeostatic downscaling in CA3 pyramidal neurons.
Figure 4. Effect of Acute Knockdown of L3mbtl1 on Homeostatic Downscaling in Hippocampal CA3 Synapses.
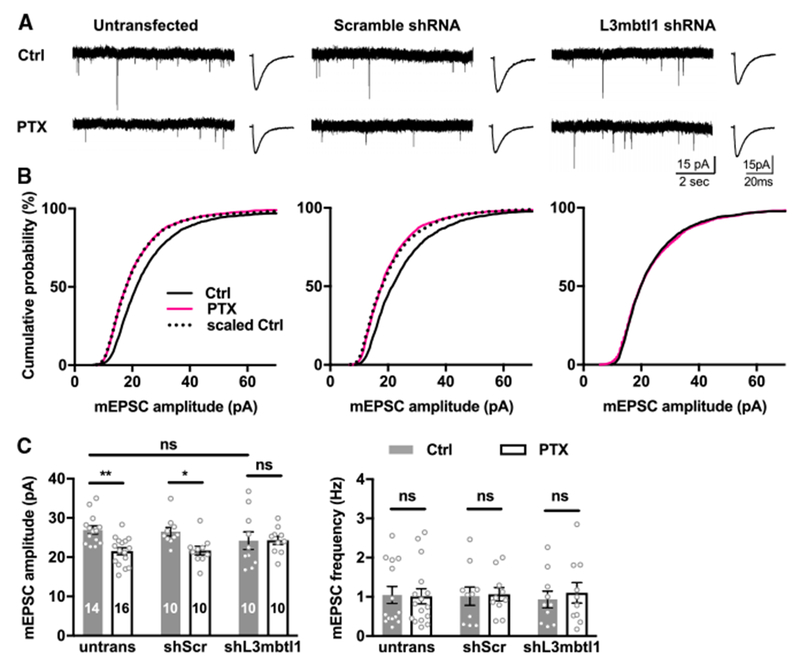
(A) Homeostatic downscaling induced by PTX treatment for 48 hr in hippocampal CA3 pyramidal neurons. Pyramidal neurons were transfected with expression vectors for shRNA (see Figures S5A and S5B) directed against scramble (middle) and L3mbtl1 (right). Left: representative mEPSC traces; right: averaged mEPSC traces. The shRNAs were transfected at DIV1 and PTX, or DMSO was applied 2 days before recording. mEPSCs were recorded from CA3 pyramidal neurons.
(B) Cumulative distribution of mEPSC amplitudes. KS tests for distributions in untransfected group: ctrl versus PTX, p = 1.8 × 10−31; scaled ctrl versus PTX, not significant. KS tests for distributions in shScr group: ctrl versus PTX, p = 2.1 × 10−26; scaled ctrl versus PTX, not significant. KS tests for distributions in shL3mbtl1 group: ctrl versus PTX, not significant.
(C) Quantification of mean mEPSC amplitudes (left) and frequencies (right).
**p < 0.01, *p < 0.05; ns, not significant; two-way ANOVA test; N = 8 mice were tested. Data shown are means ± SEM.
L3mbtl1 Occupies Gene Promoters in Hippocampus
To determine the binding loci of L3mbtl1 protein on a genome-wide scale, we performed chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq). ChIP-seq experiments were performed with antibody against endogenous L3mbtl1 in P7 hippocampus when the expression of L3mbtl1 reaches a maximal level, using KO mouse hippocampus as control (Figure 5A). Reproducible peaks were obtained from three biological replicates using MACS2 with an irreproducible discovery rate (IDR) threshold of 0.0025 as a consistency test (Table S3A). We identified 4,677 highly consistent L3mbtl1-bound regions, 93% of which are located within 5 kb up- and downstream of a transcription start site (TSS) (Figure 5C), and 47% of which are located in promoter regions (within −1 kb to 100 bp of a TSS) (Figure 5B; Table S3B), consistent with the notion that L3mbtl1 is a regulator of gene expression.
Figure 5. Genome-wide Mapping of L3mbtl1 Chromatin Binding Sites.
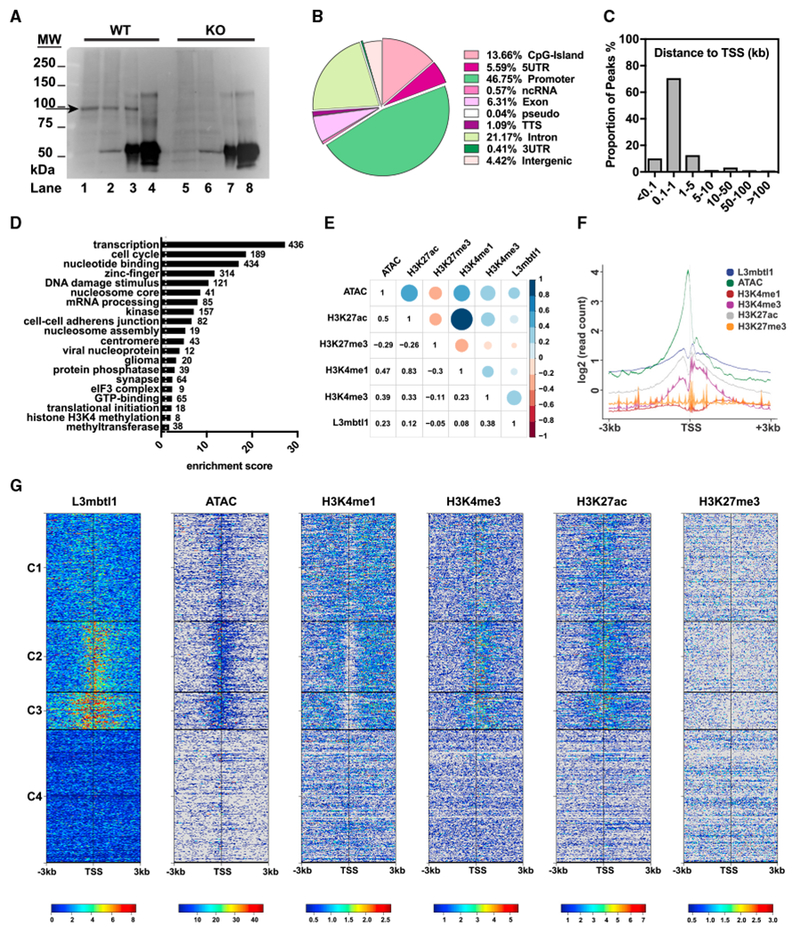
(A) Validation of antibody specificity in ChIP assays. Crosslinked chromatin prepared from wild-type or KO P7 mouse hippocampus was pulled down by Ig-L3mbtl1 or normal rabbit immunoglobulin G (IgG) and immunoblotted against L3mbtl1. Lanes 1 and 5, input lysate; lanes 2 and 6, unbound fraction; lanes 3 and 7, immunoprecipitated (IP) with Ig-L3mbtl1; and lanes 4 and 8, IP with IgG. Arrow indicates L3mbtl1 band.
(B) Distribution of 4,677 L3mbtl1-bound regions at various genomic locations. See also Figure S7. Promoters are defined as −1 kb to 100 bp of a transcription start site (TSS). TTS, transcription termination site; pseudo, pseudogenes.
(C) Distribution of L3mbtl1 binding sites around TSS. The proportion of L3mbtl1-bound regions is plotted at an increasing distance from the nearest TSS. Distance is absolute, regardless of direction (up- or downstream) from the TSS.
(D) Functional enrichment analysis of the genes obtained by ChIP-seq. Genes assigned to bound regions were tested by DAVID enrichment analysis. Clusters are sorted by enrichment score with the number of genes within each cluster labeled at the side of the bars.
(E) Correlation analysis of L3mbtl1 peaks with peaks of multiple histone PTMs and ATAC-seq data from neocortical excitatory neurons (provided by GEO: GSE63137). Peak chromosomal ranges were mapped into windows of 10 k nucleotides of the mouse genome and the pairwise Pearson correlation scores were plotted. Blue represents positive correlation, and red represents negative correlation.
(F and G) Read density distributions of L3mbtl1 and multiple histone PTMs and ATAC-seq centered on the TSS ± 3 kb of each transcript in mouse genome.(F) Averaged profiles of read densities were plotted with the log2 scale for all factors.
(G) Heat maps of read densities. All heat maps were organized into four clusters (C1, C2, C3, and C4) based on K-means clustering of L3mbtl1 ChIP-seq signal profiles.
See also Table S3.
For gene ontology analysis, bound regions within 1 kb up- and downstream from a TSS were assigned to the nearest genes. We assigned 3,188 genes as targets for L3mbtl1 (Table S3C) and the 3,000 genes with the highest peak scores were tested for functional enrichment by DAVID (Database for Annotation, Visualization, and Integrated Discovery) (Huang et al., 2009; Love et al., 2014) (Figure 5D; Table S3D). Of note, L3mbtl1 target genes were significantly enriched for gene ontology (GO) terms related to transcriptional regulation, nucleosome assembly, and other nuclear signaling, as observed for Sfmbt1 and Scml2, other members of the MBT domain-containing protein family in the cell lines (Bonasio et al., 2014; Zhang et al., 2013). We also found “synapse” among the top-20 enriched terms, which may include genes responsible for the defects in synaptic transmission and homeostatic synaptic plasticity in L3mbtl1 KO mice.
Previous studies showed that human L3mbtl1 binds to mono- and di-methylated histone lysine residues of H1bK26 and H4K20 and generally acts as a nucleosome compactor and transcriptional repressor in cell lines (Boccuni et al., 2003; Kalakonda et al., 2008; Trojer et al., 2007) and pluripotent stem cells (Perna et al., 2015). To analyze L3mbtl1 binding specificity in vivo, we investigated whether L3mbtl1 binds to chromatin regions with active or inactive chromatin marks. Using a published sequencing dataset (GEO: GSE63137) generated from mouse cortical excitatory neurons (Mo et al., 2015), we performed correlation analysis of L3mbtl1 binding sites with those of multiple histone marks (H3K27ac, H3K27me3, H3K4me1, and H3K4me3) and open chromatin sites marked by ATAC-seq signals (Figure 5E). Interestingly, L3mbtl1 binding correlates with open chromatin sites as well as potential active transcription initiation sites marked by H3K4me3 and H3K27ac. L3mbtl1, H3K4me3, and H3K27ac also showed a similar distribution pattern near the TSS (Figures 5F and 5G). Genome-wide analysis of L3mbtl1 ChIP-seq signals near the TSS revealed four clusters with distinct patterns for L3mbtl1 binding (Figure 5G), which highly correlates with signal patterns for ATAC, H3K4me3 and H3K27ac. In contrast, L3mbtl1 binding was not correlated with H3K27me3 or H3K4me1. Therefore, L3mbtl1 primarily localizes to open chromatin sites associated with active chromatin marks H3K4me3 and H3K27ac.
L3mbtl1 Regulates Genes that Control Synaptic Strength
To understand how L3mbtl1 mediates the homeostatic control of synaptic strength, we focused on a list of putative target genes that have been previously associated with synaptic scaling and synapse function (Table S1). Among 3188 L3mbtl1 target genes (Table S3C), 15 genes were identified as encoding scaling factors in Table S1. In addition, we hypothesized that genes in the synapse category (Figure 5D), the expression of which is activity dependent (Figure 1A, a threshold of 1.5-fold changes by PTX treatment with p value below 0.1) are potential targets associated with downscaling. Therefore, 12 activity-dependent genes were selected from 64 L3mbtl1-binding synaptic genes. Total RNA was isolated from primary cultures prepared from wild-type and L3mbtl1 KO mice, and qRT-PCRs were carried out to the 27 (15 plus 12) candidates to identify differentially expressed genes (Figures 6A, 6B, and S6). The expression of two genes, Ctnnb1 and Gabra2, was significantly decreased in the absence of L3mbtl1. Importantly, both genes showed L3mbtl1 binding and the presence of H3K4me3 and H3K27ac near their TSS (Figure 6C), consistent with the global binding patterns (Figure 5G). Of note, the majority of target genes were not functionally regulated by L3mbtl1 (Figure S6). This is consistent with the previous report that only ~10% of genes bound by transcription factors are functionally regulated by them (Cusanovich et al., 2014). To verify the ChIP-seq results, we confirmed the enrichment for L3mbtl1, H3K4me3 and H3K27ac at Ctnnb1 and Gabra2 promoter regions in both P7 hippocampus and cultured neurons by ChIP-qPCR (Figures 6D and 6E). We also examined the levels of L3mbtl1, H3K4me3, and H3K27ac at Ctnnb1 and Gabra2 promoter regions after 24 hr of PTX treatment in primary cultures (Figures 6D and 6E) and found that while H3K4me3 and H3K27ac were still enriched at promoter regions, L3mbtl1 was no longer enriched. Together, these findings demonstrate that L3mbtl1 occupies Ctnnb1 and Gabra2 gene promoters in an activity-dependent manner, suggesting a possible role for L3mbtl1 in regulating the activity-dependent expression of Ctnnb1 and Gabra2.
Figure 6. Identification of L3mbtl1 Target Genes.
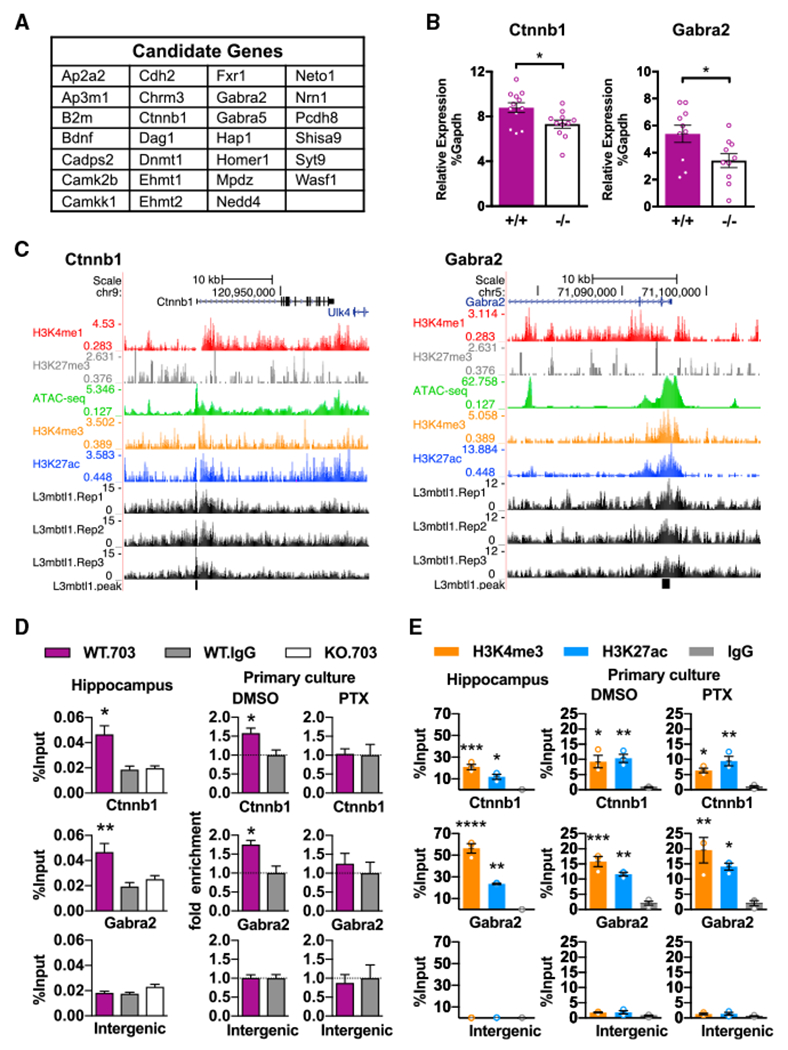
(A) List of selected putative L3mbtl1 target genes that have known functions in synaptic transmission and/or homeostatic synaptic plasticity. See also Tables S1 and S3 and Figure S6.
(B) Quantification of Ctnnb1 and Gabra2 RNA expression by qPCR. Ctnnb1 and Gabra2 expression relative to Gapdh were measured from hippocampal primary cultures prepared from wild-type (+/+) and L3mbtl1 KO (−/−) mice. **p < 0.01, *p<0.5; Student’s t test. N = 8–12 independent cultures.
(C) L3mbtl1-binding events at promoter regions of Ctnnb1 (left) and Gabra2 (right) genes obtained from ChIP-seq of P7 mouse hippocampus. ChlP-seq tracks for L3mbtl1 from wild-type samples (L3mbtl1.Rep1–3), various histone markers, and ATAC-seq tracks (GEO GSE63137) are shown. Black blocks indicate signal peaks for L3mbtl1-bound regions.
(D) L3mbtl1 binding validated by direct ChlP-qPCR assays at Ctnnb1 and Gabra2 promoter regions. ChIP was performed from P7 hippocampus (left) or primary cultures treated with DMSO (middle) or PTX (right) for 24 hr. Note that PTX treatment abolished L3mbtl1 binding in primary cultures. A one-way ANOVA test was used for the hippocampus and Student’s t test for primary culture. N = 3 biological replicates.
(E) Enrichment for H3K4me3 and H3K27ac validated by direct ChIP-qPCR assays at Ctnnb1 and Gabra2 promoter regions. ChIP was performed from P7 hippocampus (left) or primary cultures treated with DMSO (middle) or PTX (right). Note that PTX treatment did not change H3K4me3 or H3K27ac profiles at promoter regions. One-way ANOVA test. N = 3 biological replicates.
****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.5. Data shown are means ± SEM.
To address whether Ctnnb1 contributes to the effects of L3mbtl1 on synaptic scaling, we tested if knockdown of Ctnnb1 expression affects synaptic scaling and synaptic transmission. Using shRNA that induces partial KD of Ctnnb1 in wild-type neurons (Figures S5C and S5D), we showed that partial KD of Ctnnb1 was sufficient to abolish homeostatic downscaling and weaken basal excitatory synaptic transmission (Figures 7A–7C), thus mimicking the phenotype observed in L3mbtl1 KO primary neurons (Figure 2). These results suggest that Ctnnb1 mediates part of the effect of L3mbtl1 on homeostatic control of synaptic strength, but additional L3mbtl1 targets may also be involved. To test whether Ctnnb1 is sufficient to rescue the effect of L3mbtl1 loss, we transfected Ctnnb1 into L3mbtl1 KO primary neurons. Ctnnb1 transfection restored homeostatic downscaling and enhanced synaptic transmission in L3mbtl1 KO neurons (Figures 7D–7F), supporting a causal relationship between L3mbtl1 and Ctnnb1. Another L3mbtl1 target gene Gabra2 encodes the alpha-2 subunit of GABAARs. The reduced expression of the Gabra2 gene in L3mbtl1 KO neurons is likely responsible for our finding that basal inhibitory synaptic transmission is decreased in L3mbtl1 KO neurons compared to that in wild-type (Figure S4).
Figure 7. Effect of Acute Knockdown and Transfection of Ctnnb1 on Homeostatic Downscaling in Wild-Type and L3mbtl1 Knockout Neurons, Respectively.
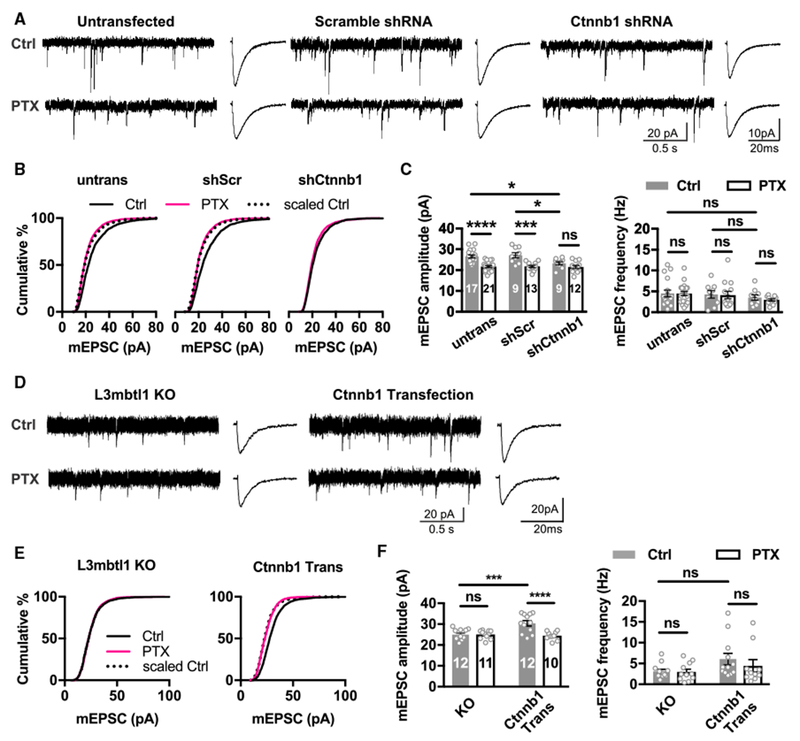
(A–C) Hippocampal primary cultures were prepared from wild-type (L3mbtl1 +/+) mice. Primary neurons were transfected with shRNA against Ctnnb1 (shCtnnb1) (see Figures S5C and S5D) for 3 days and treated with PTX or DMSO as a control for 2 days. Untransfected and those transfected with scrambled shRNA (shScr) are shown as controls.
(A) Representative mEPSC traces (left) and average mEPSC event traces (right).
(B) Cumulative distribution of mEPSC amplitudes. KS tests for distributions in untransfected group: ctrl versus PTX, p = 4.9 × 10−31; scaled ctrl versus PTX, not significant. KS tests for distributions in shScr group: ctrl versus PTX, p = 1.1 × 10−40; scaled Ctrl versus PTX, not significant. KS tests for distributions in shCtnnb1 group: ctrl versus PTX, not significant.
(C) Quantification of mean mEPSC amplitudes (left) and frequencies (right).
(D–F) Hippocampal primary cultures were prepared from L3mbtl1 −/− mice. Primary neurons were co-transfected with Ctnnb1 and EGFP (Ctnnb1 Trans) for 3 days and treated with PTX or DMSO as a control for 2 days.
(D) Representative mEPSC traces (left) and average mEPSC event traces (right) from untransfected L3mbtl1 −/− (KO) and transfected (Ctnnb1 Trans) neurons.
(E) Cumulative distribution of mEPSC amplitudes. KS tests for distributions in untransfected L3mbtl1 −/− neurons: ctrl versus PTX, not significant. KS tests for distributions in Ctnnb1 Trans group: ctrl versus PTX, p = 8.5 × 10−58; scaled ctrl versus PTX, not significant.
(F) Quantification of mean mEPSC amplitudes (left) and frequencies (right).
****p < 0.0001, ***p < 0.001, and *p < 0.05; ns, not significant; two-way ANOVA test. N = 4–5 mice. Data shown are means ± SEM.
DISCUSSION
Homeostatic synaptic scaling is one of the most important regulatory mechanisms of synaptic strength that prevents the detrimental consequences of runaway neuronal overexcitation. In the present study, we found that L3mbtl1 regulates basal excitatory and inhibitory synaptic transmission and homeostatic downscaling in excitatory synapses. Our ChIP studies identified Ctnnb1 and Gabra2 genes as activity-dependent targets of L3mbtl1 protein. Ctnnb1 expression is reduced in L3mbtl1 KO, partial KD of Ctnnb1 causes a lack of homeostatic downscaling, and transfection of Ctnnb1 into L3mbtl1 KO neurons restores homeostatic downscaling, suggesting that L3mbtl1 controls synaptic strength through Ctnnb1. Our results highlight a critical role for the activity-regulated chromatin reader molecule, L3mbtl1, in homeostatic control of synaptic strength and add to our understanding of epigenetic mechanisms of activity-dependent gene regulation.
Roles of L3mbtl1 in Synaptic Transmission and Synaptic Scaling
It has been considered that neuronal activity elevation induces a series of transcriptional regulatory events that alter chromatin structure, leading to a long-lasting effect on gene expression (Guan et al., 2002). The present study identified an activity-regulated chromatin reader, L3mbtl1, highly expressed in hippocampal neurons, as a key regulator of homeostatic synaptic scaling.
We observed differential effects of L3mbtl1 KO in dissociated primary cultures and organotypic slice cultures. Although both L3mbtl1 KO primary and organotypic neurons displayed disruption of synaptic downscaling, L3mbtl1 KO primary neurons also showed reduced basal excitatory synaptic transmission. L3mbtl1 may be a regulator for basal AMPAR-mediated synaptic strength and the failure of homeostatic downscaling could be attributed to a floor effect of a reduced number of AMPARs per synapses in primary neurons. However, arguments could be made that the activity-regulated L3mbtl1 transcript and protein levels and the activity-dependent dissociation of chromatin-bound L3mbtl1 from its target promoters strongly suggest a tightly controlled signaling pathway specifically activated during activity elevation. Indeed, both conventional KO and acute KD of L3mbtl1 in organotypic slice cultures blocked homeostatic downscaling in excitatory synapses without altering baseline AMPAR abundance, indicating a critical and cell-autonomous role for L3mbtl1 and its downstream genes in synaptic downscaling. The mechanism underlying the differential effect of L3mbtl1 KO in basal excitatory synaptic transmission in primary and organotypic slice cultures may be due to (1) the difference of recording from heterologous primary neurons and homogeneous CA3 pyramidal neurons in slice cultures and/or (2) differential basal synaptic activity between primary and organotypic slice cultures (Cingolani and Goda, 2008). Additionally, it is possible that the reduced inhibition in L3mbtl1 KO primary neurons may lead to compensatory reduction of excitatory synaptic transmission.
Ctnnb1 (β-catenin) has indispensable roles in many neurodevelopmental processes including migration and synapse formation and maturation, as a transcription factor and a synaptic scaffold molecule (Mosimann et al., 2009; Takeichi, 2007). At excitatory synaptic sites, β-catenin forms an adhesion complex with N-cadherin necessary for synapse formation and maturation (Uchida et al., 1996). β-catenin KO neurons exhibited reduced basal excitatory synaptic transmission and a lack of homeostatic scaling (Okuda et al., 2007). Goda and colleagues illustrated the importance of interactions between β-catenin and N-cadherin on basal excitatory synaptic transmission and homeostatic synaptic plasticity. In this study, we showed that Ctnnb1 is necessary and sufficient for L3mbtl1-mediated regulation of synaptic strength. Whether additional L3mbtl1 target genes are involved warrants further study.
The composition of GABAAR subunits changes during development. The expression of Gabra2 is high at postnatal day 0 and decreases during development (Fritschy et al., 1994). We observed a reduced basal inhibitory synaptic transmission in L3mbtl1 KO neurons compared with that in wild-type that can be attributed to the reduced expression of Gabra2 in L3mbtl1 KO neurons.
L3mbtl1-Mediated Transcriptional Regulation
The three MBT domains in L3mbtl1 protein compact chromatin by binding to mono- and dimethylated histone marks, including H1bK26, H3K9 and H4K20 (Li et al., 2007; Min et al., 2007; Sims and Rice, 2008; Trojer et al., 2007). In heterologous cell lines, L3mbtl1 forms a complex with heterochromatin protein 1γ (HP1γ) and retinoblastoma protein (pRB), and contributes to repression of E2F target genes (Trojer et al., 2007). In this study, we identified the in vivo binding sites of L3mbtl1 in the brain. Our analysis of genomic occupancy suggested that L3mbtl1 localizes to open chromatin sites associated with histone marks for active chromatin. Two genes Ctnnb1 and Gabra2, bound by L3mbtl1 at their promoter regions, showed reduced transcript levels in the absence of L3mbtl1. Taken together with previous reports, the data suggest that L3mbtl1 acts as a transcriptional activator or repressor depending on the gene targets and cell types. We also observed that the binding of L3mbtl1 to the promoters of these genes was abolished by PTX treatment, indicating that upon neuronal activity elevation, L3mbtl1 protein dissociates from chromatin of target genes. Therefore, we suggest that L3mbtl1 regulates basal expression of the Ctnnb1 and Gabra2 genes, and further, that the activity-dependent dissociation of L3mbtl1 from chromatin may allow the recruitment of other protein complexes required for gene expression upon activity elevation.
The dissociation between L3mbtl1 protein and chromatin complexes is triggered by DNA DSBs followed by degradation via the ubiquitin-mediated proteasomal pathway in heterologous cell lines (Acs et al., 2011). Importantly, DNA DSBs are triggered by epileptic insults, amyloid β, learning and memory, and naive visual stimuli in the brain, indicating that DSBs are a part of the activity-dependent events in vivo and can be events upstream of L3mbtl1 degradation leading to the expression of synaptic downscaling (Crowe et al., 2006, 2011; Suberbielle et al., 2013). The mechanism of activity-dependent downregulation of L3mbtl1 mRNA is not clear, but Ca2+ influx through NMDARs and L-type Ca2+ channels may play a critical role in the induction of activity-dependent transcriptional repression (Naranjo and Mellström, 2012).
Our current knowledge on the recruiting mechanisms of L3mbtl1 to its target sites is still limited. Histone mark H4K20me has been shown to promote L3mbtl1 binding in heterologous cell lines (Kalakonda et al., 2008); however, the interaction between histone and L3mbtl1 is too weak to be considered solely responsible for recruitment (Trojer and Reinberg, 2008). More likely, synergistic interactions with other proteins may be required to recruit L3mbtl1 to endogenous loci. To test the possibility that L3mbtl1 interacts with additional transcriptional factors, we searched for DNA motifs enriched in L3mbtl1 binding sites. Among the top DNA consensus motifs, we found one motif matching the E2F transcription factor consensus sequence (TTTTCGCG) (Figure S7). It has been shown that L3mbtl1 occupies E2F target sites in heterologous cell lines (Trojer et al., 2007), likely through binding to methylated Rb protein via an MBT domain (Saddic et al., 2010). This suggests a model whereby the recruitment of L3mbtl1 to at least a subset of promoter regions in vivo is facilitated by E2F/Rb complexes. Given that our ChIP-seq data highlight distinct neuronal gene targets for L3mbtl1 in neurons, a tissue-specific and cell-type-specific recruitment mechanism for L3mbtl1 is highly likely and warrants further investigation.
We have identified 12 neuronal activity-sensitive chromatin regulators, including L3mbtl1, out of 246 chromatin regulator genes. It is interesting to address whether the protein levels of the remaining 11 genes parallel transcriptional changes and alter downstream gene expression. Three genes, Hdac11, Jarid2, and Cbx6, are of particular interest in homeostatic downscaling because of their prominent expression in neurons (Zhang et al., 2014). Elucidating the roles of these other chromatin regulators in synaptic plasticity coupled with our results presented here will provide a comprehensive view of epigenetic regulation of gene expression in homeostatic synaptic scaling.
EXPERIMENTAL PROCEDURES
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Massachusetts Medical School. L3mbtl1 mutant mice were generated previously (Qin et al., 2010) and backcrossed with C57BL/6 for four generations. C57BL/6 mice were also used for primary cultures and ChIP qPCRs. Both sexes were used.
RNA Extraction and qPCR
Total RNA was extracted from hippocampus or hippocampal primary cultures using RNAqueous Micro Kits (Ambion) and reverse transcribed using High Capacity RNA-to-cDNA Kits (Applied Biosystems). qRT-PCR was performed on an ABI 7500 Fast Sequence Detection System (Applied Biosystems). For RNA expression analysis, TaqMan assays (Applied Biosystems) were used with Gapdh as a reference gene (Supplemental Experimental Procedures). For ChIP-qPCR, EpiTect ChIP qPCR assays (QIAGEN) were used. All reactions were performed with RT2 SYBR Green, following the manufacturer’s recommended cycling conditions, and subjected to melting curve analysis. All changes of gene expression were determined using the 2−ΔΔCT method (Futai et al., 2013; Livak and Schmittgen, 2001).
RNA-Seq
Sequencing libraries were prepared in accordance with the ScriptSeq v2 protocol following rRNA depletion (RiboZero, Illumina) and run on single-end 50-bp modules in Illumina HiSeq 2000. For detailed RNA-seq and analysis procedures, see the Supplemental Experimental Procedures.
ChIP Sequencing
Libraries from three wild-type samples and three L3mbtl1 KO samples were prepared from 10-ng ChIP’d DNA each using a NEBNext Ultra DNA Library Prep Kit (NEB) and subjected to 50-bp single-end Illumina sequencing. For detailed ChIP-seq and analysis procedures, see the Supplemental Experimental Procedures.
In Situ Hybridization
Fluorescein- or digoxigenin (DIG)-labeled cRNA probes were used for in situ hybridization. cRNA probes were synthesized as described previously (Yamasaki et al., 2010). For detailed procedures, see the Supplemental Experimental Procedures.
Primary and Slice Culture Preparation
Primary hippocampal cultures were prepared from early postnatal (P0–1) mice as described previously (Brewer et al., 1993) with some modifications (Futai et al., 2013). Organotypic hippocampal slice cultures were prepared from P6-7 mice as previously described (Futai et al., 2007, 2013). Neurons were transfected on DIV 1–2 via a biolistic gene gun (BioRad) using gold particles coated with plasmid DNA, per the manufacturer’s protocol.
Electrophysiology
Whole-cell recordings were performed from primary hippocampal cultures (DIV 14–16) and organotypic hippocampal slice cultures (DIV 6–8) as previously described (Futai et al., 2007, 2013). For detailed recording and analysis procedures, see the Supplemental Experimental Procedures.
Statistical Analyses
Results are reported as mean ± SEM. Student’s t test and one-way and two-way ANOVA with Sidak’s post hoc test were applied to normally distributed data. Kolmogorov-Smirnov (KS) tests were used for testing multiplicative scaling of miniature E/IPSCs. Statistical significance was set at p < 0.05 for the Student’s t test and ANOVA and at p < 10−4 for the KS test because the sample number of the miniature E/IPSC was large (Kim et al., 2012).
Supplementary Material
Highlights.
A chromatin reader, L3mbtl1, is down regulated by neuronal activity elevation
L3mbtl1 regulates homeostatic downscaling in excitatory synapses
Ctnnb1 is one of the target genes of L3mbtl1 that mediates synaptic scaling
ACKNOWLEDGMENTS
We thank Drs. Thomas G. Fazzio, Paul D. Gardner, Carlos Lois, and Andrew R. Tapper for valuable discussions. We also thank Ms. Naoe Watanabe and Mr. Kurtis N. McCannell for skillful technical assistance. This work was supported by grants from the Whitehall Foundation (20120844 to K.F.), the Japan Foundation for Pediatric Research (to K.F.), the Naito Foundation (to M.U.), Grants-in-Aid for Scientific Research (15K06732 to M.U.), and the NIH (R01NS085215 to K.F.; R01MH104341 to S.A.; and P50MH096890 to S.A. and H.H.).
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq and ChIP-seq raw and processed data reported in this paper is GEO: GSE104802. The accession number for the original immunoblotting and confocal images reported in this paper is Mendeley Data: https://doi.org/10.17632/s5m23x8ttb.1.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.028.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, and Dantuma NP (2011). The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol 18, 1345–1350. [DOI] [PubMed] [Google Scholar]
- Adams-Cioaba MA, and Min J (2009). Structure and function of histone methylation binding proteins. Biochem. Cell Biol 87, 93–105. [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, and Nimer SD (2003). The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J. Biol. Chem 278, 15412–15420. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, and Reinberg D (2010). MBT domain proteins in development and disease. Semin. Cell Dev. Biol 21, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, and Reinberg D (2014). Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife 3, e02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, and Price PJ (1993). Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res 35, 567–576. [DOI] [PubMed] [Google Scholar]
- Cingolani LA, and Goda Y (2008). Differential involvement of beta3 integrin in pre- and postsynaptic forms of adaptation to chronic activity deprivation. Neuron Glia Biol 4, 179–187. [DOI] [PubMed] [Google Scholar]
- Crowe SL, Movsesyan VA, Jorgensen TJ, and Kondratyev A (2006). Rapid phosphorylation of histone H2A.X following ionotropic glutamate receptor activation. Eur. J. Neurosci 23, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe SL, Tsukerman S, Gale K, Jorgensen TJ, and Kondratyev AD (2011). Phosphorylation of histone H2A.X as an early marker of neuronal endangerment following seizures in the adult rat brain. J. Neurosci 31, 7648–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Pavlovic B, Pritchard JK, and Gilad Y (2014). The functional consequences of variation in transcription factor binding. PLoS Genet 10, e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Paysan J, Enna A, and Mohler H (1994). Switch in the expression of rat GABAA-receptor subtypes during postnatal development: an immunohistochemical study. J. Neurosci 14, 5302–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K, Kim MJ, Hashikawa T, Scheiffele P, Sheng M, and Hayashi Y (2007). Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci 10, 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K, Doty CD, Baek B, Ryu J, and Sheng M (2013). Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J. Neurosci 33, 3612–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, and Kandel ER (2002). Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 111, 483–493. [DOI] [PubMed] [Google Scholar]
- Gurvich N, Perna F, Farina A, Voza F, Menendez S, Hurwitz J, and Nimer SD (2010). L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc. Natl. Acad. Sci. USA 107, 22552–22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ibata K, Sun Q, and Turrigiano GG (2008). Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron 57, 819–826. [DOI] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, et al. (2008). Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27, 4293–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J,Tsien RW, and Alger BE (2012). An improved test for detecting multiplicative homeostatic synaptic scaling. PLoS ONE 7, e37364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Queenan BN, Rozeboom AM, Bellmore R, Lim ST, Vicini S, and Pak DT (2013). Mossy fiber-CA3 synapses mediate homeostatic plasticity in mature hippocampal neurons. Neuron 77, 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, and Patel DJ (2007). Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25,402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, and Arrowsmith CH (2007). L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol 14, 1229–1230. [DOI] [PubMed] [Google Scholar]
- Mitra A, Mitra SS, and Tsien RW (2011). Heterogeneous reallocation of presynaptic efficacy in recurrent excitatory circuits adapting to inactivity. Nat. Neurosci 15, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, and Basler K (2009). Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol 10, 276–286. [DOI] [PubMed] [Google Scholar]
- Naranjo JR, and Mellström B (2012). Ca2+-dependent transcriptional control of Ca2+ homeostasis. J. Biol. Chem 287, 31674–31680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, Yu LM, Cingolani LA, Kemler R, and Goda Y (2007). beta-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc. Natl. Acad. Sci. USA 104, 13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F, Gurvich N, Hoya-Arias R, Abdel-Wahab O, Levine RL, Asai T, Voza F, Menendez S, Wang L, Liu F, et al. (2010). Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q-polycythemia vera. Blood 116, 2812–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F, Vu LP, Themeli M, Kriks S, Hoya-Arias R, Khanin R, Hricik T, Mansilla-Soto J, Papapetrou EP, Levine RL, et al. (2015). The polycomb group protein L3MBTL1 represses a SMAD5-mediated hematopoietic transcriptional program in human pluripotent stem cells. Stem Cell Reports 4, 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo K, and Goda Y (2010). Unraveling mechanisms of homeostatic synaptic plasticity. Neuron 66, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Van Buren D, Huang HS, Zhong L, Mostoslavsky R, Akbarian S, and Hock H (2010). Chromatin protein L3MBTL1 is dispensable for development and tumor suppression in mice. J. Biol. Chem 285, 27767–27775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA,West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, and Sage J (2010). Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem 285, 37733–37740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanzenbacher CT, Sambandan S, Langer JD, and Schuman EM (2016). Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EY, Jiang Y, Mao W, Futai K, Hock H, and Akbarian S (2015). Cognition and mood-related behaviors in L3mbtl1 null mutant mice. PLoS ONE 10, e0121252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, and Rice JC (2008). PR-Set7 establishes a repressive trans-tail histone code that regulates differentiation. Mol. Cell. Biol 28, 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, and Mucke L (2013). Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci 16, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Sasaki T, Matsumoto W, Matsuki N, and Ikegaya Y (2010). Circuit topology for synchronizing neurons in spontaneously active networks. Proc. Natl. Acad. Sci. USA 107, 10244–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci 8, 11–20. [DOI] [PubMed] [Google Scholar]
- Trojer P, and Reinberg D (2008). Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle 7, 578–585. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. (2007). L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915–928. [DOI] [PubMed] [Google Scholar]
- Turrigiano G (2011). Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu. Rev. Neurosci 34, 89–103. [DOI] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, and Takeichi M (1996). The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol 135, 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Matsui M, and Watanabe M (2010). Preferential localization of muscarinic M1 receptor on dendritic shaft and spine of cortical pyramidal cells and its anatomical evidence for volume transmission. J. Neurosci 30, 4408–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, and Reinberg D (2013). SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev 27, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


